The CDK Pef1 and protein phosphatase 4 oppose each other for regulating cohesin binding to fission yeast chromosomes
Figures
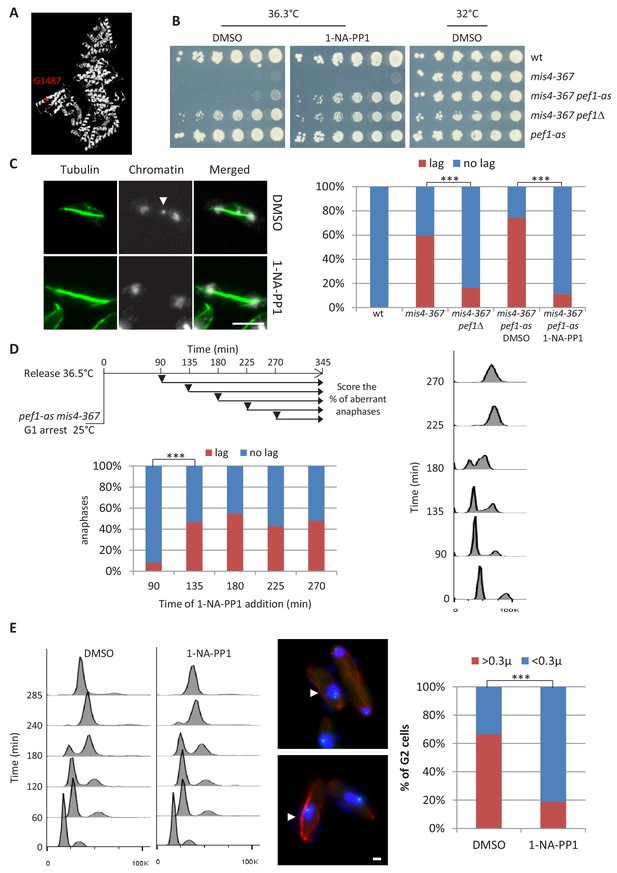
Inhibition of Pef1 kinase activity suppressed Mis4G1487D cohesion and chromosome segregation defects.
(A) The mis4-367 allele results in a G1487D substitution within the last HEAT repeat of Mis4. (B) Cell growth assay showing that inhibition of Pef1 kinase activity suppresses mis4-367-thermosensitive growth phenotype. (C) Inhibition of Pef1 kinase activity suppresses mis4-367 chromosome segregation defects. Cells were cultured at 36.5°C for a complete cell cycle. Lagging chromatids appear as DAPI-stained material (arrow) along the anaphase spindle (tubulin staining in green). Bar = 5 µm. ***p<0.0001 two-sided Fisher’s exact test (Figure 1—source data 1). (D) Pef1 inhibition must occur before S phase onset to rescue mis4-367 chromosome segregation. Cells were arrested in G1 by nitrogen starvation, released into the cell cycle at 36.5°C and 1-NA-PP1 added at the indicated time points (arrows). Cell cycle progression was followed by measurement of DNA content. Anaphase cells with lagging chromatids were scored at the 345 min time point. ***p<0.0001 two-sided Fisher’s exact test (Figure 1—source data 1). (E) Pef1 kinase inhibition improved sister-chromatid cohesion. Cells were arrested in G1 by nitrogen starvation, released into the cell cycle at 36.5°C with or without 1-NA-PP1. Cells were harvested after DNA replication (285 min) and processed for FISH using a centromere two-linked probe. Distance between FISH signals was measured in G2 cells, as judged by DNA content and the interphase array of microtubules. Bar = 1 µm. ***p<0.0001 two-sided Fisher’s exact test (Figure 1—source data 1).
-
Figure 1—source data 1
Statistical tests.
- https://cdn.elifesciences.org/articles/50556/elife-50556-fig1-data1-v2.xlsx
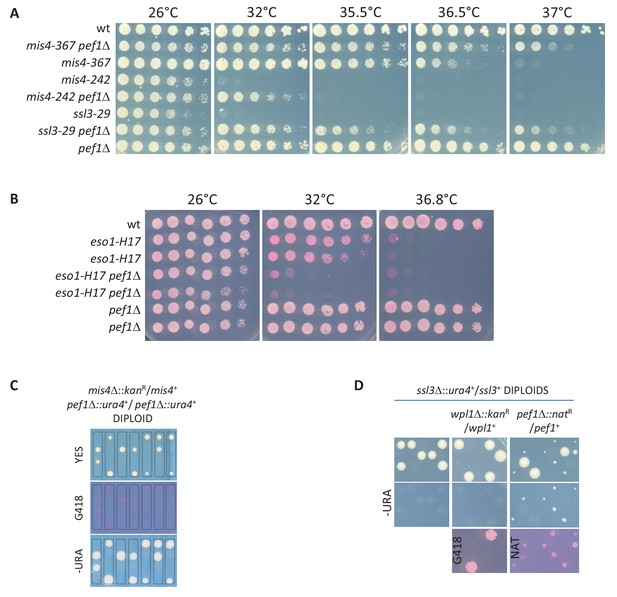
pef1 genetic interactions.
(A) Deletion of the pef1 gene alleviates the growth defects of cohesin loader mutants. (B) Pef1 ablation exacerbates the thermosensitive growth phenotype of the cohesin acetyl-transferase mutant eso1-H17. (C) Deletion of the pef1 gene does not bypass Mis4 requirement. Tetrad dissection from a mis4Δ::kanR/mis4+ pef1Δ::ura4+/ pef1Δ::ura4+diploid strain. Among the four spores of a tetrad, only two form viable colonies. All viable colonies are mis4+. (D) Deletion of pef1 allows cell survival in the absence of the ssl3 gene. The wpl1 or pef1 gene was deleted in a ssl3Δ::ura4+/ssl3+ diploid strain. The resulting diploids were sporulated and tetrads dissected. In an otherwise wild-type (left), or wpl1Δ/wpl1+ (middle) background, Ura+ (ssl3Δ) progeny was not observed. By contrast, small growing Ura+ NatR (ssl3Δ pef1Δ) colonies were frequent among the progeny from the pef1Δ/pef1+ diploid (right).
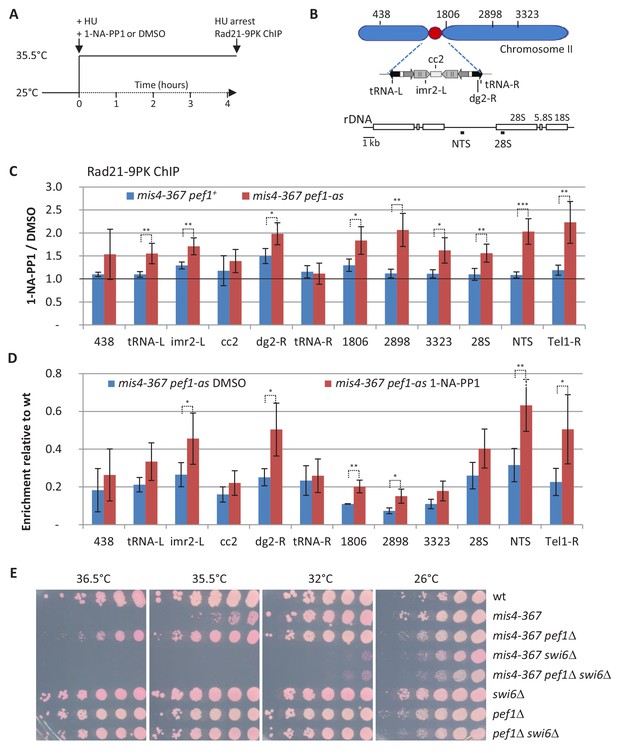
Inhibition of the CDK Pef1 in mis4-367 increased Rad21 binding to S phase chromosomes.
(A) Scheme of the experiment. Hydroxyurea (HU) was added to 12 mM at the time of the temperature shift along with 1-NA-PP1 or solvent alone (DMSO). Cells were collected after 4.25 hr. The S phase arrest was confirmed by DNA content analysis (Figure 2—figure supplement 1). (B) Schematics showing the loci analyzed by ChIP-qPCR. (C) The effect of 1-NA-PP1 treatment on Pef1-as is shown by the ratio 1-NA-PP1/DMSO (red) for each site analyzed. The ratios in a pef1+ background (blue) estimate the off target effects of the inhibitor. Ratios were calculated from the ChIP data shown in Figure 2—figure supplement 1. Bars indicate mean ± SD from four ratios. ***p≤0.001, **p≤0.01, *p≤0.05, by two-tailed, unpaired t-test with 95% confidence interval (Figure 2—source data 1). (D) Rad21 binding relative to wild-type. The ratios highlight the recovery of Rad21 binding upon inhibition of the CDK relative to wild-type levels. Ratios were calculated from the data shown in Figure 2—figure supplement 1. Bars indicate mean ± SD from four ratios. **p≤0.01, *p≤0.05, by two-tailed, unpaired t-test with 95% confidence interval (Figure 2—source data 1). (E) Cell growth assay showing that pef1Δ does not suppress mis4-367 thermosensitive phenotype in the absence of the swi6 gene.
-
Figure 2—source data 1
Raw ChIP data and t-tests.
- https://cdn.elifesciences.org/articles/50556/elife-50556-fig2-data1-v2.xlsx
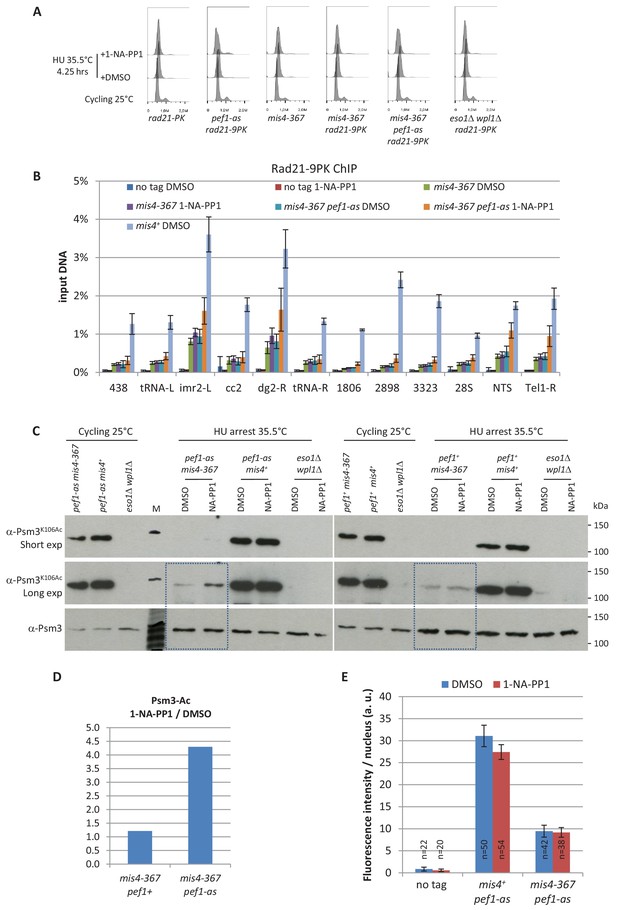
DNA content analysis, raw ChIP data, Psm3 acetylation and nuclear spreads from HU-arrested cells.
(A) DNA content analyses. The drift in the 1C peak to the right in HU-arrested cells is due to an increase in the mitochondrial DNA content as the cells elongate (Sazer and Sherwood, 1990). (B) Rad21-9PK ChIP-qPCR results expressed as % of input DNA. The ‘no tag’ control estimates background enrichment. Bars indicate mean ± SD from 4 ChIPs (Figure 2—source data 1). (C) Effect of Pef1 kinase inhibition on Psm3 acetylation. Total protein extracts were analyzed by western blotting using the indicated antibodies. M: molecular weight markers. The boxes indicate the lanes used for signal quantification. (D) Quantification of Psm3 acetylation in mis4-367 pef1-as cells at the restrictive temperature. Band intensities were measured from the blots shown in panel C. Psm3-Ac was normalized to Psm3 and the ratios 1-NA-PP1/DMSO were calculated. (E) The inhibition of Pef1 kinase activity did not induce a global change in the total amount of chromatin-bound Rad21. The amount of chromatin-bound Rad21-9PK per nucleus was measured by nuclear spreads and indirect immunofluorescence using anti-PK antibodies. The bars represent the mean fluorescence intensity per nucleus + /- the 95% confidence interval.
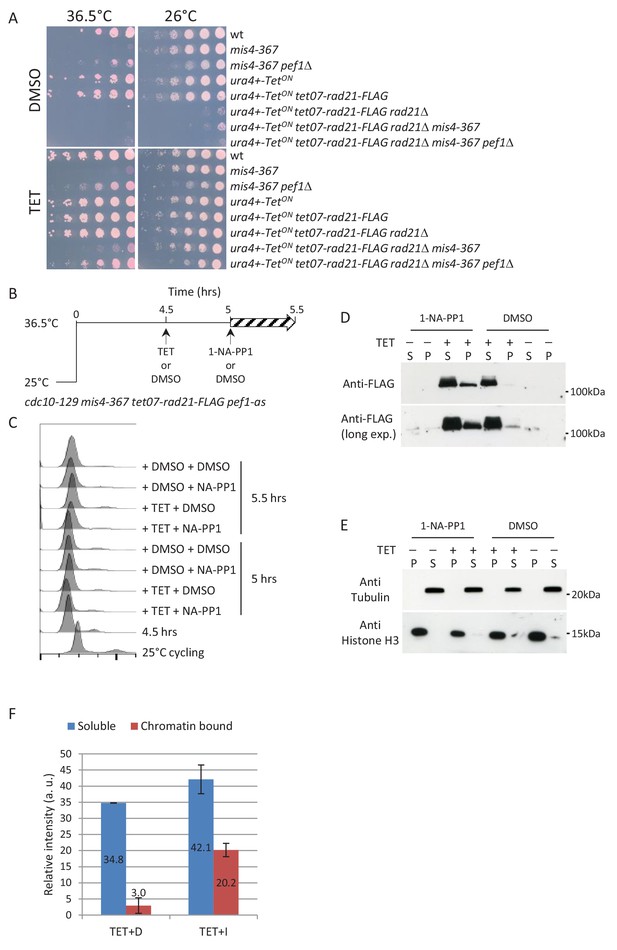
Inhibition of Pef1 kinase in G1-arrested mis4-367 cells allows neo-synthesized Rad21 to bind chromatin.
(A) The tetracycline (TET) inducible tet07-rad21-FLAG construct can substitute for the endogenous rad21 gene. The last two lanes show that pef1Δ suppresses mis4-367 ts phenotype when tet07-rad21-FLAG is the sole source of Rad21. (B) Experimental scheme. Cells cultured in EMM2 medium were arrested in G1 by the cdc10-129 mutation. After 4.5 hr tet07-rad21-FLAG was induced by the addition of TET or left un-induced (DMSO). Pef1-as was inhibited 30 min later and samples collected after 30 min. (C) DNA content analysis. (D) Western blot analysis of Rad21-FLAG in the chromatin (P) and soluble (S) fractions. (E) Fractionation controls. Anti-tubulin and anti-Histone H3 antibodies were used as markers for the soluble (S) and chromatin (P) fractions, respectively. (F) Rad21-FLAG signals were quantified for the TET samples from the long and short exposure blots shown in (D). The bars represent the mean relative band intensities + /- SD.
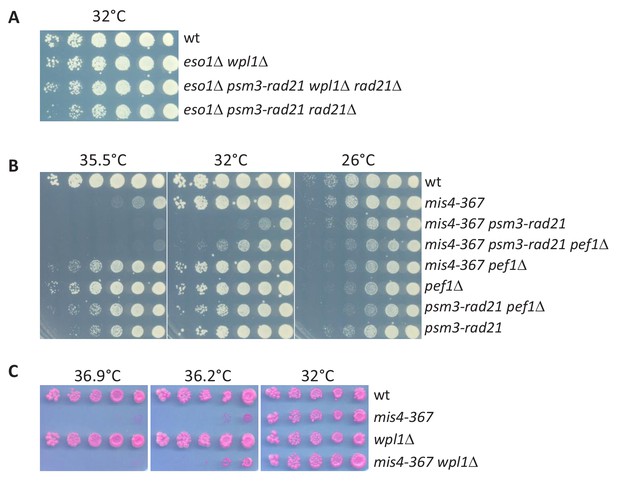
Pef1 acts independently from the Psm3/Rad21 interface.
(A) Deletion of wpl1 or a psm3-rad21 gene fusion allows cell survival in the absence of the otherwise essential eso1 gene. (B) The psm3-rad21 gene fusion does not suppress the mis4-367 thermosensitive growth defect. However deletion of the pef1 gene improves growth in this genetic setup. (C) Deletion of the wpl1 gene does not suppress mis4-367 growth defect.
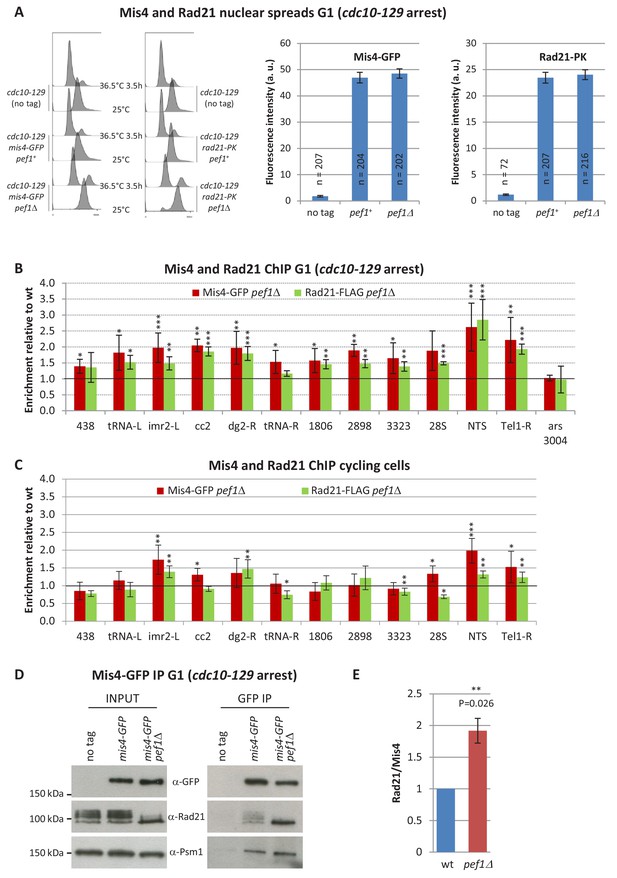
Pef1 ablation affects Rad21 and Mis4 binding to G1 chromosomes.
(A) Mis4-GFP and Rad21-PK binding to whole G1 nuclei. Cells were cultured at 36.5°C to induce the cdc10-129 arrest and collected after 3.5 hr. G1 arrest was monitored by DNA content analysis. Mis4-GFP and Rad21-PK binding to whole nuclei were measured by nuclear spreads and indirect immunofluorescence. The graphs show the mean fluorescence intensity per nucleus + /- the 95% confidence interval. (B) Mis4-GFP and Rad21-FLAG ChIP from G1-arrested cells. Raw ChIP data (Figure 4—figure supplement 1) were normalized to wild-type levels. Bars indicate mean ± SD, n = 4. A two-tailed, unpaired t-test was used to assess enrichment over wild-type levels. ***p≤0.001, **p≤0.01, *p≤0.05 with 95% confidence interval (Figure 4—source data 1). (C) Rad21-FLAG and Mis4-GFP ChIP from cycling cells. ChIP data (Figure 4—figure supplement 1) were normalized to wild-type levels. Bars indicate mean ± SD, n = 4. ***p≤0.001, **p≤0.01, *p≤0.05, by two-tailed, unpaired t-test with 95% confidence interval (Figure 4—source data 1). (D) Pef1 ablation increased Rad21 binding to its loader. Mis4-GFP was immuno-purified from G1 (cdc10-129) protein extracts and co-purifying proteins were analyzed by western blotting with the indicated antibodies. (E) Quantification of Rad21 in Mis4-GFP IPs. Band intensities were measured and the ratios Rad21/Mis4 were normalized to wt. Bar = mean+/-SD from four biological replicates. **p≤0.01 by one sample t test with 95% confidence interval.
-
Figure 4—source data 1
Raw ChIP data and t-tests.
- https://cdn.elifesciences.org/articles/50556/elife-50556-fig4-data1-v2.xlsx
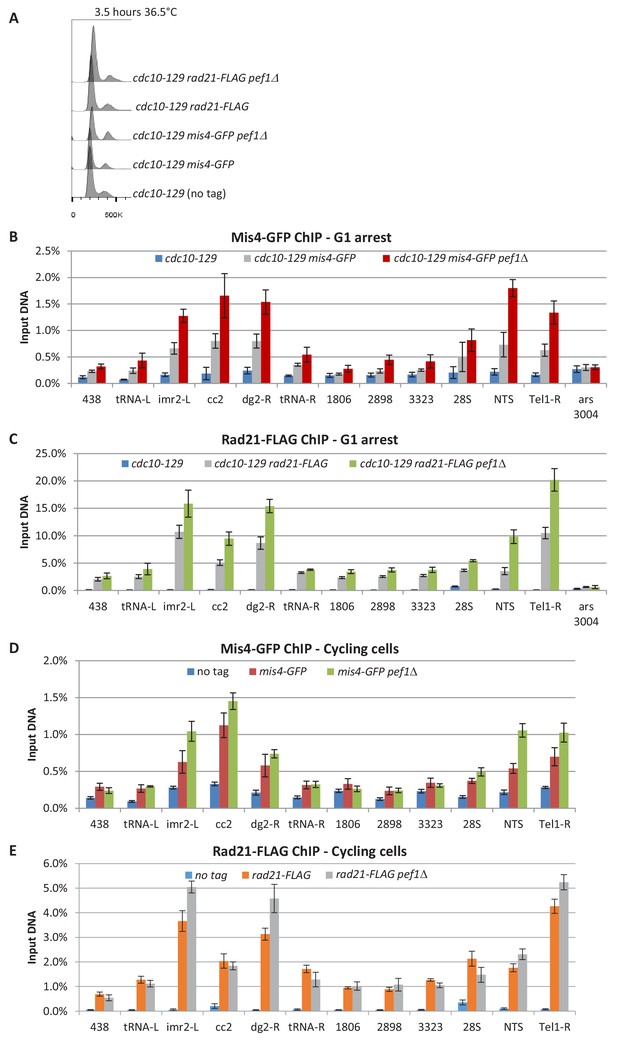
Mis4-GFP and Rad21-FLAG ChIP from G1 and cycling cells.
(A) Cells were cultured at 36.5°C to induce the cdc10-129 arrest and collected for ChIP after 3.5 hr. G1 arrest was monitored by DNA content analysis. (B) Mis4-GFP ChIP, cdc10-129 arrest. (C) Rad21-FLAG ChIP, cdc10-129 arrest. (D) Mis4-GFP ChIP, cycling cells. (E) Rad21-FLAG ChIP, cycling cells. Bars indicate mean ± SD from 4 ChIP (Figure 4—source data 1).
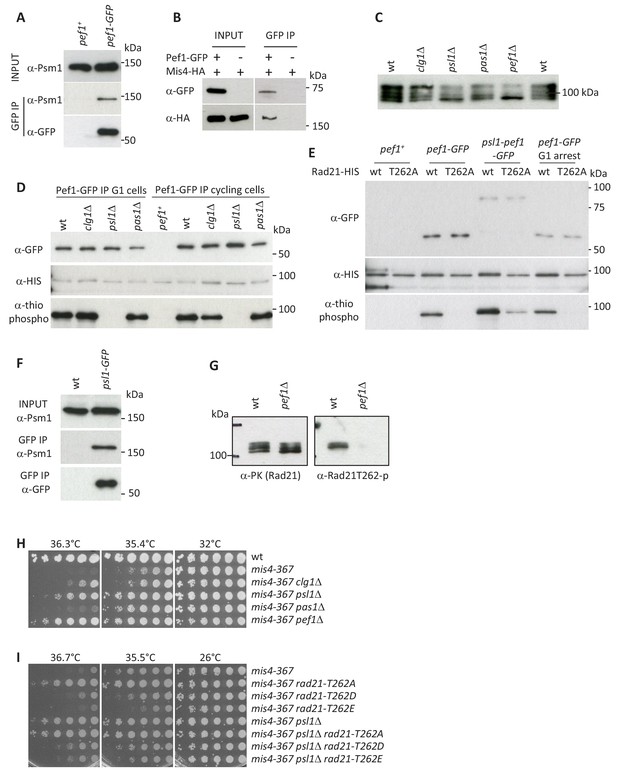
Pef1 phosphorylates Rad21.
(A,B) Pef1 co-immunoprecipitates cohesin (A) and the cohesin loader Mis4 (B) from total protein extracts. (C) Western blot analysis of total protein extracts from cycling cells probed with anti-Rad21 antibodies. (D) In vitro kinase assays. Pef1-GFP immuno-purified (IP) from cycling or G1 (cdc10-129) cells was incubated with in vitro translated Rad21-HIS in the presence of ATPγS and the proteins analyzed by western blotting. Phosphorylated products were detected using an anti-thiophosphate ester antibody. (E) In vitro kinase assays. Rad21-T262A prevents Rad21 phosphorylation by Pef1. The fusion protein Psl1-Pef1-GFP phosphorylates Rad21. (F) Psl1 co-immunoprecipitates Psm1 from total protein extracts (cycling cells). (G) Rad21-PK was immuno-purified from cycling cells and probed by western blotting with the indicated antibodies. (H,I) Growth assays for suppression of the ts growth defect of mis4-367.
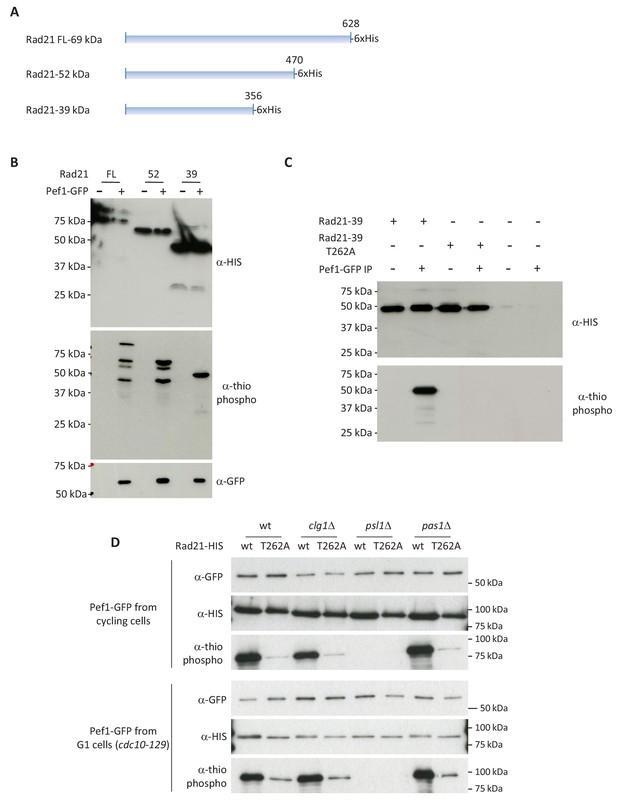
Mapping the Pef1 phosphorylation site within Rad21.
(A) Truncated forms of Rad21 used in the kinase assays. (B–C) In vitro kinase assays using Pef1-GFP purified from cycling cells. The reaction products were analyzed by western blotting with the indicated antibodies. Note that all Rad21 forms migrate slower than predicted from their calculated molecular weight. The shortest Rad21 derivative (Rad21-39) is efficiently phosphorylated by Pef1. The T262A substitution abrogates in vitro phosphorylation of Rad21-39 by Pef1. (D) Full length Rad21-T262A is a poor Pef1 substrate.
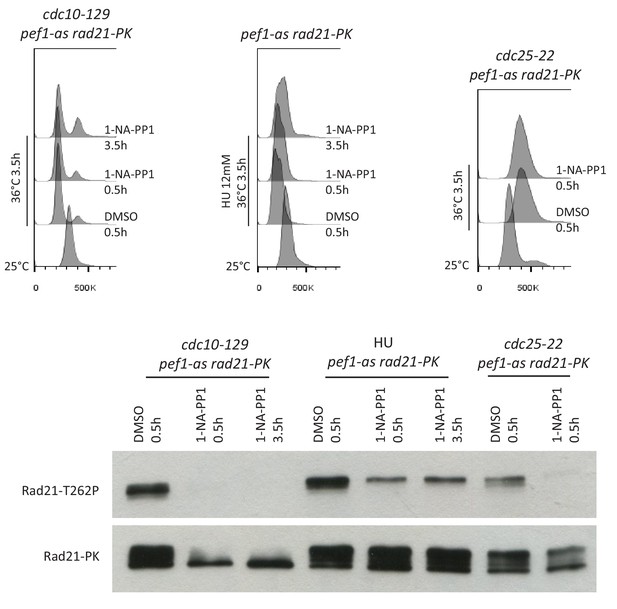
Rad21-T262 phosphorylation in G1, S and G2-arrested cells.
Exponentially growing cells at 25°C were shifted to 36°C and collected after 3.5 hr. For the S-phase arrest, 12 mM HU was added at the time of the temperature shift. Pef1-as inhibition was done by adding 1-NA-PP1 either from the beginning of the experiment (1-NA-PP1 3.5 hr) or 0.5 hr before cells were collected (1-NA-PP1 0.5 hr). Cell cycle arrest was monitored by DNA content analysis. The drift of the peaks to the right for the 36°C samples is due to an increase in the mitochondrial DNA content (Sazer and Sherwood, 1990). Rad21-PK was immuno-purified from total protein extracts and probed by western blotting with anti Rad21-T262p and anti-PK antibodies.
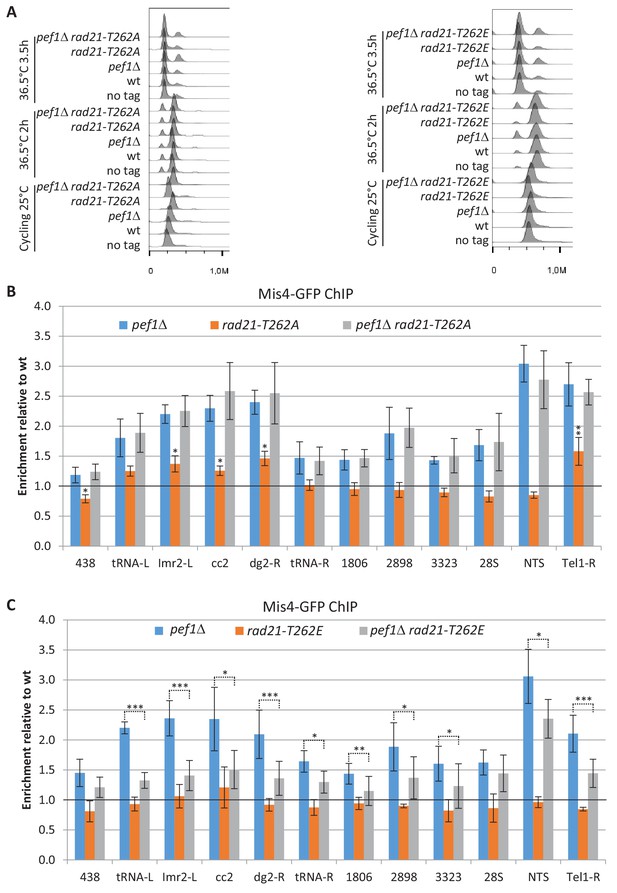
Rad21-T262 phosphorylation modulates Mis4 binding to G1 chromosomes.
(A) DNA content analysis. Cultures of the indicated strains were shifted to 36.5°C to induce the cdc10-129 arrest and cells collected for ChIP after 3.5 hr. (B,C) Mis4-GFP ChIP relative to wild-type. Bars indicate mean ± SD, n = 4. ***p≤0.001, **p≤0.01, *p≤0.05, by two-tailed, unpaired t-test with 95% confidence interval (Figure 6—source data 1).
-
Figure 6—source data 1
Raw ChIP data and t-tests.
- https://cdn.elifesciences.org/articles/50556/elife-50556-fig6-data1-v2.xlsx
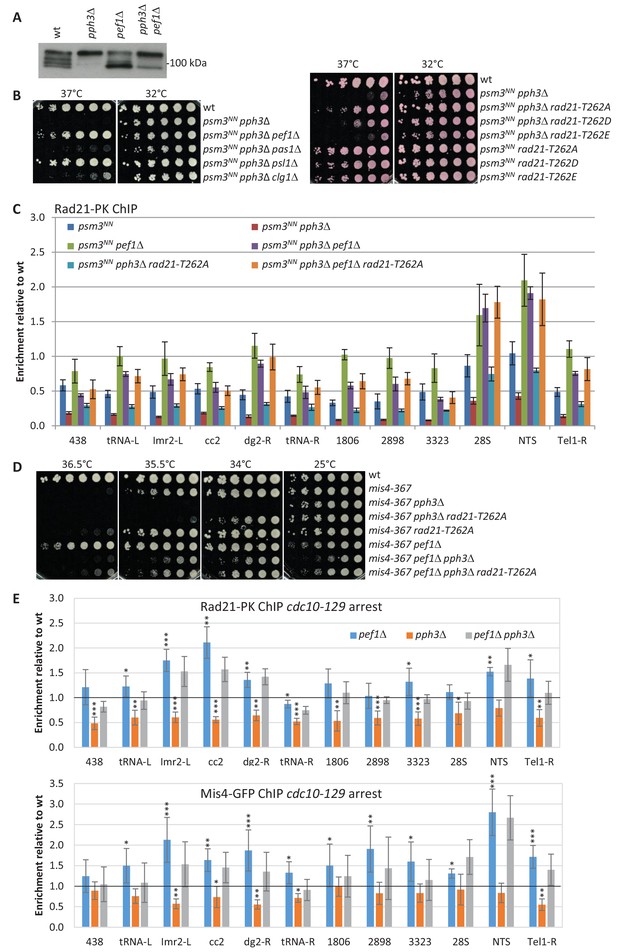
Pef1 and PP4 oppose each other for regulating Rad21 binding to chromosomes.
(A) Western blot analysis of total protein extracts from cycling cells probed with anti-Rad21 antibodies. (B) Cell growth assays showing that the ts growth defect of psm3NN pph3Δ is efficiently rescued by pef1 and psl1 deletion mutants (left) and rad21-T262A (right). (C) Rad21-ChIP after one complete cell cycle at 36°C. Data are expressed relative to wild-type. Bars indicate mean ± SD from four ratios. t-tests are shown in Figure 7—figure supplement 1 and Figure 7—source data 1. (D) Cell growth assays showing that pef1Δ, rad21-T262A and pph3Δ display opposite genetic interactions with mis4-367. (E) Rad21 and Mis4 ChIP relative to wild-type in cdc10-129-arrested cells. Bars indicate mean ± SD from four ratios (Figure 4—source data 1).
-
Figure 7—source data 1
Raw ChIP data and t-tests.
- https://cdn.elifesciences.org/articles/50556/elife-50556-fig7-data1-v2.xlsx
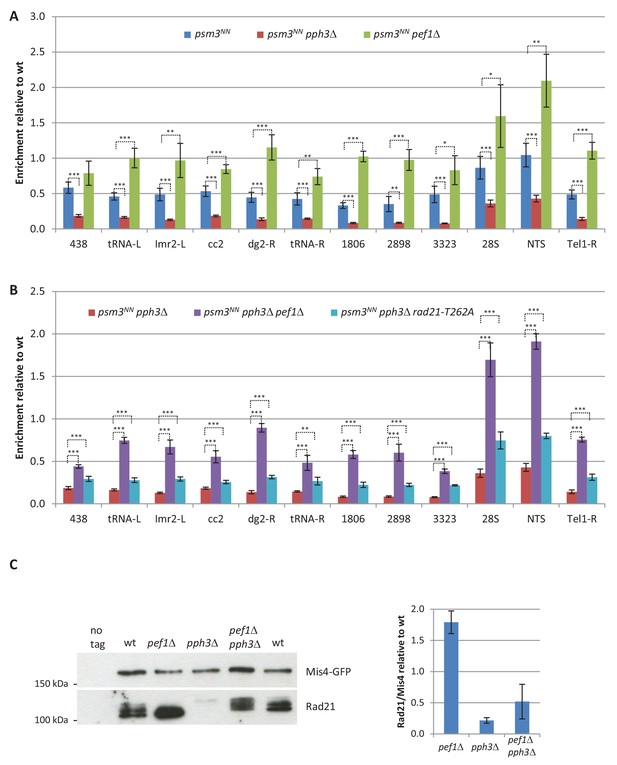
Pairwise comparisons of ChIP data from Figure 7C and quantification of Rad21 in Mis4 immunoprecipitates.
(A,B) Pairwise comparisons of ChIP data from Figure 7C. Data are shown in two separate graphs for clarity. Bars indicate mean ± SD, n = 4. ***p≤0.001, **p≤0.01, *p≤0.05, by two-tailed, unpaired t-test with 95% confidence interval (Figure 7—source data 1). (C) Mis4-GFP was immuno-purified from G1 (cdc10-129) protein extracts and co-purifying proteins were analyzed by western blotting. Band intensities were measured and the ratios Rad21/Mis4 were normalized to wt. Bar = mean+/-SD from two biological replicates.
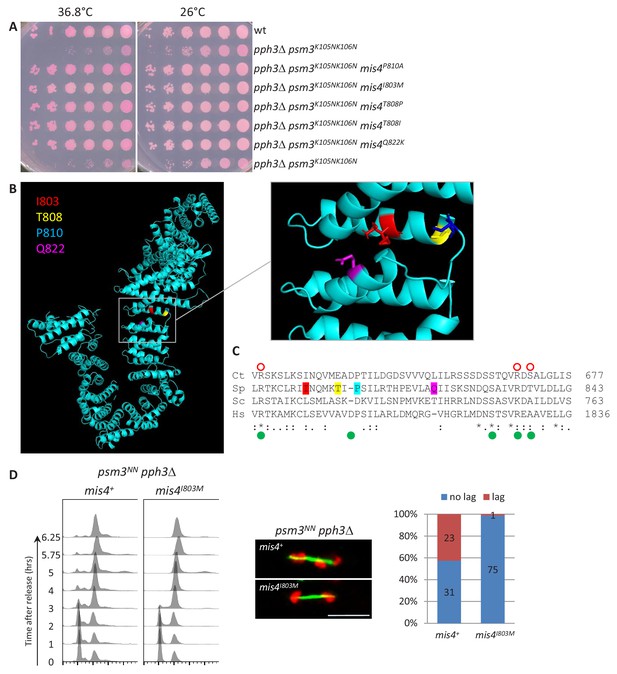
Mis4I803M suppresses pph3Δ psm3NN chromosome segregation defects.
(A) Cell growth assay. The mis4 mutations restore pph3Δ psm3NN growth at elevated temperature (B) Model structure of Mis4 showing the location of the mutated residues. (C) Sequence alignment of Mis4 with Scc2 proteins from other species. The mutated residues in Mis4 are colored as in (B). Ct Scc2 residues required for Scc1 binding are indicated by open red circles and Hs Scc2 residues mutated in Cornelia de Lange syndrome are indicated by green dots as in Kikuchi et al. (2016). Ct, Chaetomium thermophilum; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe, Hs, Homo sapiens. (D) Mis4I803M efficiently suppressed pph3Δ psm3NN chromosome segregation defects. Cells were arrested in G1 by nitrogen starvation at 25°C and released at 37°C. Progression in the cell cycle was monitored by DNA content analysis. Cells from the 5.75 and 6.25 time points were processed for DNA and tubulin staining to score the number of anaphase cells with lagging chromatids. Bar = 5µm.
Additional files
-
Supplementary file 1
Key Resources Table.
- https://cdn.elifesciences.org/articles/50556/elife-50556-supp1-v2.docx





