MicroRNA-934 is a novel primate-specific small non-coding RNA with neurogenic function during early development
Figures
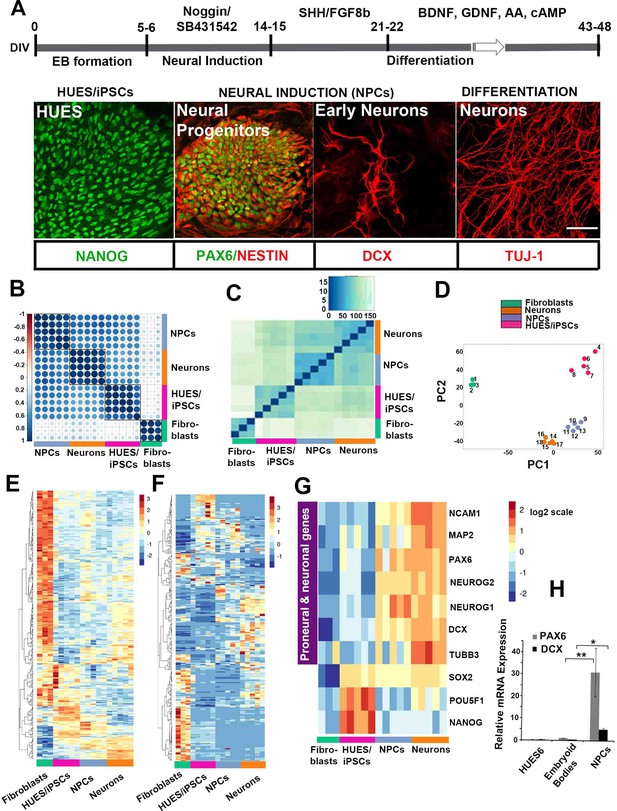
Directed neural differentiation of hESC/iPSC and RNA/small RNA sequencing at distinct stages of differentiation.
(A) Schematic representation of directed neural differentiation of hESCs/iPSCs. Embryoid body (EB) formation (5–6 DIV) is followed by dual SMAD inhibition for neural induction and NPC generation (14–15 DIV). Neuronal differentiation then proceeds until 43–48 DIV in the presence of indicated factors. hESCs are positive for the pluripotency marker Nanog. At 12 DIV, characteristic rosettes comprising NPCs double-positive for Pax6 and Nestin are evident while DCX-positive neuroblasts/early born neurons are also seen. βΙΙΙ-tubulin (TUJ-1)-positive neurons are predominantly detected at 48 DIV. Scale bar = 40 μm; (B) RNA expression correlation map incorporating the 300 RNA genes with highest standard deviations across all samples; groups exhibit distinct expression patterns and within-group uniformity. Darker blue colors depict stronger positive correlations between samples, as calculated by Pearson’s correlation coefficient. (C) Heatmap depicting similarity between samples based on expression of the 50 most expressed miRNAs. Darker blue colors denote higher similarity. (D) Graph depicting the first two principal components (PC1, PC2) of the top 1000 miRNAs with the highest variance across all samples. All groups exhibit distinct expression patterns and within-group uniformity. Samples: Fibroblasts (green): 1, Fetal; 2. Control (C1); 3, PD-1; hESCs/iPSCs (magenta): 4, hESCs; 5, C1 iPSCs; 6, C2 iPSCs; 7, PD1 iPSCs; 8, PD2 iPSCs; iPSC-derived NPCs (purple): 9, hESC-NPCs; 10, C1 NPCs ; 11, C2 NPCs ; 12, PD1 NPCs; 13, PD2 NPCs; iPSC-derived neurons (orange): 14, hESC-neurons ; 15, C1 neurons ; 16, C2 neurons; 17, PD1 neurons; 18, PD2 neurons. (E) Heatmap depicting Z scores of log2-transformed expression levels of the 300 genes with highest variance across all samples. (F) Heatmap depicting Z scores of log2-transformed expression levels of the 250 miRNAs with highest variance across all samples. (G) Heatmap showing that expression of proneural and neuronal markers starts at the stage of neural induction. (H) RT-qPCR analysis shows up-regulation of Pax6 and DCX mRNA at NPC stage (n = 3, for Pax6 31.1+10.1 at the NPC stage; p=0.006 compared to HUES6, and for DCX 5+0.58 at the NPC stage; p=0.01 compared to HUES6). Bars and error bars represent mean values and the corresponding SEMs; 0.01<*p<0.05; 0.001<**p<0.01.
-
Figure 1—source data 1
qRT PCR data for PAX6 and DCX relative expression.
- https://cdn.elifesciences.org/articles/50561/elife-50561-fig1-data1-v2.xlsx
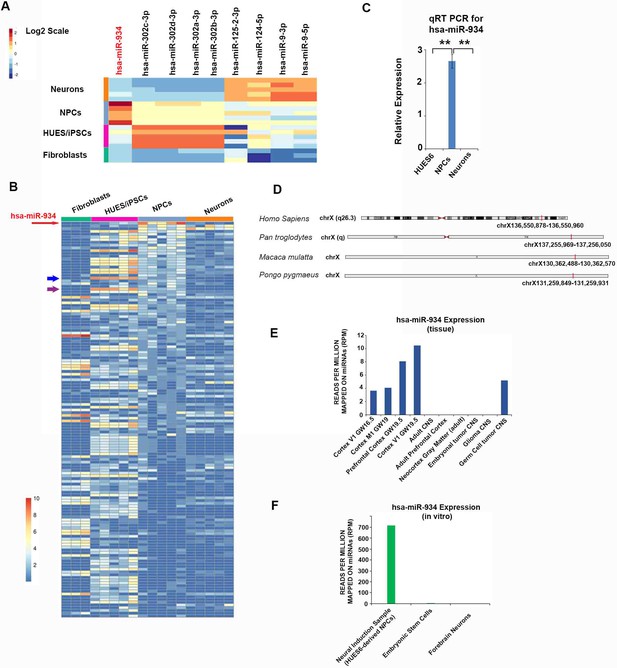
miR-934 is specifically expressed during progenitor expansion and early neuron generation.
(A) Heatmap profiling the expression of selected characteristic miRNAs across directed neural differentiation of hESCs/iPSCs to NPCs and neurons, demonstrating also the high expression of miR-934 during neural induction (Fibroblasts vs NPCs; p=0.0004, iPSCs vs NPCs; p=0.0001, Neurons vs NPCs; p=3.5×10−6). Warmer colors signify higher expression Z scores of the log2-transformed expression values. (B) Heatmap profiling the expression (log2; reads per million) of 144 miRNAs with stage specificity (tau >0.7) which are sorted based on median expression in NPCs. miR-934 is the highest expressed miRNA in NPCs and clearly segregated, exhibiting a stage-specific expression pattern. Red arrow denotes miR-934 ranking on the top, while warmer and cooler colors signify higher and lower expression values, respectively. The blue and purple arrows indicate hsa-miR-302d-5p and hsa-miR-302b-5p, respectively, which are selected in the HUES/iPSCs stage. (C) RT-qPCR analysis of hESCs and their NPC- and neuronal derivatives confirms the highly segregated expression of miR-934 at the neural induction stage (2.6+0.2, n = 3, p=0.006). (D) Chromosomal location of miR-934 in human and non-human primate species (adapted from UCSC Genome Browser. (E, F) Graphs incorporating miRNA expression data from uniformly-analyzed small RNA-Seq libraries (detailed in Supplementary file 4) corresponding to human fetal and adult neural tissue (E) and cellular samples with reference to our NPCs data (F). miR-934 expression shows selective expression in samples associated with human cortical development in vivo and neural induction in vitro. Bars and error bars represent mean values and the corresponding SEMs; 0.001<**p<0.01.
-
Figure 2—source data 1
qRT PCR data for hsa-miR-934 relative expression.
- https://cdn.elifesciences.org/articles/50561/elife-50561-fig2-data1-v2.xlsx
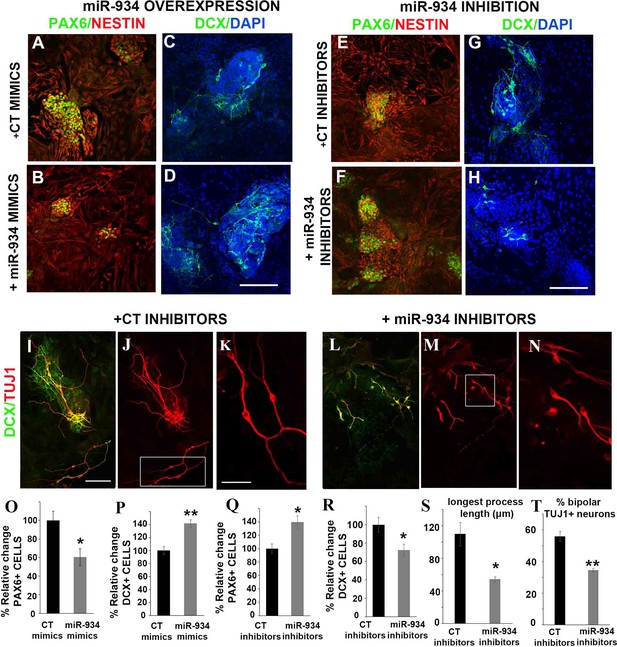
Perturbation of miR-934 affects the balance between Pax6+ progenitors and DCX+ neuroblasts and impacts on the morphological transition of newly born neurons.
(A–H) Representative confocal images of hESC-derived cultures at neural induction stage (DIV12) following immunolabeling for Pax6 (green) and Nestin (red) or for doublecortin (DCX, green), upon miR-934 overexpression or inhibition, as indicated. Cell nuclei were visualized with Hoechst (blue). Scale bar = 100 μm. (O, P) Overexpression of miR-934 resulted in a reduction of the percentage of PAX6+ progenitors out of all cells in culture (CT mimics: 100 ± 9.9 and miR934 mimics: 60.6 ± 9.2, p=0.04, n = 3) and an increase of the percentage DCX+ neuroblasts out of all cells in culture (CT mimics: 100 ± 6.1 and miR934 mimics: 141.8 ± 5.6, p=0.007, n = 3). (Q, R) Inhibition of miR-934 has the opposite effect (for PAX6+% cell quantification: CT inhibitors 100 ± 7.2 and miR934 inhibitors 139 ± 9.6, p=0.01, n = 5; for DCX+% cell quantification: CT mimics 100 ± 7.6 and miR934 inhibitors 72.2 ± 6.5, p=0.02, n = 5–6). (I–J, L–M) Representative confocal images following immunolabeling for DCX (green) and TUJ1 (red) of control cultures at neural induction (DIV12) or upon miR-934 inhibition. Scale bar = 100 μm. TUJ1+ cells in white insets in (J, M) are shown at higher magnification in (K, N), respectively. Scale bar = 50 μm. Measurements performed were based on TUJ1 expression: Inhibition of miR-934 during neural induction affects the morphology of newly-born neurons leading to (S) reduction of the length of the longest TUJ1+ neurite (μm): for control inhibitors (CT) 109.78 ± 14. 2 vs. 54.53 ± 2.8 for miR-934 inhibitors; p=0.02, n = 4, 280 cells and (T) to decreased number of TUJ1+ neurons exhibiting a bipolar phenotype (% bipolar neurons over total neurons analyzed for control CT inhibitors: 55.9 ± 3.1 vs. 34.4 ± 1.1 for miR-934 inhibitors; p=0.003, n = 4, 160 cells). Bars and error bars represent mean values and the corresponding SEMs; 0.01<*p<0.05; 0.001<**p<0.01.
-
Figure 3—source data 1
Data on estimation of % Relative PAX+ and DCX+ cells.
- https://cdn.elifesciences.org/articles/50561/elife-50561-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Data on measurements of Longest Process in TUJ1+ neurons and of % of bipolar TUJ1+ neurons.
- https://cdn.elifesciences.org/articles/50561/elife-50561-fig3-data2-v2.xlsx
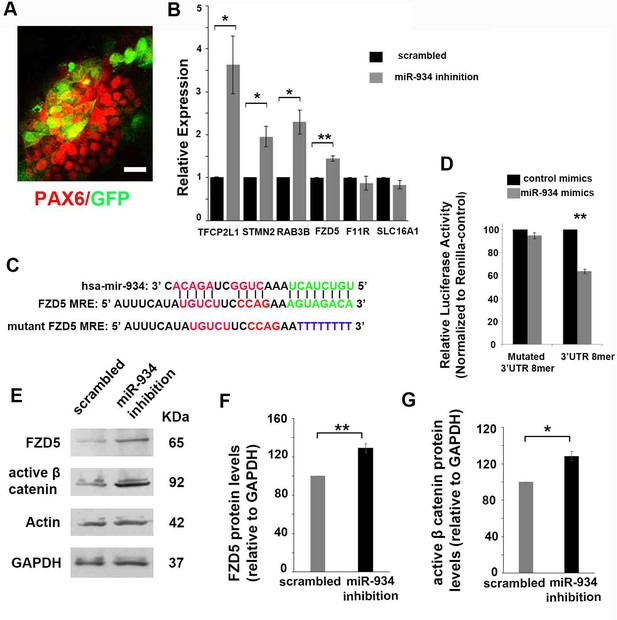
Identification of miR-934 targets during neural induction.
(A) Confocal image showing PAX6+ neural progenitors (red) at DIV 15 transduced with miRZip lentivector (GFP fluorescence, green) for sustained inhibition of miR-934. Scale bar: 20 μm. (B) qRT-PCR data showing upregulation of miR-934 targets upon sustained miR-934 inhibition: TFCP2L1 (scrambled control 1+0.006 vs miR934 inhibition 3.63+0.67, p=0.03, n = 4), STMN2 (scrambled control 1+0.003 vs miR934 inhibition 1.95+0.23, p=0.02, n = 4), RAB3B (scrambled control 1+0.003 vs miR934 inhibition 2.29+0.28, p=0.01, n = 4), and FZD5 (scrambled control 1,004+0005 vs miR934 inhibition 1,44+0,061, p=0,005, n = 4). No changes were observed in the mRNA expression of F11R or SLC16A1 (p=0,18 for SLC16A1and p=0,47 for F11R, n = 4). (C) Sequence complementarity of miR-934 with the 8-mer binding site in Fzd5 3'-UTR region (green) and corresponding mutated site (blue), used in luciferase activity assay. (D) Estimation of the suppression of luciferase activity in HEK293T cells upon co-transfection with miR-934 mimics and the MRE (8-mer binding site) reporter construct. Suppression of luciferase activity was released upon mutation of the FZD5 8mer binding site (p=0.002, n = 3). (E–G) Sustained inhibition of miR-934 results in increased protein levels of FZD5 (p=0.007, n = 4) (E, F) and active β-catenin (p=0.01, n = 4) (E, G). Bars and error bars represent mean values and the corresponding SEMs; 0.01<*p<0.05; 0.001<**p<0.01.
-
Figure 4—source data 1
qRT PCR data for expression of miR-934 predicted targets upon sustained miR934 inhibition.
- https://cdn.elifesciences.org/articles/50561/elife-50561-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Data on luciferase activity in HEK293T cells upon co-transfection with miR-934 mimics and the MRE (8-mer binding site) reporter construct.
- https://cdn.elifesciences.org/articles/50561/elife-50561-fig4-data2-v2.xlsx
-
Figure 4—source data 3
Data on western blot results for FZD5 and b-catenin proteins following miR934 Sustained inhibition.
- https://cdn.elifesciences.org/articles/50561/elife-50561-fig4-data3-v2.xlsx
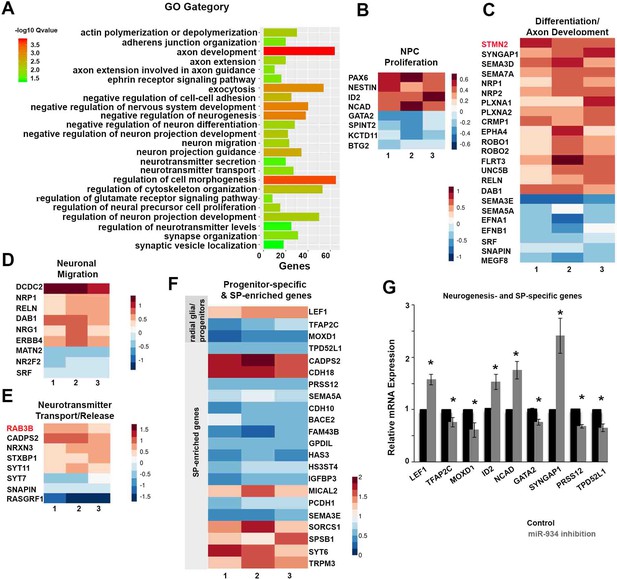
Sustained Inhibition of miR-934 induces global expression changes in genes associated with NPC proliferation and neuronal differentiation.
(A) GO enrichment analysis of differential gene expression data following sustained inhibition of miR-934. (B–E) Heatmaps presenting the log2-fold change of molecules involved in the indicated biological processes following miR-934 inhibition, as determined by GO enrichment analysis, from three independent experiments 1, 2 and 3. miR-934 targets STMN2 and RAB3B are depicted in red. (F) Heatmap presenting the log2-fold change of progenitor-specific and SP-enriched genes. Warmer colors indicate higher fold changes. (G) Graph showing qRT-PCR validation data for genes affected by miR-934 inhibition (n = 3–4): LEF1 (scrambled control 1+0.0003 vs miR934 inhibition 1.57+0.09, p=0.009), TFAP2C (scrambled control 1+0.0006 vs miR934 inhibition 0.75+0.08, p=0.04), MOXD1 (scrambled control 1+0.0002 vs miR934 inhibition 0.6+0.14, p=0.04), ID2 (scrambled control 1+0.01 vs miR934 inhibition 1.52+0.14, p=0.03), NCAD (scrambled control 1+0.002 vs miR934 inhibition 1.75+0.17, p=0.01), GATA2 (scrambled control 1+0.01 vs miR934 inhibition 0.75+0.06, p=0.01), SYNGAP1 (scrambled control 1+0.0004 vs miR934 inhibition 2.4+0.33, p=0.02), PRSS12 (scrambled control 1+0.0002 vs miR934 inhibition 0.67+0.03, p=0.002) and TPD52L1 (scrambled control 1+0.0002 vs miR934 inhibition 0.65+0.07, p=0.03). Bars and error bars represent mean values and the corresponding SEMs; 0.01<*p<0.05.
-
Figure 5—source data 1
qRT PCR data for expression of genes affected by miR-934 inhibition.
- https://cdn.elifesciences.org/articles/50561/elife-50561-fig5-data1-v2.xlsx
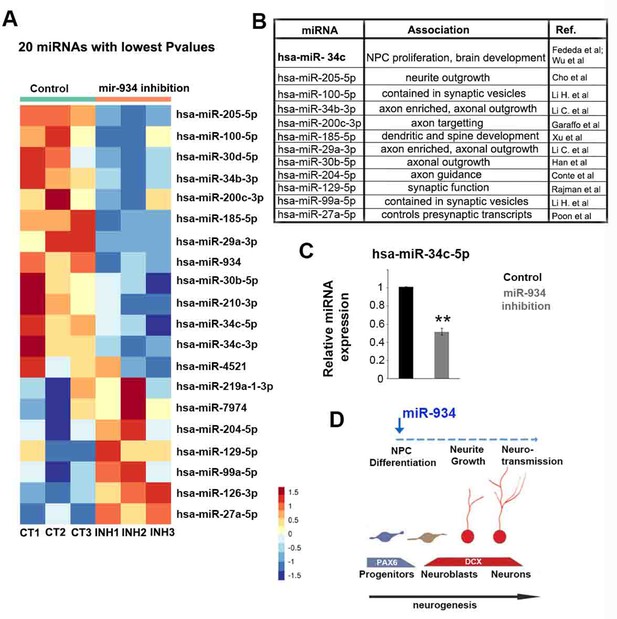
miRNA expression profiling following sustained inhibition of miR-934.
(A) Heatmap showing the 20 miRNAs with lowest p-values at differential expression analysis following miR-934 sustained inhibition. (B) Table describing the association of differentially expressed miRNAs with axon/dendrite development and/or synapse formation. miRNA associations taken from Fededa et al., 2016; Wu et al., 2014; Cho et al., 2013; Conte et al., 2014; Garaffo et al., 2015; Han et al., 2015; Li et al., 2019; Li et al., 2015; Poon et al., 2016; Rajman et al., 2017; Xu et al., 2013. (C) qRT-PCR showing downregulation of mir34c-5p following miR-934 sustained inhibition (scrambled control 1+0.005 vs miR934 inhibition 0.51+0.03, n = 3, p=0.005). Bars and error bars represent mean values and the corresponding SEMs; 0.001<**p<0.01. (D) Sketch depicting the neurogenic function of miR-934 also affecting subsequent neuronal differentiation processes as reflected in large-scale changes in the expression of genes and miRNAs associated with neurogenesis.
-
Figure 6—source data 1
qRT-PCR data for expression of miR-34c-5p following miR-934 sustained inhibition.
- https://cdn.elifesciences.org/articles/50561/elife-50561-fig6-data1-v2.xlsx
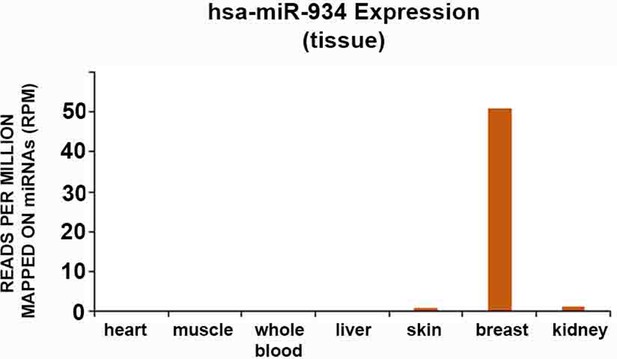
Graph incorporating miRNA expression data from uniformly-analyzed small RNA-Seq libraries of non-neural human tissue (detailed in Supplementary file 3).
Association was noted only in one case (breast).
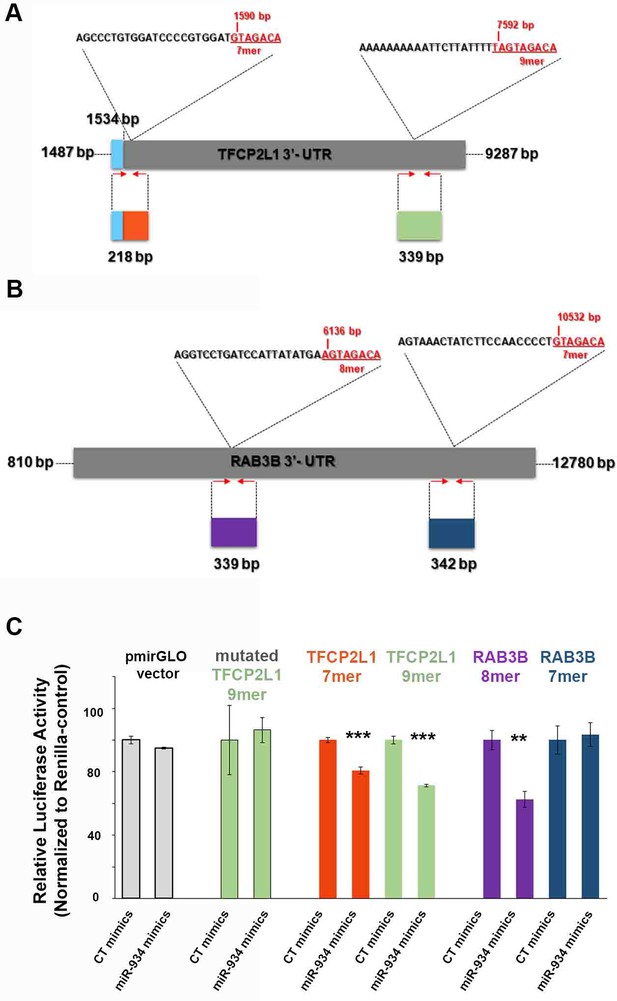
Identification of binding sites for miR-934 on the 3'-UTR of TFCP2L1 and RAB3B.
(A) Schematic representation of TFCP2L1 3’-UTR depicting in red fonts the seed-binding sequences of the two predicted MREs for miR-934 with their respective positions. To examine mechanistically the inhibition exerted by miR-934 on TFCP2L1 expression, a luciferase reporter system was used to evaluate independently the binding of miR-934 on each of the 2 MREs which contain either a 7mer, or a 9-mer site. Shown in color code are the sequences and sizes of TFCP2L1 3’-UTR regions containing the 2 MREs, as they were cloned into the pmirGLO vector [orange: 7mer (blue box represents the last 47 base pairs at the 3’ end of the human TFCP2L1 coding region); green: 9mer]. Arrows indicate primer sites for cloning. Fragments of the 3'-UTR of TFCP2L1 containing each of the two possible binding domains for miR-934 (218 bp, including the 7mer sequence; and 339 bp including the 9-mer sequence) were cloned separately into the 3'-UTR of the dual luciferase reporter construct pmirGLO. (B) Schematic representation of RAB3B 3’-UTR depicting in red fonts the seed-binding sequences of the two predicted MREs for miR-934 with their respective positions. To examine mechanistically the inhibition exerted by miR-934 on RAB3B expression, a luciferase reporter system was used to evaluate independently the binding of miR-934 on each of the 2 MREs, which contain either a 8mer or a 7mer site. Shown in color code are the sequences and sizes of RAB3B 3’-UTR regions containing the 2 MREs, as they were cloned into the pmirGLO vector (purple: 8mer; dark blue: 7mer). Arrows indicate primer sites for cloning. Fragments of the 3'-UTR of RAB3B containing each of the two possible binding domains for miR-934 (339 bp, including the 8mer sequence; and 342 bp including the 7-mer sequence) were cloned separately into the 3'-UTR of the dual luciferase reporter construct pmirGLO. (C) Upon co-transfection of HEK293T cells with miR-934 mimics and the different TFCP2L1 and RAB3B reporter constructs, luciferase activity was suppressed in both constructs for TFCP2L1 (p=0.0002 for 7mer and p=0.0007 for 9mer) and only in the construct containing the 8mer site sequence for RAB3B (p=0.002). By contrast luciferase activity was not affected upon transfection of control constructs (pmiRGLO vector and the mutated TFCP2L1 9mer) in the presence of miR934 mimics. Color code as in (A) and (B).
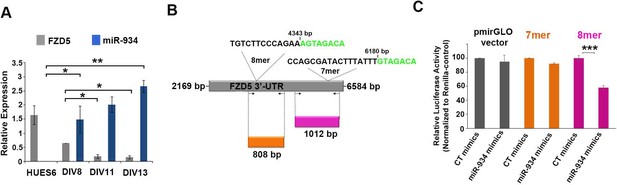
Identification of an 8mer-seed sequence as binding site for miR-934 on the 3'-UTR of Fzd5.
(A) RT-qPCR analysis at different time points of neural induction, showing up-regulation of miR-934 and parallel down-regulation of the mRNA of its predicted target, Fzd5. Using the DIANA-microT-CDS target prediction algorithm, two putative MREs were identified on the 3'-UTR of Fzd5. (B) Schematic representation of Fzd5 3’-UTR depicting in green characters the seed-binding sequences of the two predicted MREs for miR-934 with their respective positions. To examine mechanistically the inhibition exerted by miR-934 on Fzd5 expression, a luciferase reporter system was used to evaluate independently the binding of miR-934 on each of the 2 MREs which contain either a 7mer (binding score: 0.032), or an 8-mer (binding score: 0.005) site. Shown in color code are the sequences and sizes of Fzd5 3’-UTR regions containing the 2 MREs, as they were cloned into the pmirGLO vector (orange: 7mer; magenta: 8mer). Arrows indicate primer sites for cloning. Fragments of the 3'-UTR of Fzd5 containing each of the two possible binding domains for miR-934 (1012 bp, including the 7mer sequence; and 808 bp including the 8-mer sequence) were cloned separately into the 3'-UTR of the dual luciferase reporter construct pmirGLO. (C) Upon co-transfection of HEK293T cells with miR-934 mimics and the different Fzd5 reporter constructs, luciferase activity was only suppressed in the construct (808 bp) containing the 8mer site sequence (p=0.000008). Color code as in (A).
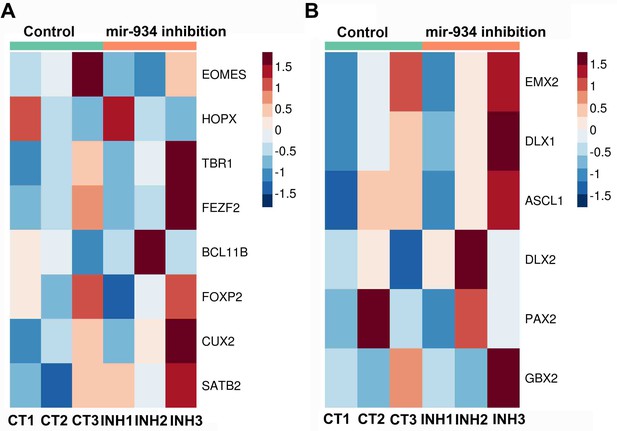
Heatmaps showing the expression of genes displaying no significant differences following miR-934 sustained inhibition.
(A) Genes associated with intermediate progenitors, outer radial glia and cortical plate neurons. (B) Genes associated with regional identities during brain development.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (H. sapiens) | hESC line HUES6 | Cowan CA, Klimanskaya I, McMahon J, Atienza J, Witmyer J, Zucker JP, et al. Derivation of embryonic stem-cell lines from human blastocysts. The New England journal of medicine. 2004;350 (Muñoz-Sanjuán and Brivanlou, 2002):1353–6. | RRID:CVCL_B19 | Cell line (H. sapiens) |
| Chemical compound, drug | Knockout Serum Replacement (KSR) | Life Technologies Thermo Fisher | Life Technologies, Thermo Fisher 10828–010 | |
| Chemical compound, drug | KnockOut DMEM medium | Life Technologies Thermo Fisher | Life Technologies, Thermo Fisher 10829–018 | |
| Chemical compound, drug | matrigel matrix | BD biosciences | BD 354234 | See Materials and methods |
| Chemical compound, drug | Noggin | R and D Systems | R and D 6057 NG-25 | See Materials and methods |
| Chemical compound, drug | SB431542 | Tocris Biosciences | Tocris Biosciences 1614 | See Materials and methods |
| Commercial assay or kit | RNA Clean and Concentrator kit | Zymo Research | Zymo Research R1018 | |
| Recombinant DNA reagent | pmiR-GLO reporter vector | Promega | Promega E1330 | |
| Recombinant DNA reagent | miRZip lentivector-based anti-microRNA system | Systems Biosciences | https://systembio.com/products/mirna-and-lncrna-research-tools/mirzip-knockdown-vectors/mirzip-anti-mirna-constructs/ | |
| Chemical compound, drug | Lipofectamine 2000 | Life Technologies/Thermo | Life Technologies11668–019 | |
| Sequenced-based reagent | miR-934 mimics | Exiqon/Qiagen | http://www.exiqon.com/ls/Pages/ProductDetails.aspx?ProductId=k1WVIuhVVQH848C13pCVyN5WGs82ItaE%2flSNAGCdK1%2bEWkr83WchdvRAbgzMNpIk3WDIx9O1eBJZUcYGe81LzgTaw2jJcGjXgRYNy58buck%3d&CategoryType=2yRAQGj8eprRK3%2bx8lE0zg%3d%3d | |
| Sequenced-based reagent | miR-934 inhibitors | Dharmacon | https://horizondiscovery.com/products/gene-modulation/knockdown-reagents/mirna/PIFs/miRIDIAN-microRNA-Hairpin-Inhibitor?nodeid=mirnamature-mimat0004977 | |
| Antibody | mouse monoclonal anti-PAX6 | Developmental Studies Hybridoma Bank | RRID:AB_528427 | Dilution (1:50) See Materials and methods |
| Antibody | goat polyclonal anti-doublecortin (DCX) | Santa Cruz | Sc-8067 | Dilution (1:100) See Materials and methods |
| Antibody | rabbit polyclonal anti-Frizzled 5 | Abcam | Ab75234 | Dilution (1:1000) See Materials and methods |
| Antibody | rabbit monoclonal anti-active β-catenin | Cell signaling | Cell signaling, 19807 | Dilution (1:1000) See Materials and methods |
| Software, algorithm | ImageJ | ImageJ (http://imagej.nih.gov/ij/) | RRID:SCR_003070 | |
| Sequenced-based reagent | RT-qPCR primers | This paper | See Supplementary file 7 |
Additional files
-
Supplementary file 1
Supplemental table 1 showing the 144 miRNAs across all differentiation stages demonstrating tissue specificity index tau >0.7.
- https://cdn.elifesciences.org/articles/50561/elife-50561-supp1-v2.docx
-
Supplementary file 2
Supplemental table 2 presenting The 10 most highly expressed miRNAs in NPCs demonstrating tissue specificity index tau >0.7.
- https://cdn.elifesciences.org/articles/50561/elife-50561-supp2-v2.docx
-
Supplementary file 3
Supplemental table 3 showing All Blastn results with evalue <1 for a query of the mature hsa-miR-934 sequence against all mature miRNAs in miRBase v22.1.
- https://cdn.elifesciences.org/articles/50561/elife-50561-supp3-v2.docx
-
Supplementary file 4
Supplemental table 4 presenting IDs and description of the small RNA-Seq human libraries of human tissue and cell data included in the analysis for miR934 expression.
- https://cdn.elifesciences.org/articles/50561/elife-50561-supp4-v2.docx
-
Supplementary file 5
Supplemental table 5 showing all the predicted mRNA targets for miRNA-934.
- https://cdn.elifesciences.org/articles/50561/elife-50561-supp5-v2.docx
-
Supplementary file 6
Supplemental table 6 presenting the 3’ UTR binding sites for miR934 on its four identified targets (adapted from Diana microT-CDS).
- https://cdn.elifesciences.org/articles/50561/elife-50561-supp6-v2.docx
-
Supplementary file 7
Supplemental table 7 listing the sequences of primers and oligos used in this study.
- https://cdn.elifesciences.org/articles/50561/elife-50561-supp7-v2.docx
-
Supplementary file 8
Supplemental table 8 showing information on progenitor-specific and SP-enriched genes affected by sustained inhibition of miR-934.
- https://cdn.elifesciences.org/articles/50561/elife-50561-supp8-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/50561/elife-50561-transrepform-v2.pdf





