A large genomic insertion containing a duplicated follistatin gene is linked to the pea aphid male wing dimorphism
Figures
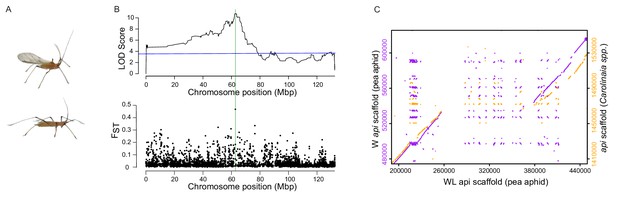
Linkage and association mapping of api.
(A) Winged (top) and wingless (bottom) males. (B) Top: QTL analysis revealed the presence of a single QTL peak on the X chromosome as assembled by Li et al. (2019). The blue line indicates p=0.01 determined by 1000 permutations of the phenotype data relative to the genotype data. Further RFLP analysis of recombinant F2 individuals localized api to a ~ 190 kb region, which is highlighted by a green, vertical line. Bottom: Genetic differentiation between winged and wingless males using FST values calculated from 20 kb windows across the X chromosome. (C) Dot plots showing areas of similarity among our newly assembled pea aphid genomic scaffolds containing api (either W:winged or WL:wingless, as indicated) and the scaffold containing api from a species in the Carolinaia genus.

Wingless, but not winged reads, map to the wingless-specific region.
Shown is the read depth of pool-seq data from alfalfa pea aphids (winged: blue, wingless: red) mapping against the reconstructed WL api scaffold. The wingless-specific sequence and recombination mapping defined api region (‘api’) are indicated.
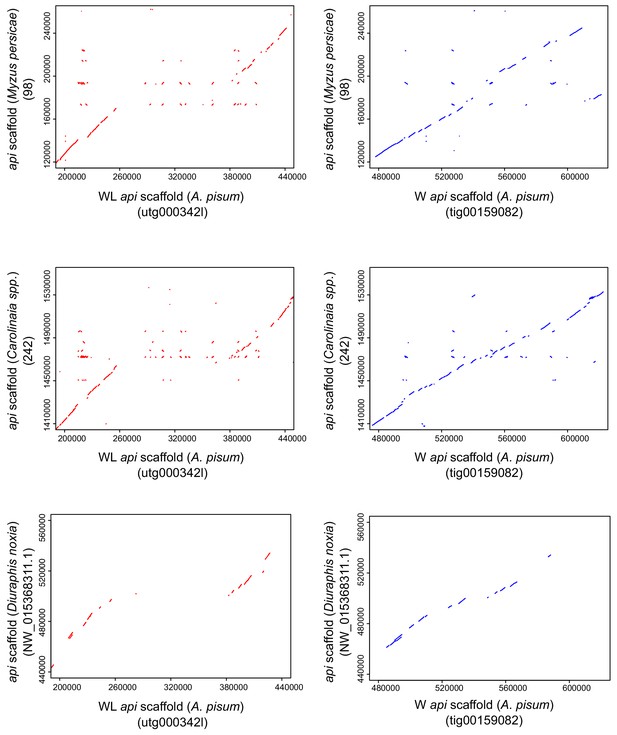
Similarity of the wingless-specific region to putatively homologous regions in the peach-potato aphid (Myzus persicae), the Carolinaia spp. aphid, and the Russian wheat aphid (Diuraphis noxia).
Dot plots of the api homologous regions: the pea aphid WL api scaffold compared to the corresponding scaffolds of other aphid species (indicated in parentheses); the pea aphid W api scaffold compared to the corresponding scaffolds of other aphid species (indicated in parentheses).

The male wing-dimorphic pea aphid (Acyrthosiphon pisum, arrow) is embedded in a clade of mainly monomorphic winged species.
Red species names indicate only wingless males in that species; blue names indicate only winged males in that species; orange names indicate both winged and wingless males in that species; black names indicate lack of phenotype data for that species. Male phenotype was determined from the online resource aphidsonworldsplants.info, which combines information from multiple sources (Blackman and Eastop, 2000; Blackman and Eastop, 1994). Phylogeny is reproduced from Hardy et al. (2015), with their maximum-likelihood and parsimony bootstrap values shown on the left and right of slashes, respectively, and Bayesian posterior probabilities shown below branches.
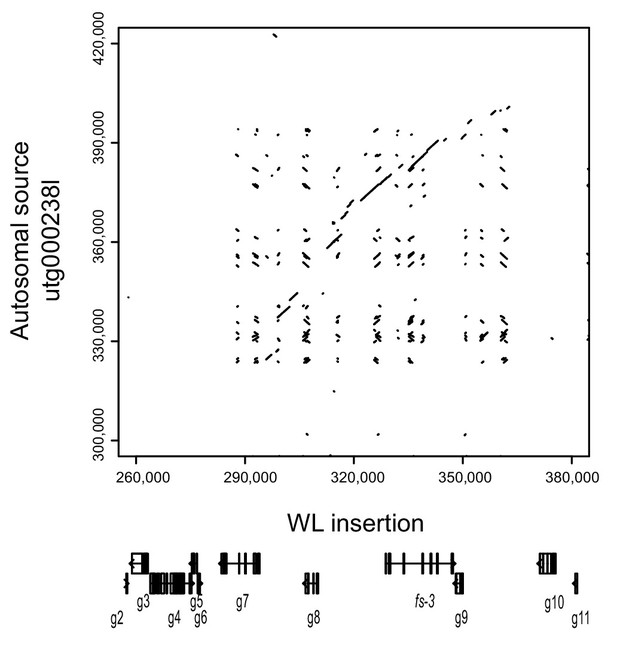
Similarity of the wingless-specific region to an autosomal scaffold containing the fs-2 gene.
Dot plots of the WL insertion region compared to the autosomal source in the pea aphid genome.
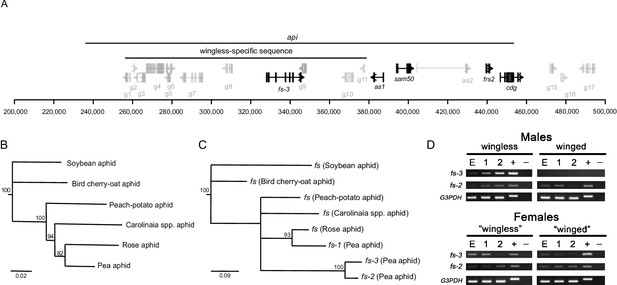
An expressed follistatin gene duplicate in the wingless-specific insertion.
(A) Gene models at the api locus, with distances shown in bp on the x-axis. The recombination mapping defined api region and the wingless-allele specific insertion are indicated. ORFs inside the wingless-specific insertion are our own annotation, while those outside the insertion are those of the genome v3.0 (Li et al., 2019). Repetitive ORFs and those outside of the api region are shown in gray. ORFs with evidence for expression either from RT-PCR or RT-qPCR are shown in black. (B) Phylogenetic tree of the aphids studied here: the soybean aphid (Aphis glycines), the bird cherry-oat aphid (Rhopalosiphum padi), the peach-potato aphid (Myzus persicae), an unidentified Carolinaia genus species, the rose aphid (Macrosiphum rosae), and the pea aphid (Acyrthosiphon pisum). The maximum likelihood tree was constructed from 2,378 bp of DNA sequence from elongation factor 1α, 18S rRNA, 12S rRNA, cytochrome b, and cytochrome c oxidase subunit I collected from Genbank. (C) Maximum likelihood tree of the different copies of follistatin (fs) nucleotide sequences. Nodes with bootstrap values < 80 are collapsed. (D) RT-PCR analysis of fs-3 and fs-2 gene expression across different developmental stages (E: embryo cDNA, 1: 1st instar nymph cDNA, 2: 2nd instar nymph cDNA; +: genomic DNA, -: no cDNA) of wingless males, winged males, daughters of crowded adult wingless females (‘winged’ females, which are predominantly winged females), daughters of uncrowded adult wingless females (‘wingless’ females, which are predominantly wingless female samples). Females here are heterozygous for the api locus, and thus have fs-3 as indicated by the positive band in the female gDNA lanes. Data for the G3PDH gene are provided as a positive control.
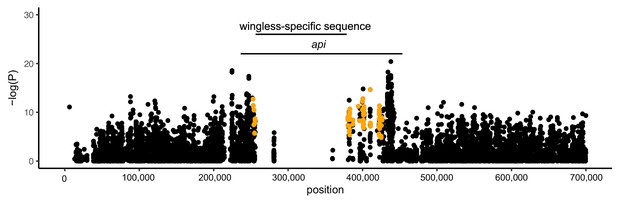
Association between SNPs at api and the male wing phenotype.
Points show the association between SNPs and the male phenotype, illustrated as the -log(P) from a Fisher’s exact test between SNPs and the male wing phenotype using the alfalfa biotype pool-seq data. Orange points are SNPs that are perfectly segregating with the phenotype in the across-biotype data.
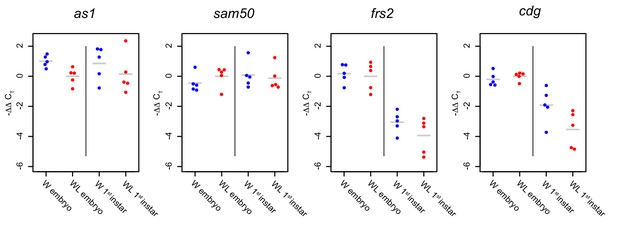
Genes in the insertion-adjacent region are not differentially expressed as measured by RT-qPCR analysis.
Each gene was measured in five biological replicates collected from winged and wingless male embryos and first instar nymphs. Each point represents a replicate. Blue indicates winged and red indicates wingless males. Y-axes show the ΔCT values for each sample subtracted from the average ΔCT value of wingless embryos (-ΔΔCT): higher values therefore represent stronger relative gene expression. Short gray horizontal lines show means.
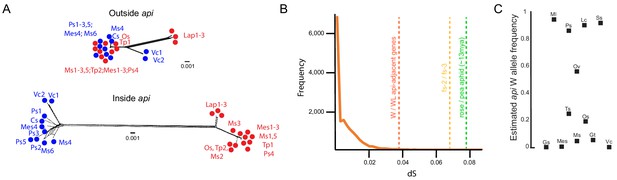
The polymorphism predates radiation within the species complex.
(A) Tree networks, built with SplitsTrees, based on positions ~ 1,235 kb-1,245kb (‘Outside api’) or ~379 kb–388 kb (‘Inside api’) on the WL api scaffold. Winged individuals are colored in blue and wingless alleles in red. Abbreviations: Cs: Cytisus scoparius, Lap: Lathyrus pratensis, Mes: Melilotus spp., Ms: Medicago sativa, Os: Ononis spinosa, Ps: Pisum sativum, Tp: Trifolium pratense, and Vc: Vicia cracca. (B) Neutral divergence times, as measured by dS. Orange line shows the dS distribution for 16,367 genes, derived from comparing individuals Lap1 and Ms3. Dotted vertical lines indicate the dS values for the average of the three api-adjacent genes (as1, sam50, frs2) when winged and wingless individuals are compared, the comparison of fs-2 and fs-3, and the average rose to pea aphid dS. (C) Allele frequency estimates based on SNP frequency analysis of read counts in pool-seq data for each of the different pea aphid biotypes. Biotypes that overlap with data in (A) are shown with the same abbreviations as in (A), but some biotypes are unique to analyses in (A) or (C). Additional abbreviations: Gs: Genista sagittalis, Gt: Genista tinctoria, Lc: Lotus corniculatus, Ml: Medicago lupulina, Ov: Onobrychis viciifolia, Ss: Securigera spp., and Ts: Trifolium spp.
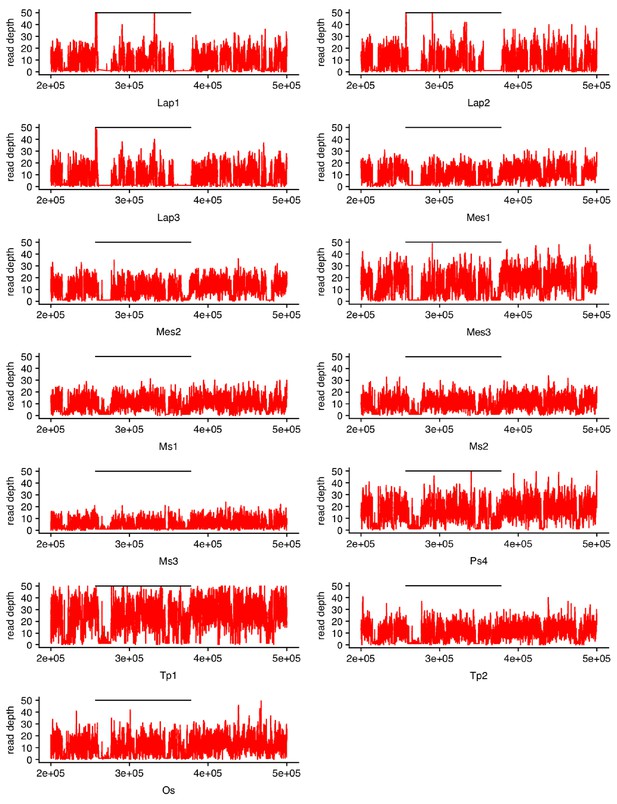
Reads from wingless individuals of different biotypes map to the wingless-specific region.
The x-axis shows position along the api wingless scaffold. Read depth, on the y-axis, differs among individuals. A solid black line indicates the wingless-specific region. Abbreviations: Mes: Melilotus officinalis, Tp: Trifolium pratense, Lap: Lathyrus pratensis, Ps: Pisum sativum, Ms: Medicago sativa, Os: Ononis spinosa.

Reads from winged individuals of different biotypes do not map to the wingless-specific region.
The x-axis shows position along the api wingless scaffold. Read depth is absolute, and differs among individuals. A solid black line indicates the wingless-specific region. Abbreviations: Mes: Melilotus officinalis, Tp: Trifolium pratense, Lap: Lathyrus pratensis, Ps: Pisum sativum, Ms: Medicago sativa, Os: Ononis spinosa, Mes: Melilotus suaveolens, Vc: Vicia cracca, Cs: Cytisus scoparius..
Additional files
-
Source data 1
Genotypes of F2 individuals used for QTL analysis.
- https://cdn.elifesciences.org/articles/50608/elife-50608-data1-v1.xlsx
-
Source data 2
Fst values for winged versus wingless pool-seq comparisons.
- https://cdn.elifesciences.org/articles/50608/elife-50608-data2-v1.xlsx
-
Source data 3
Data from RT-qPCR analysis.
- https://cdn.elifesciences.org/articles/50608/elife-50608-data3-v1.xlsx
-
Source data 4
Alignment of different biotype individuals outside of the api region.
- https://cdn.elifesciences.org/articles/50608/elife-50608-data4-v1.xlsx
-
Source data 5
Alignment of different biotype individuals inside the api region.
- https://cdn.elifesciences.org/articles/50608/elife-50608-data5-v1.xlsx
-
Source data 6
Between biotype dS comparisons.
- https://cdn.elifesciences.org/articles/50608/elife-50608-data6-v1.xlsx
-
Supplementary file 1
ORFs inside and directly outside of the api region.
- https://cdn.elifesciences.org/articles/50608/elife-50608-supp1-v1.xlsx
-
Supplementary file 2
Synonymous and non-synonymous substitution rates based on codon-aligned nucleotide sequences.
- https://cdn.elifesciences.org/articles/50608/elife-50608-supp2-v1.xlsx
-
Supplementary file 3
Collection locations for winged and wingless males used in the pool-seq study.
- https://cdn.elifesciences.org/articles/50608/elife-50608-supp3-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/50608/elife-50608-transrepform-v1.docx





