GLI transcriptional repression regulates tissue-specific enhancer activity in response to Hedgehog signaling
Figures
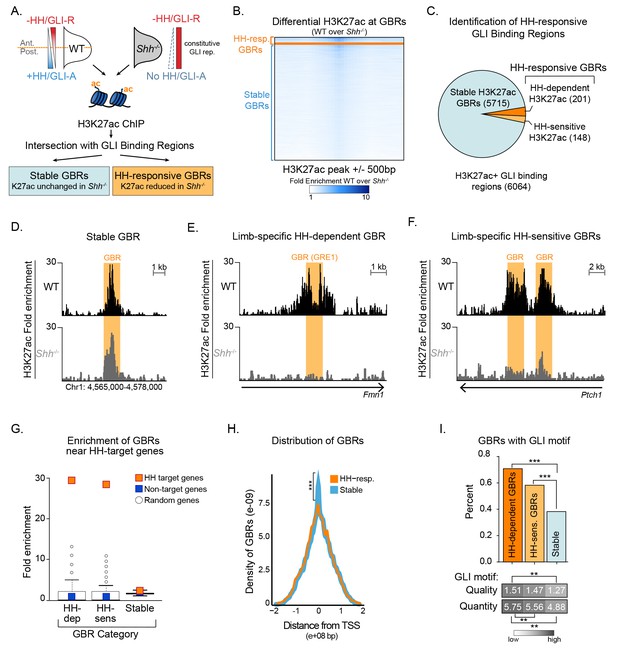
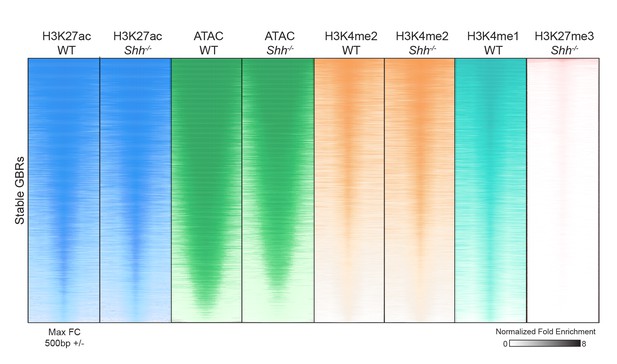
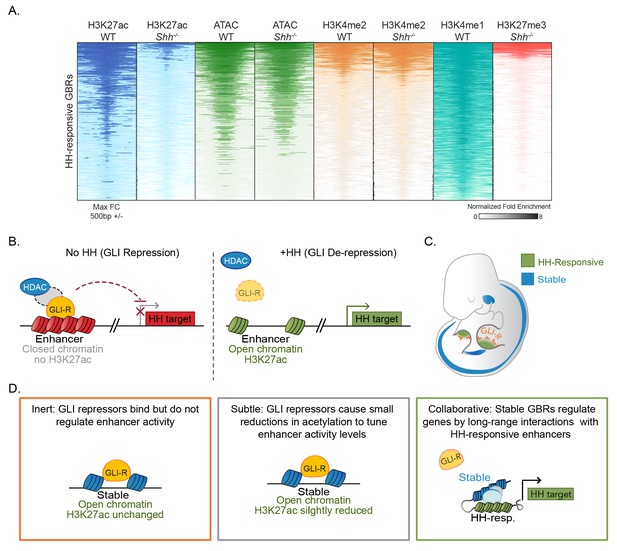
Hedgehog signaling regulates acetylation of H3K27 at a subset of GLI binding regions.
(A) Pipeline for identifying different categories of GLI bound regions (GBRs). (B) Heatmap depicting differential H3K27ac enrichment in WT over Shh-/- limb buds for HH-responsive and Stable GBRs. (C) Classification of GBR categories from E10.5 GBRs with H3K27ac in WT limbs. (D-F). H3K27ac enrichment in WT and Shh -/- is shown across a representative genomic region near a Stable GBR (D), and biologically validated HH-responsive GBRs: a HH-dependent GBRs, GRE1, at the HH target gene Gremlin 1 (Grem1) (Li et al., 2014) (E) and HH-sensitive GBRs shown to regulate limb-specific expression of the HH target Ptch1 (Lopez-Rios et al., 2014) (F). (G) HH-dependent GBRs, HH-responsive GBRs and Stable GBRs are significantly enriched (2 kb upstream- 1 kb downstream of TSS) near HH target genes compared to randomly chosen genes (p=0, p=0 and p=0, respectively, permutation test based on 1000 permutations). (H) Proportional distribution of Stable and HH-responsive GBRs arounds transcription start sites (TSS), indicating significant enrichment of Stable GBRs at TSS compared to HH-responsive GBRs (p=2.55e-40, Fisher's exact test, two sided). (I) Both HH-dependent and HH-sensitive GBRs have significantly more GLI motifs than Stable GBRs (top)(p=2.2e-16 and p=8.00e-06; one-sided proportional test). HH-dependent and HH-sensitive GBRs containing GLI motifs have significantly higher quality of GLI motifs than Stable GBRs (Quality score; p=5.03e-13 and p=5.98e-08; one-sided Wilcoxon test) and significantly more motifs per GBR within HH-dependent GBRs than Stable GBRs (Quantity score; p=5.92e-06; one-sided Wilcoxon test). See Figure 1—figure supplement 1, Figure 1—source data 1, Figure 1—source data 2, Figure 1—source data 3, Figure 1—source data 4.
-
Figure 1—source data 1
Endogenous GLI3-Flag ChIP-seq analyzed data and called peaks.
GLI3 binding regions with called peaks with a false discovery rate (FDR) < 0.05 from two biological replicates of E10.5 (32–35S) forelimbs. Rank ordered coordinates, peak length, log2 fold change (log2FC) and FDR are listed for each peak.
- https://cdn.elifesciences.org/articles/50670/elife-50670-fig1-data1-v1.xlsx
-
Figure 1—source data 2
WT vs Shh-/- H3K27ac ChIP-seq analyzed data and called peaks.
H3K27ac called peaks with a FDR < 0.05 from two biological replicates from WT and Shh-/- E10.5 forelimbs. For each peak, the assigned Peak ID, coordinates, peak type, fold change normalized to input for WT and Shh-/- samples and fold change of WT over Shh-/- are listed. Additional tabs include sorted datasets for sub-classifications. Tabs containing GBRs indicate intersections with GLI binding regions.
- https://cdn.elifesciences.org/articles/50670/elife-50670-fig1-data2-v1.xlsx
-
Figure 1—source data 3
H3K4me1 ChIP-seq analyzed data and called peaks from GSE86690.
H3K4me1 called peaks with a false discovery rate (FDR) < 0.05 from two biological replicates of E10.5 WT forelimbs. Note that this is a reanalysis of a publicly available ENCODE dataset (see methods).
- https://cdn.elifesciences.org/articles/50670/elife-50670-fig1-data3-v1.xlsx
-
Figure 1—source data 4
Motifs uncovered from HH-responsive enhancers.
Table showing the top 20 motifs uncovered from de novo motif analysis on HH-responsive GBRs. The enrichment is relative to matched genomic controls. Note that ‘HH_resp_2’ is the only motif with an enrichment value of greater than two and corresponds with a known GLI binding motif.
- https://cdn.elifesciences.org/articles/50670/elife-50670-fig1-data4-v1.pdf
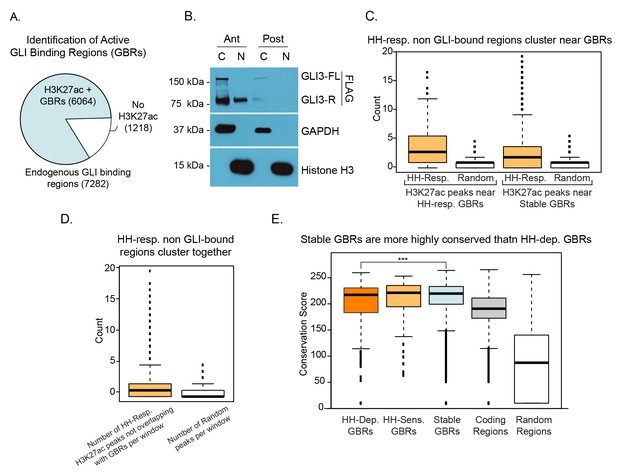
Nuclear localization of GLI3 and properties of GLI binding regions.
(A) Intersection of endogenous GLI3 binding and H3K27ac in E10.5 WT limb buds. (B) Western blots from anterior and posterior E10.5 limb buds indicating the distribution of endogenous GLI3-FLAG in cytoplasmic and nuclear fractions (C = cytoplasmic fraction, N = nuclear fraction; Ant = Anterior forelimb, Post = Posterior forelimb) (n = 3). (C) Hedgehog-responsive enhancers that are not bound by GLI are clustered near GLI binding regions. Box plot indicates the proximity of HH-responsive H3K27ac peaks that are not bound by GLI to either HH-Responsive GBRs or Stable GBRs compared to random peaks. For both HH-responsive and stable GBRs, the number of HH-Responsive non-GBR H3K27ac peaks is significantly larger than the number of random regions (Wilcoxon-test p-value=0). (D) HH-responsive peaks not bound by GLI3 are clustered together. The genome was split into 100,000 base-pair non-overlapping windows and the number of HH-responsive H3K27ac peaks that are not bound by GLI3 were counted as well as the number of random peaks. Only windows that overlapped with at least one HH-responsive H3K27ac peak or random peak were considered. The two counts are significantly different (Wilcoxon-test p-value=0). The dark black line indicates the median. The lower boundary of the box indicates the first quantile, while the upper boundary of the third box is the third quantile. The circles indicate outliers. (E) Box plot showing the conservation scores for different classes of GBRs. The conservation scores correspond to phastCons values linearly scaled from 0 to 255. HH-responsive GBRs have significantly lower conservation scores than stable GBRs (p-value=0.0001134492, one sided Wilcoxon test). None of the other pairs of GBRs are significantly different from each other. ‘Coding regions’ represent conservation scores for all protein coding genes in the mouse mm10 genome while ‘Random regions’ represent conservation scores for a set of 1000 random genomic loci that do not overlap with any gene. The dark black line indicates the median. The lower boundary of the box indicates the first quantile, while the upper boundary of the third box is the third quantile. The circles indicate outliers.
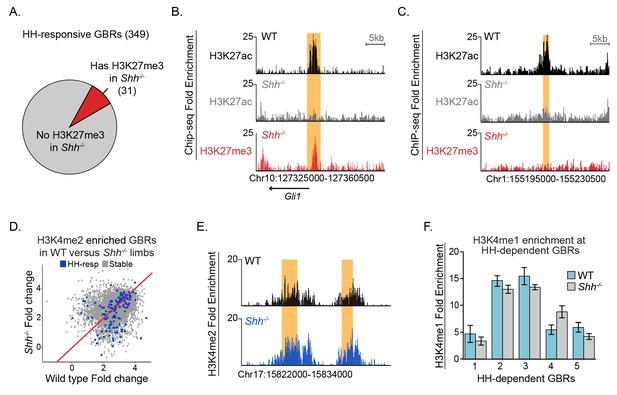
Most HH-responsive GBRs are not regulated by Polycomb repression and retain markers of poised enhancers.
(A) Chart depicts HH-responsive GBRs that contain enrichment for the PRC2 marker H3K27me3 in Shh-/- limb buds (n = 2). (B) Tracks depicting a HH-responsive region in Gli1 with differential H3K27ac enrichment in WT and Shh-/- limb buds and H3K27me3 enrichment in Shh-/- limb buds. (C) Tracks depicting a representative HH-dependent GBR that also lacks H3K27me3. (D) Scatter plot for H3K4me2 enrichment of Stable and HH-responsive GBRs from WT and Shh-/- limb buds (n = 2). No GBRs show significant changes in di-methylation of H3K4 between WT and Shh-/-. (E) Representative track showing comparable levels of H3K4me2 enrichment for a HH-responsive GBR in WT and Shh-/- limb buds. (F) Quantitative-PCR assays indicating H3K4me1 ChIP enrichment in WT and Shh-/- limb buds at HH-dependent GBRs (n = 2). See Figure 2—figure supplement 1, Figure 2—source data 1, Figure 2—source data 2, Figure 2—source data 3.
-
Figure 2—source data 1
Shh-/- H3K27me3 ChIP-seq analyzed data and called peaks.
H3K27me3 called peaks with a FDR < 0.05 from two replicates of Shh-/- E10.5 forelimbs. For each peak, the assigned Peak ID, coordinates, log2 fold change normalized signal to input. Additional tab includes H3K27me3 peaks that overlap with GLI3 binding regions; the GBR sub-classifications are specified.
- https://cdn.elifesciences.org/articles/50670/elife-50670-fig2-data1-v1.xlsx
-
Figure 2—source data 2
Hedgehog responsive genes with H3K27me3 enrichment.
The first column indicates genes previously identified as differentially expressed between Shh-/- and WT E10.5 limb buds (Lewandowski et al., 2015). The second column indicates the fold enrichment of H3K27me3 at the promoter compared to Input with the adjusted P-value indicated in the third column. The fourth column indicates whether the gene has a HH-dependent GBR (indicated by one and yellow shading) within the same presumptive TAD (Dixon et al., 2012). There are 22 HH-dependent target genes out of 80 HH-responsive genes.
- https://cdn.elifesciences.org/articles/50670/elife-50670-fig2-data2-v1.pdf
-
Figure 2—source data 3
WT vs Shh-/- H3K4me2 ChIP-seq analyzed data and called peaks.
H3K4me2 called peaks with a FDR < 0.05 from two replicates from WT and Shh-/- E10.5 forelimbs. For each peak, the assigned Peak ID, coordinates, peak type, fold change normalized to input for WT and Shh-/- samples and fold change of WT over. Shh-/- are listed. Additional tabs include sorted files for each peak type. Under the ‘GLI3 binding’ column, ‘TRUE’ implies overlap with a GBR, while ‘FALSE’ indicates no overlap.
- https://cdn.elifesciences.org/articles/50670/elife-50670-fig2-data3-v1.xlsx
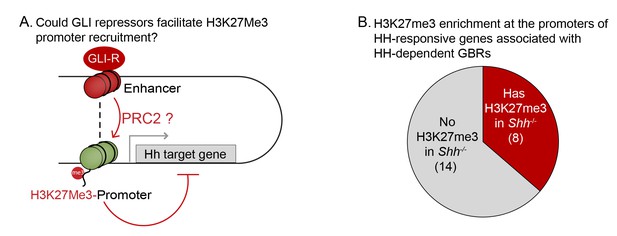
H3K27Me3 enrichment at the promoters of GLI target genes.
(A) Schematic illustrating a hypothetical mechanism by which GLI repressors bound to distal enhancers could facilitate the deposition of PRC2-marked H3K27Me3 at the promoters of target genes. (B) H3K27Me3 enrichment within the promoters of 22 HH responsive genes that also have HH-dependent GBRs (Figure 2—source data 2) was determined as for the enhancers except that the reads were summed in gene promoters instead of peak regions within a window spanning from 1500 bp upstream to 500 bp downstream of the transcriptional start site. H3K27Me3 enrichment was present in the promoters of 8/22 target genes.
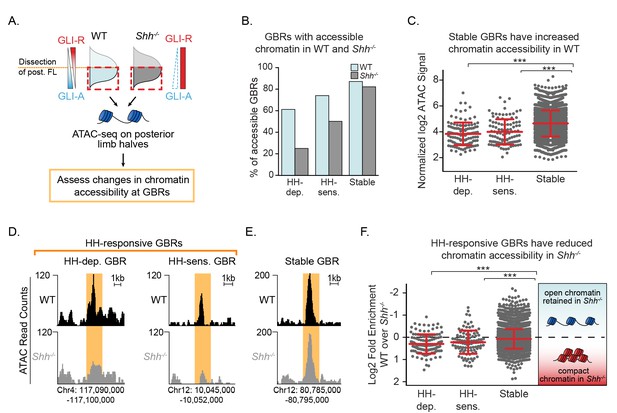
Chromatin accessibility is reduced in the absence of Hedgehog signaling.
(A) ATAC-seq pipeline for single pairs of dissected posterior halves of forelimbs (n = 2). ATAC peaks, signifying accessible chromatin regions were intersected with Stable GBRs and HH-responsive GBRs. (B) Many HH-responsive GBRs that are accessible in WT limb buds are inaccessible Shh-/- limb buds, while the accessibility of Stable GBRs remains largely unchanged. (C) Plot of log2 normalized signal in chromatin accessibility in WT limbs indicating that Stable GBRs are more accessible than HH-dependent and HH-responsive GBRs (p=3.98e-19, p=9.21e-11; Wilcoxon rank sum test). Each data point represents a single GBR and red bars indicate the median, upper and lower quartiles. D-E. Representative ATAC-seq peaks showing lack of accessibility in Shh-/- limb buds at HH-responsive GBRs (D), but not in Stable GBRs (E, F) Plot of log2 fold changes in chromatin accessibility in the presence and absence of HH signaling. HH-responsive GBRs are significantly less accessible than Stable GBRs (Stable vs. HH-sensitive. p=0.001; Stable vs. HH-dependent p=4.99e-09; Wilcoxon rank sum test). See Figure 3—source data 1.
-
Figure 3—source data 1
WT vs Shh-/- ATAC Seq analyzed data and called peaks.
Coordinates for all ATAC peaks in the WT group that overlap with GBRs are listed. ‘Shh_ATAC_peak’ identifies the corresponding id# for that peak in the Shh-/- data, and if a peak is not present in the Shh-/- samples, it is marked as NA. A column for each GBR type identifies which GBR type a given ATAC peak overlaps with. The number indicates the peak ID. If a peak region does not overlap with the type of peak in that list, it will be marked as NA. The normalized log2 transformed signals are shown for each sample in addition to the ‘average’ signal across all samples. The ‘t’ statistic calculates the difference in signals between WT and Shh-/- by taking into consideration fold-change and variance among samples. A positive t statistic values indicate a peak is more accessible in WT than Shh-/- and a negative t statistic indicates higher accessibility in Shh-/-. The ‘p.value’ is obtained from a moderated t-test using limma. The ‘p.value.adj’ is the adjusted p-value (FDR) using the Benjamini-Hochberg procedure.
- https://cdn.elifesciences.org/articles/50670/elife-50670-fig3-data1-v1.xlsx
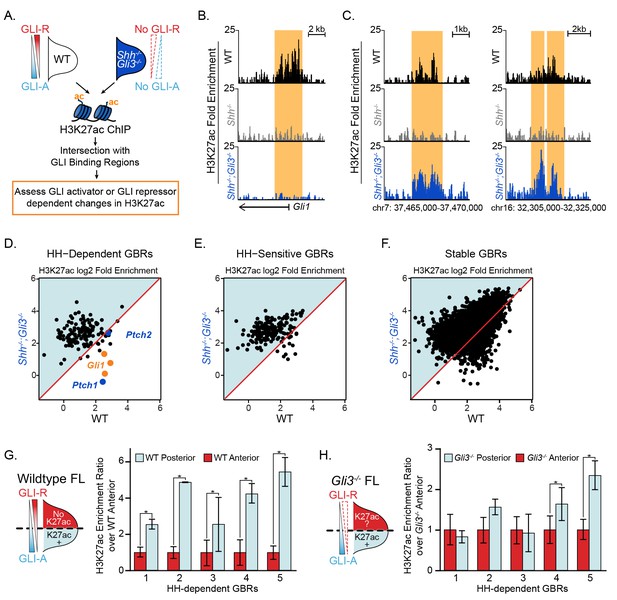
GLI de-repression activates most HH-responsive enhancers.
(A) Shh-/-;Gli3-/- H3K27ac ‘MicroChIPs’ on single pairs of E10.5 forelimbs (33–34S) Shh-/-;Gli3-/- and WT littermate controls (n = 2, respectively). (B) A HH-responsive GBR near Gli1 which requires GLI activator for H3K27ac enrichment. (C) Representative examples of HH-responsive GBRs, activated by loss of GLI repressor that do not require GLI activator. (D-F) Scatter plot of H3K27ac enrichment of HH-dependent, HH-sensitive and Stable GBRs in WT and Shh-/-;Gli3-/- limbs. Each dot represents a single GBR. The p-values indicate a significant enrichment of acetylation in Shh-/-;Gli3-/- among all GBR classes (p-values: HH-dependent = 2.26e-08, HH-sensitive = 5.41e-11, Stable = 3.4e-185;Wilcoxon-rank sum tests). (G-H) E10.5 WT and Gli3-/- limb buds were dissected into anterior and posterior halves as indicated and selected HH-dependent GBRs were tested for H3K27ac enrichment by quantitative PCR in each fraction (n = 4). HH-dependent GBRs have higher ratios of posterior to anterior H3K27ac enrichment in WT limb buds (G), while many HH-dependent GBRs have equal ratios of posterior to anterior H3K27ac enrichment in Gli3-/- limb buds (H) (n = 3) (asterisks indicate p<0.05, paired T-test). See Figure 4—source data 1.
-
Figure 4—source data 1
WT vs Shh-/-;Gli3-/- H3K27ac MicroChIP-seq analyzed data and called peaks.
H3K27ac called peaks with a FDR < 0.05 from two replicates of WT, Shh-/- and Shh-/-;Gli3-/- E10.5 (33–34S) forelimbs. Separate tabs for each genotype include peak coordinates and log2 fold change over input. Additional tabs include a peak summary and differential analysis of WT vs. Shh-/-;Gli3-/-. Differential analysis tab lists peak coordinates, peak type, fold change normalized to input for WT and Shh-/-;Gli3-/- samples and fold change of WT over Shh-/-;Gli3-/-.
- https://cdn.elifesciences.org/articles/50670/elife-50670-fig4-data1-v1.xlsx
-
Figure 4—source data 2
MicroChIP H3K27ac enrichment in Shh-/-;Gli3-/- limb buds at HH-responsive GBRs with H3K27me3 in Shh-/- limbs.
List of 31 HH-responsive GBRs with H3K27me3 enrichment in Shh-/- limb buds and their enrichment of H3K27ac in Shh-/-;Gli3-/-. 20 of the 31 regions were detected in the H3K27ac MicroChIP analyses, those not detected are noted. For the 20 regions detected, they are classified as having H3K27ac in Shh-/-;Gli3-/- as ‘present/increased’ or ‘absent/decreased’ compared to WT controls.
- https://cdn.elifesciences.org/articles/50670/elife-50670-fig4-data2-v1.xlsx
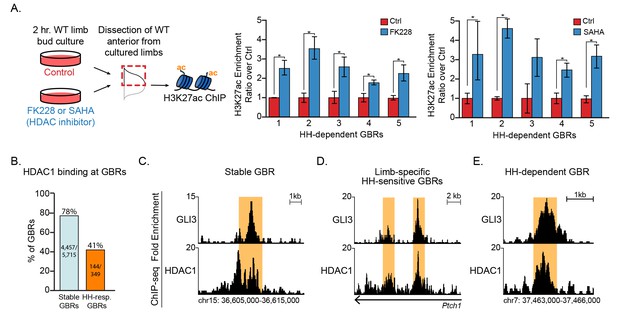
HDACs regulate H3K27ac at HH-responsive GBRs.
(A) Inhibition of HDACs using 250 nM of FK228 or 20 μM SAHA in cultured limb buds for two hours resulted in significant increases of H3K27ac enrichment at HH-dependent GBRs from anterior cultured limb buds compared to DMSO control anterior limbs (FK228 n = 4; SAHA n = 5; asterisks indicate p<0.05, paired T-test). (B) HDAC1 binding at Stable and HH-responsive GBRs (n = 4). (C-E) HDAC1 at GLI3 binding regions, shown at a representative Stable GBR (C), limb-specific HH-sensitive GBRs near the HH target genes Ptch12 (Lopez-Rios et al., 2014) (D), and a HH-dependent GBR, (region also shown in Figure 4C) (E, D) See Figure 5—figure supplement 1, Figure 5—source data 1.
-
Figure 5—source data 1
HDAC1 ChIP-seq analyzed data and called peaks.
HDAC1 binding regions with called peaks with a false discovery rate (FDR) < 0.05 from four biological replicates of E11.5 (40–44S) pooled forelimbs and hindlimbs. Rank ordered coordinates, peak length, log2 fold change (log2FC) and FDR are listed for each peak.
- https://cdn.elifesciences.org/articles/50670/elife-50670-fig5-data1-v1.xlsx
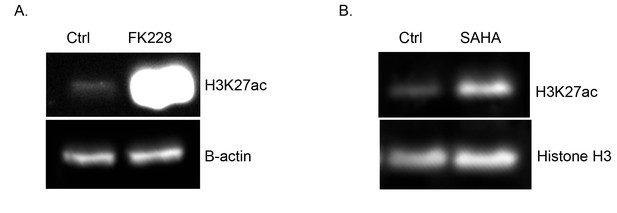
H3K27ac is increased upon HDAC inhibition.
Western blot of cultured limb buds treated with 0.4% DMSO or the HDAC inhibitors, FK228 (250 nM, n = 2) (A) or SAHA (20 μM, n = 2) (B), for 2 hr showing increased overall levels of H3K27 acetylation. Approximately, 20 pairs of E10.5 forelimbs were lysed and 15 μg of protein was loaded on gel. Note that these are whole limb buds rather than anterior and posterior fractions shown in Figure 5.
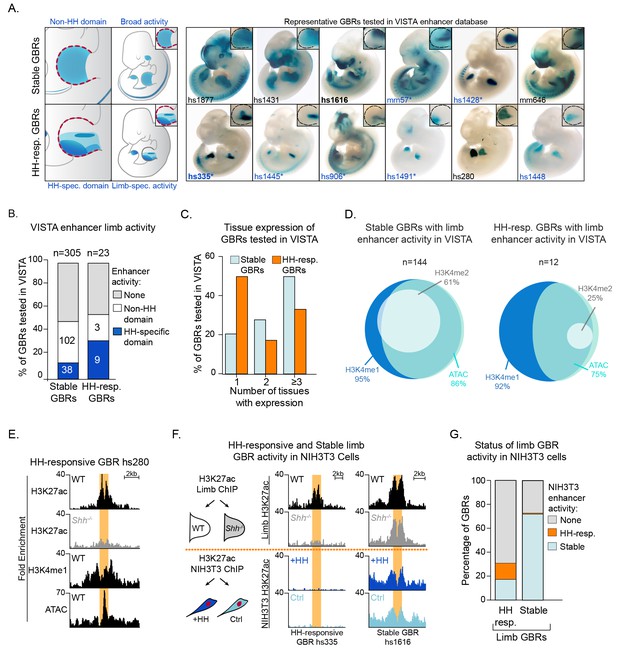
Hedgehog-responsive GBRs have tissue-specific enhancer activity within HH-specific domains.
(A) Enhancers with annotated limb activity in VISTA corresponding to representative HH-responsive GBRs (bottom) and Stable GBRs (top) with limbs magnified and outlined in insets. Limb buds containing HH-specific domains of enhancer activity are indicated by an asterisk. (B) Chart indicating total number of VISTA enhancers tested for HH-responsive and Stable GBRs, the numbers of enhancers for each category and their limb enhancer activity. (C) Chart delineating the percentage of HH-responsive and Stable limb enhancers that drive expression in one or more tissues. (D) Venn Diagram of enhancer marks H3K27ac, H3K4me1, H3K4me2 and ATAC, in Stable and HH-responsive GBRs tested in VISTA that drive expression in the limb. GBRs, are by definition are marked by H3K27ac. (E) Enrichment of enhancer markers at a representative HH-responsive GBR tested in VISTA (hs280, Figure 6A). (F) Schematic of NIH3T3 H3K27ac ChIP treated with and without the HH agonist purmorphamine (+HH) and the activity of representative HH-responsive and Stable limb GBRs in response to HH activation in limb and NIH3T3 cells (n = 2). (G) Graph indicating how the acetylation status of HH-responsive and Stable limb GBRs responds to HH signaling in HH-responsive NIH3T3 cells. See Figure 6—source data 1; Figure 6—source data 2.
-
Figure 6—source data 1
Stable and HH-responsive GLI binding regions with limb enhancer activity in the VISTA dataset.
Tab 1. Columns indicate VISTA enhancer IDs, coordinates, number of tissues with limb enhancer activity as annotated by the VISTA database (Visel et al., 2007) and its corresponding GBR category. Tabs 2–5. Normalized signal and called peaks for H3K27ac, ATAC, H3K4me1 and H3K4me2 in WT, and Shh-/- limbs if applicable, is listed for all HH-responsive GBRs (Tab 2- normalized signal; Tab 3-called peaks) and Stable GBRs (Tab 4- normalized signal, Tab 5-called peaks) that were shown to drive limb activity, as tested in the VISTA database. For called peaks in Tabs 3, 5, ‘NO’ denotes not a significantly called peak, ‘YES’ denotes a called peak.
- https://cdn.elifesciences.org/articles/50670/elife-50670-fig6-data1-v1.xlsx
-
Figure 6—source data 2
NIH3T3 H3K27ac ChIP-seq analyzed data and called peaks.
H3K27ac called peaks with a FDR < 0.05 from two replicates of purmorphamine (‘pm’) treated or DMSO control NIH3T3 cells. For each peak, the assigned Peak ID, coordinates, peak type, fold change normalized to input for purmorphamine treated and control samples, and fold change of purmorphamine treated over control are listed. Additional tabs include sorted files for each peak type.
- https://cdn.elifesciences.org/articles/50670/elife-50670-fig6-data2-v1.xlsx
Model for GLI transcriptional repression.
(A) Summary of enhancer status at HH-responsive GBRs. (B) In the absence of HH, GLI repressors bind to enhancers for HH target genes, limiting their accessibility and, directly or indirectly, recruiting an HDAC complex that de-acetylates Histone H3K27, inactivating the enhancer. In the presence of HH signaling, GLI de-repression and loss of associated HDAC activity result in increased accessibility, the accumulation of H3K27ac and gene transcription. (C) Schematic showing tissue-restricted activity of HH-responsive GBRs within HH-responsive gene expression domains. (D) Possible roles for Stable GBRs in HH transcriptional regulation.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (M. musculus) | Gli3Xt-J Gli3+/- | Jackson Laboratory | Jackson Cat# 000026, MGI Cat# 2169581, RRID:MGI:2169581 | Obtained from the Laboratory of Dr. Andy McMahon |
| Genetic reagent (M. musculus) | Shhtm1amc Shh+/- | Jackson Laboratory | Jackson Cat# 003318, MGI Cat# 3584154, RRID:MGI:3584154 | Obtained from the Laboratory of Dr. Andy McMahon |
| Genetic reagent (M. musculus) | Gli3FLAG | Laboratory of Dr. Andy McMahon | Obtained from the Laboratory of Dr. Andy McMahon | |
| Genetic reagent (M. musculus) | Swiss Webster Wildtype | Charles River | Charles River Cat# NCI 551 IMSR Cat# TAC:sw, RRID:IMSR_TAC:sw | |
| Cell line | NIH 3T3 | ATCC | Cat# CRL-6442, RRID:CVCL_0594 | Used for conventional ChIP-seq |
| Antibody | Anti-H3K27ac (mouse mono-clonal) | Diagenode | Diagenode Cat# C15200184, RRID:AB_2713908 | Used for conventional ChIP-seq |
| Antibody | Anti-H3K27ac (rabbit polyclonal) | Abcam | Abcam Cat# ab4729, RRID:AB_2118291 | Used for conventional ChIP-qPCRs |
| Antibody | Anti-H3K27ac (rabbit polyclonal) | Diagenode | Diagenode Cat# C15410196, RRID:AB_2637079 | Used for conventional MicroChIP-seq |
| Antibody | Anti-H3K27me3 (rabbit polyclonal) | Abcam | Abcam Cat# Ab195477, RRID:AB_2819023 | Used for conventional ChIP-seq |
| Antibody | Anti-H3K4me1 (rabbit polyclonal) | Millipore | Millipore Cat# 07–436, RRID:AB_310614 | Used for conventional ChIP-qPCRs |
| Antibody | Anti-H3K4me2 (rabbit polyclonal) | Millipore | Millipore Cat# 07–030, RRID:AB_11213050 | Used for conventional ChIP-seq |
| Antibody | Anti-M2 FLAG (mouse monoclonal) | Sigma | Sigma-Aldrich Cat# F3165, RRID:AB_259529 | Used for conventional ChIP-seq and WB (1:4000) |
| Antibody | Anti-HDAC1 (rabbit polyclonal | Abcam | Abcam Cat# ab7028, RRID:AB_305705 | Used for conventional ChIP-seq |
| Antibody | Anti-Histone H3 (rabbit polyclonal | Cell Signaling Technology | Cell Signaling Technology Cat# 4499, RRID:AB_10544537 | Used for WB (1:4000) |
| Antibody | Anti-GAPDH (rabbit polyclonal | Cell Signaling Technology | Cell Signaling Technology Cat# 5174, RRID:AB_10622025 | Used for WB (1:1000) |
| Antibody | Anti-B-actin (rabbit polyclonal | Cell Signaling Technology | Cell Signaling Technology Cat# 8457, RRID:AB_10950489 | Used for WB (1:2000) |
| Antibody | Donkey-anti-mouse | Jackson Immuno-Research | Jackson ImmunoResearch Labs Cat# 715-035-150, RRID:AB_2340770 | Used for WB (1:5000) |
| Antibody | Donkey-anti-rabbit | Jackson Immuno-Research | Jackson ImmunoResearch Labs Cat# 711-005-152, RRID:AB_2340585 | Used for WB (1:5000) |
| Antibody | Dynabeads M-280 Sheep Anti-Mouse IgG | Invitrogen, Thermo Fisher Scientific | Thermo Fisher Scientific Cat# 11201D, RRID:AB_2783640 | |
| Antibody | Dynabeads M-280 Sheep Anti-Rabbit IgG | Invitrogen, Thermo Fisher Scientific | Thermo Fisher Scientific Cat# 11203D, RRID:AB_2783009 | |
| Chemical compound, drug | Purmorphamine | Stemgent | Stemgent Cat# 04–0009 | Used in cell culture (400 nM) |
| Chemical compound, drug | SAHA | Selleckchem | Selleckchem Cat# MK0683 | Used in limb bud culture (20 μM) |
| Chemical compound, drug | FK228 | Selleckchem | Selleckchem Cat# S3020 | Used in limb bud culture (250 nM) |
| Commercial Assay or Reagent | SensiFAST SYBR-LoROX | Bioline | Bioline Cat# BIO-94020 | |
| Commercial Assay or Reagent | NEBNext DNA Library Prep Master Mix Set for Illumina | New England Biolabs | NEB Cat# E6040L, E7645L | |
| Commercial Assay or Reagent | Agencourt AMPure XP | Beckman Coulter | Beckman Coulter Cat# A63881 | |
| Commercial Assay or Reagent | True MicroChIP Kit | Diagenode | Diagenode Cat# C01010130 | |
| Commercial Assay or Reagent | MicroPlex Library Prep Kit | Diagenode | Diagenode Cat# C05010012 | |
| Commercial Assay or Reagent | Liberase | Roche | Roche Cat# 05401119001 | Cell dissociation (100 µg/mL) |
| Software, Tools | MACS version 2.1.0 | (Zhang et al., 2008https://github.com/taoliu/MACS | MACS, RRID:SCR_013291 | |
| Software, Tools | limma | (Ritchie et al., 2015) | LIMMA, RRID:SCR_010943 | http://bioconductor.org/packages/release/bioc/html/limma.html |
| Software, Tools | R statistical software | (R Development Core Team, 2014) | R Project for Statistical Computing, RRID:SCR_001905 | https://www.r-project.org/ |
| Software, Tools | CisGenome | (Ji et al., 2008) | CisGenome, RRID:SCR_001558 | http://www.biostat.jhsph.edu/~hji/cisgenome/ |
| Database, Tools | JASPAR motif database | (Khan et al., 2018) | JASPAR, RRID:SCR_003030 | http://jaspar.genereg.net/ |
| Database, Tools | Transfac motif database | (Matys et al., 2006) | TRANSFAC, RRID:SCR_005620 | http://gene-regulation.com/pub/databases.html |
| Database, Tools | VISTA enhancer browser | (Visel et al., 2007) | VISTA Enhancer Browser, RRID:SCR_007973 | https://enhancer.lbl.gov/ |