Partial loss of CFIm25 causes learning deficits and aberrant neuronal alternative polyadenylation
Figures
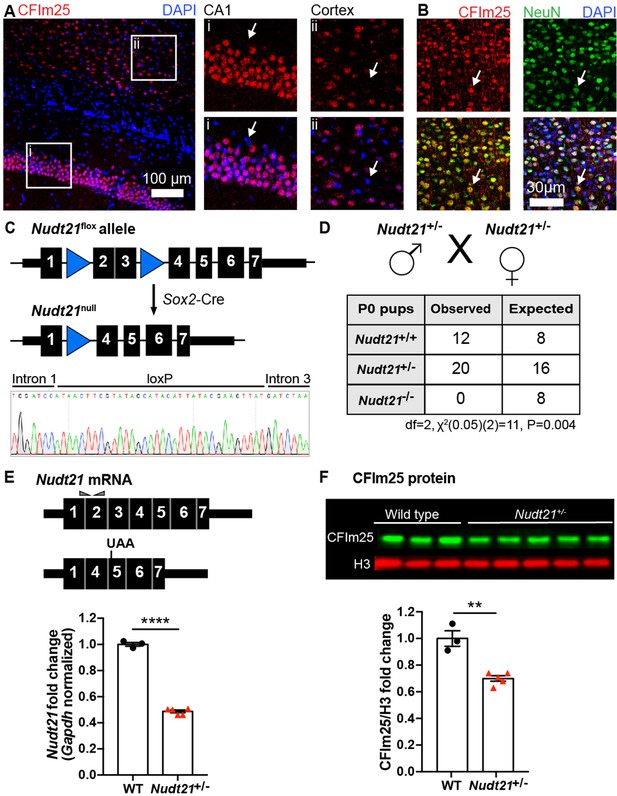
Nudt21 heterozygotes have 50% less Nudt21 mRNA in their neurons, but only 30% reduced CFIm25 protein.
(A) Immunofluorescence showing CFIm25 expression in the nuclei of some cells in the (i) mouse hippocampus CA1 region and (ii) cortex. Arrows indicate nuclei that do not have CFIm25. (B) Immunofluorescence of the mouse cortex showing colocalization of CFIm25 and the neuronal marker NeuN. Arrows indicate a nucleus with CFIm25 and NeuN expression. (C) Schematic of the floxed and recombined Nudt21 alleles (top), and sequencing showing successful recombination (bottom). (D) Observed and expected offspring counts born with each possible genotype from Nudt21+/- mating pairs. No homozygous Nudt21 null offspring were born, when eight would be expected if loss of Nudt21 did not affect survival: p=0.004, analyzed by two-tailed, chi-square test. (E) Schematic of wild-type and recombined Nudt21 mRNA (top). Triangles indicate RT-qPCR primer binding sites, and UAA shows site of induced premature stop codon after recombination. RT-qPCR analysis shows expected 50% reduction of whole-brain, Gapdh-normalized, wild-type Nudt21 mRNA in five-week-old mice with one wild-type Nudt21 allele and one recombined, null allele (bottom): p<0.0001, n = 3–5/genotype. (F) Western blot image comparing five-week-old Nudt21+/- mice CFIm25 protein levels with their WT littermates (top). Western blot analysis showing ~30% reduction of H3-normalized CFIm25 protein levels in Nudt21+/- mice: p=0.0012, n = 3–5/genotype. We confirmed that CFIm25 does not regulate H3. For all charts, error bars indicate SEM. All data analyzed by unpaired, two-tailed t-test unless otherwise stated. **p<0.01; ****p<0.0001. Weights of the heterozygous animals are shown in Figure 1—figure supplement 1.
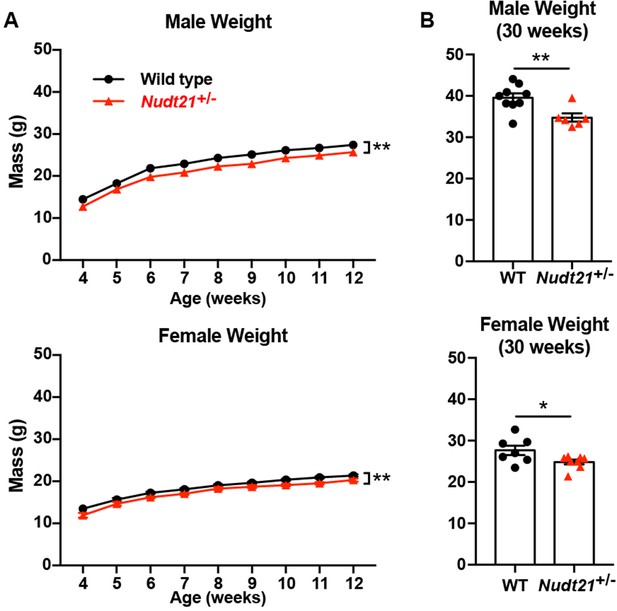
Nudt21+/-mice weigh less.
(A) Male (top) and female (bottom) Nudt21+/- mice weigh less than their wild-type littermates from weening to twelve weeks: p=0.002 (males) and p=0.004 (females), analyzed by two-way repeated measures ANOVA (genotype*week). n = 15–26/genotype. (B) Reduced weight persists up to at least 30 weeks in male (top) and female (bottom) Nudt21+/- mice: p=0.008 (males) and p=0.04 (females), analyzed by unpaired, two-tailed t-test. n = 6–9/genotype. Error bars indicate SEM. *p<0.05; **p<0.01.
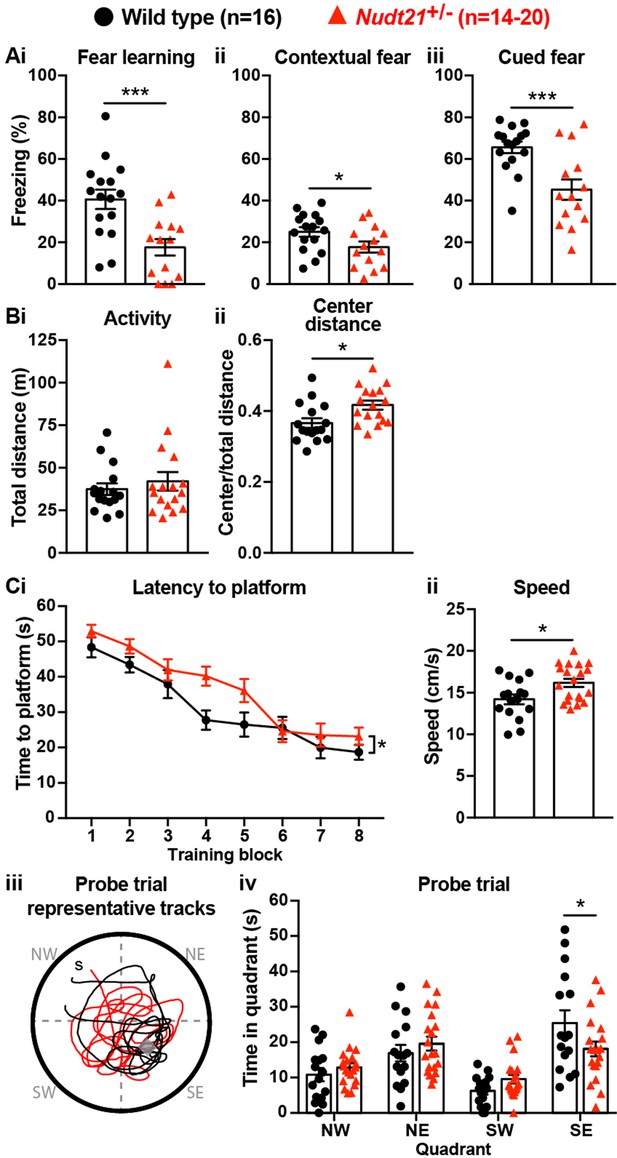
Partial loss of Nudt21-encoded CFIm25 protein causes learning deficits.
(A) Nudt21+/- mice have conditioned fear learning deficits: fear learning, p=0.0009; contextual fear, p=0.045; cued fear, p=0.0009. (B) The open field assay shows that Nudt21+/- mice are no more active (i) but spend relatively more time in the center of the open field (ii), indicating reduced anxiety: p=0.01. (C) Nudt21+/- mice have spatial learning deficits in the Morris water maze. (i) They take longer to find the hidden platform during the training blocks (p=0.026, two-way, repeated measures ANOVA), (ii) despite swimming faster and farther (p=0.013). (iii and iv) When the hidden platform is removed in the probe trial, they spend less time in the quadrant that previously had the platform (p=0.039, Sidak’s multiple comparisons test). For all assays, mice were between 30–40 weeks of age. Error bars indicate SEM. Representative tracks are from the animal with the median result in each genotype. All data were analyzed by unpaired, two-tailed t-test unless otherwise stated. *p<0.05; ***p<0.001.
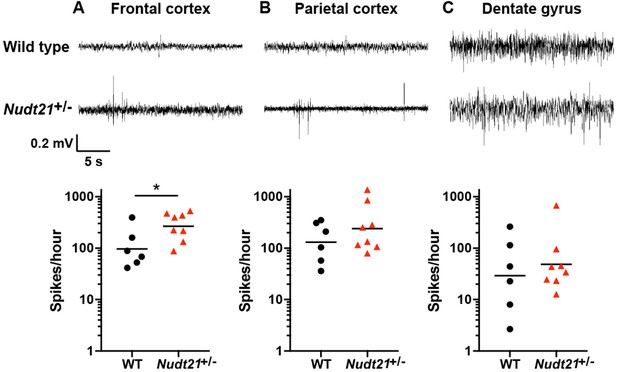
Nudt21+/-mice have increased cerebral spike activity.
(A) 57-week-old Nudt21+/- mice have significantly increased spike activity in the frontal cortex by EEG (p=0.029), but not in the parietal cortex (B) and dentate gyrus (C). Representative traces are on top and spike count summaries below. All data analyzed by two-tailed Mann-Whitney test. Central tendency lines show the geometric mean. *p<0.05.

Nudt21 heterozygotes have altered hippocampal alternative polyadenylation.
(A)(i) Representative poly(A)-ClickSeq (PAC-seq) track showing altered alternative polyadenylation in Ddx6: n = 3/treatment. Peaks are 3′ end sequencing reads. Each peak indicates a different mRNA isoform with a different cleavage and polyadenylation site. Bracketed numbers show counts per million; kb stands for kilobases. (ii) Schemata of the three Ddx6 mRNA isoforms identified by PAC-seq. Triangles show the binding sites of the qPCR primers used to validate the PAC-seq results. The gray primers detect all the Ddx6 mRNA isoforms, whereas the green primers only detect the isoform with the longest 3′ UTR. (iii) Gapdh-normalized RT-qPCR quantification showing relatively less of the long isoform of Ddx6 in the hippocampi of 46-week-old Nudt21+/- mice compared to their wild-type littermates. Error bars indicate SEM. Data analyzed by unpaired, two-tailed t-test. p=0.0003. (B) Nudt21+/- and wild-type mice separate by principle component analysis (PCA) of poly(A) site usage. (C) Volcano plot showing relative mRNA length change in Nudt21+/- mice compared to their wild-type littermates. The horizontal, dashed line shows Padjusted = 0.05, n = 3/genotype. (D) Nudt21+/- and wild-type mice segregate by PCA of the proteome by quantitative mass spectrometry. (E) Mass spectrometry quantification of protein level fold change for genes with significantly altered APA (Padjusted <0.05). Source files for the PAC-seq and mass spectrometry quantification data are available in Figure 4—source data 1. ***p<0.001.
-
Figure 4—source data 1
RNA length and protein level changes in genes with misregulated APA in Nudt21+/-mice.
Alternative polyadenylation (APA), likelihood ratio (lr), degrees of freedom (df), mass spectrometry (MS), fold change (FC).
- https://cdn.elifesciences.org/articles/50895/elife-50895-fig4-data1-v1.xlsx

Human embryonic stem cell derived neurons infected with shRNA targeting NUDT21 have 30% less CFIm25 protein.
(A) Schematic of NUDT21 mRNA with qPCR primers (top) and RT-qPCR quantification of NUDT21 mRNA levels in human embryonic stem cell (ESC)-derived neurons infected with scrambled shRNA (shScramble) or shRNA targeting NUDT21 (shNUDT21). The neurons infected with shNUDT21 have a 40% reduction of GAPDH-normalized NUDT21: p=0.0009, n = 3–4/treatment. (B) Western blot image and quantification showing a 30% reduction of H3-normalized CFIm25 protein in shNUDT21-infected neurons: p=0.0005, n = 4/treatment. We confirmed that CFIm25 does not regulate H3. ***p<0.001.
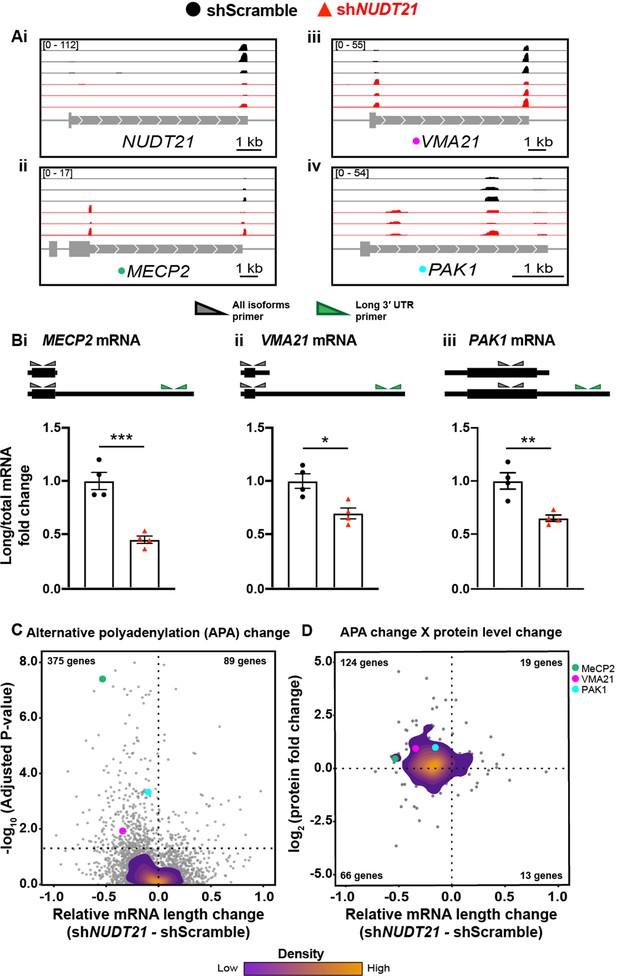
NUDT21 depletion induces aberrant alternative polyadenylation and altered protein levels.
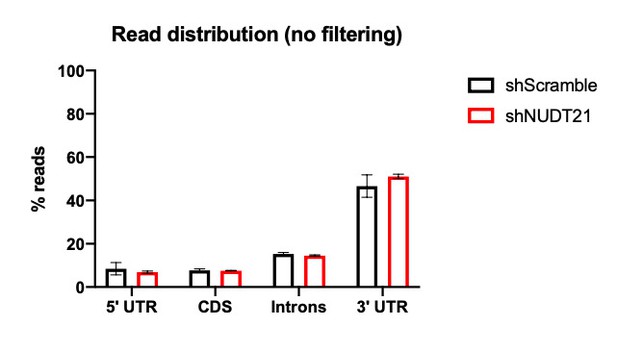
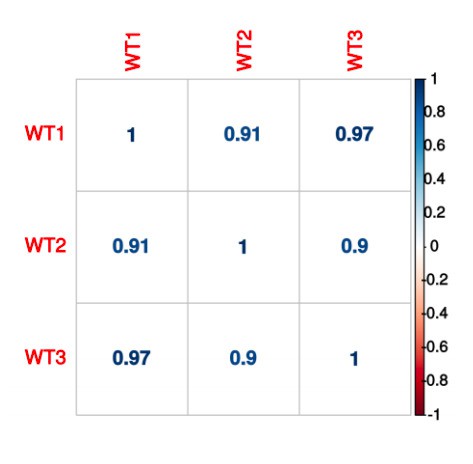
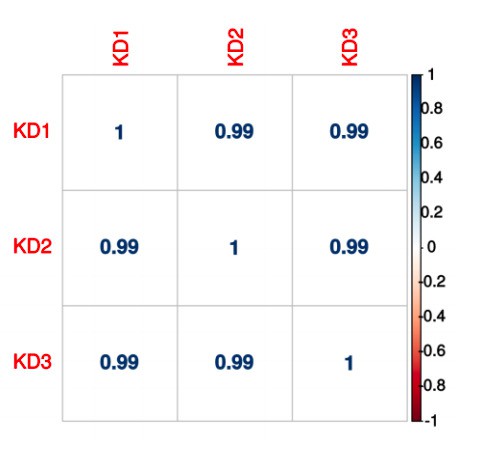
(A)(i) Poly(A)-ClickSeq (PAC-seq) track showing reduced NUDT21 and (ii-iv) PAC-seq tracks showing altered alternative polyadenylation in example genes MECP2, VMA21, and PAK1. n = 3/treatment. Peaks are 3′ end sequencing reads. Multiple peaks in a gene indicate multiple mRNA isoforms with different cleavage and polyadenylation sites. Bracketed numbers show counts per million; kb stands for kilobases. (B) Schemata of the mRNA isoforms identified by PAC-seq for MECP2, VMA21, and PAK1 (top). Triangles show the binding sites of the qPCR primers used to validate the PAC-seq results. The gray primers detect all mRNA isoforms for the target gene, whereas the green primers only detect the isoform with the longest 3′ UTR. Gapdh-normalized RT-qPCR quantification showing relatively less of the long isoforms in the shNUDT21-infected neurons (bottom) for MECP2, p=0.0007 (i), VMA21, p=0.01 (ii), and PAK1, p=0.005 (iii). Error bars indicate SEM. Data analyzed by unpaired, two-tailed t-test. (C) Volcano plot showing relative mRNA length change in shNUDT21-infected neurons compared to shScramble-infected controls. The horizontal, dashed line shows Padjusted = 0.05, n = 3/treatment. NUDT21 loss in neurons predominantly results in shorter mRNAs (p<0.0001, two-tailed chi-square test). >90% of reads are in 3′ UTRs (Figure 6—figure supplement 1A). Principle component analysis (PCA) shows sample separation by treatment (Figure 6—figure supplement 1B). Distal cleavage sites of NUDT21-regulated mRNAs are enriched for the CFIm25 binding motif, UGUA, in mRNAs that shorten after NUDT21 knockdown, but not in non-target mRNAs (Figure 6—figure supplement 1C). (E) Mass spectrometry quantification of protein level fold change for genes with significantly altered APA (Padjusted <0.05). mRNA shortening predominantly results in increased protein levels (p<0.0001, two-tailed conditional chi-square test). Source files for the PAC-seq and mass spectrometry quantification data are available in Figure 6—source data 1. *p<0.05; **p<0.01; ***p<0.001.
-
Figure 6—source data 1
RNA length and protein level changes in genes with misregulated APA following NUDT21 inhibition in human neurons.
Alternative polyadenylation (APA), likelihood ratio (lr), degrees of freedom (df), first polyadenylation site (F), middle/center polyadenylation site (M), last polyadenylation site (L) mass spectrometry (MS), fold change (FC), intellectual disability (ID), probability of loss of function intolerance (pLI).
- https://cdn.elifesciences.org/articles/50895/elife-50895-fig6-data1-v1.xlsx
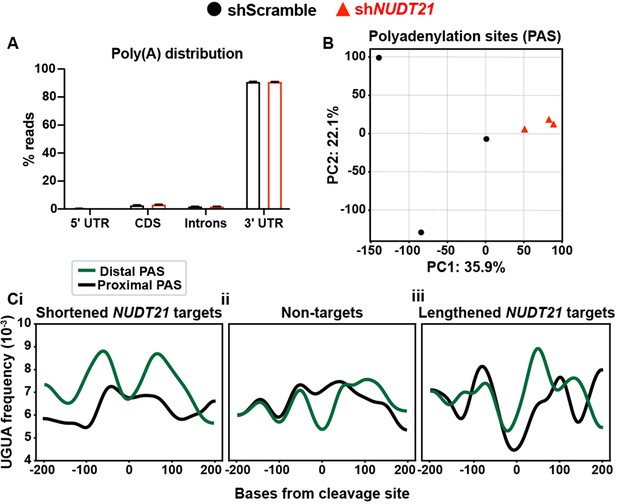
Accessory analyses support PAC-seq validity.
(A) >90% of filtered reads map to the 3′ UTR. (B) Control and shNUDT21-infected human neurons segregate by principle component analysis (PCA) of alternative polyadenylation. (Ci) The CFIm25 binding motif, UGUA, is enriched upstream of the distal cleavage and polyadenylation site (PAS) of NUDT21-regulated mRNAs that shorten after NUDT21 knockdown. (ii) There is almost no upstream UGUA frequency difference between proximal and distal cleavage sites in mRNAs not affected by NUDT21 loss. (iii) There is a slight UGUA enrichment upstream of the proximal cleavage site in NUDT21-regulated mRNAs that lengthen after NUDT21 loss.
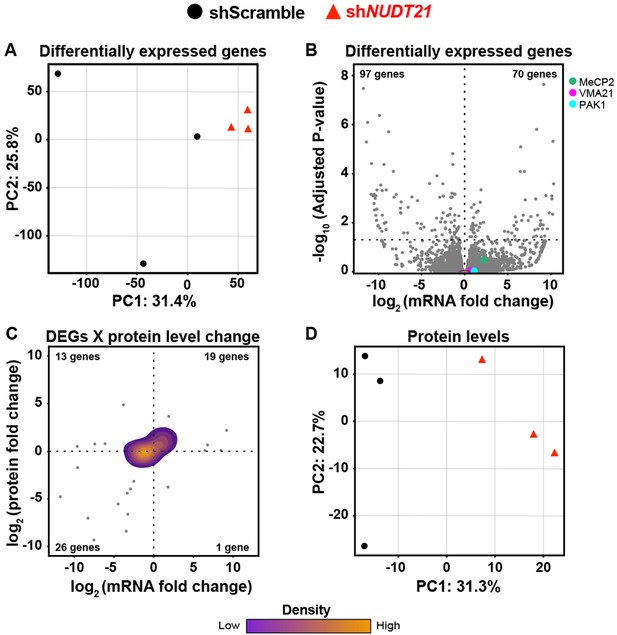
Partial loss of NUDT21 function in neurons leads to downstream transcriptomic and proteomic dysregulation.
(A) Control and shNUDT21-infected human neurons separate by principle component analysis (PCA) of differentially expressed genes (DEGs). (B) Volcano plot showing DEGs in shNUDT21-infected neurons compared to shScramble-infected controls. The horizontal, dashed line shows Padjusted = 0.05, n = 3/treatment. (C) Mass spectrometry quantification of protein level fold change for genes with significantly altered DEGs (Padjusted <0.05). (D) Control and shNUDT21-infected human neurons segregate by PCA of the proteome. Source files for the differential gene expression and mass spectrometry quantification data are available in Figure 7—source data 1.
-
Figure 7—source data 1
Differential gene expression following NUDT21 inhibition in human neurons.
Differentially expressed gene (DEG), fold change (FC), mean expression of a gene across all samples (baseMean), log(FC) standard error (lfcSE), differential test statistic (stat), intellectual disability (ID), probability of loss of function intolerance (pLI).
- https://cdn.elifesciences.org/articles/50895/elife-50895-fig7-data1-v1.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Mus musculus) | Nudt21 | NCBI Gene ID: 68219 | ||
| Gene (Homo sapiens) | NUDT21 | NCBI Gene ID: 11051 | ||
| Strain, strain background (M. musculus; male and female) | C57BL/6J | The Jackson Laboratory | IMSR Cat# JAX:000664, RRID:IMSR_JAX:000664 | |
| Genetic reagent (M. musculus) | Sox2-Cre | PMID:14595839 | IMSR Cat# JAX:008454, RRID:IMSR_JAX:008454 | |
| Genetic reagent (M. musculus) | Nudt21flox/flox | PMID:30830875 | RRID:MGI:6385511 | Dr. Eric Wagner (University of Texas Medical Branch) |
| Genetic reagent (M. musculus) | Nudt21+/- | This paper | RRID:MGI:6385902 | Line maintained in the H. Zogbhi lab |
| Cell line (H. sapiens) | HEK293T | ATCC | ECACC Cat# 12022001, RRID:CVCL_0063 | |
| Cell line (H. sapiens) | WA09 | WiCell | RRID:CVCL_9773 | |
| Antibody | Anti-CFIm25 (rabbit polyclonal) | Proteintech | Proteintech Cat# 10322–1-AP, RRID:AB_2251496 | 1:50 |
| Antibody | Anti-NeuN (mouse monoclonal) | Millipore | Millipore Cat# MAB377, RRID:AB_2298772 | 1:50 |
| Antibody | Anti-CFIm25 (mouse monoclonal) | Santa Cruz Biotechnology | Santa Cruz Biotechnology Cat# sc-81109, RRID:AB_2153989 | 1:500 |
| Antibody | Anti-H3 (rabbit monoclonal) | Cell Signaling Technology | Cell Signaling Technology Cat# 4499, RRID:AB_10544537 | 1:10,000 |
| Recombinant DNA reagent | pGIPZ | Open Biosystems | shRNA backbone | |
| Sequence-based reagent | Nudt21 forward | This paper | qPCR primer | TACATCCAGCAGACCAAGCC |
| Sequence-based reagent | Nudt21 reverse | This paper | qPCR primer | AATCTGGCTGCAACAGAGCT |
| Sequence-based reagent | Gapdh forward | This paper | qPCR primer | GGCATTGCTCTCAATGACAA |
| Sequence-based reagent | Gapdh reverse | This paper | qPCR primer | CCCTGTTGCTGTAGCCGTAT |
| Sequence-based reagent | NUDT21 forward | This paper | qPCR primer | CTTCAAACTACCTGGTGGTG |
| Sequence-based reagent | NUDT21 reverse | This paper | qPCR primer | AAACTCCATCCTGACGACC |
| Sequence-based reagent | GAPDH forward | This paper | qPCR primer | CGACCACTTTGTCAAGCTCA |
| Sequence-based reagent | GAPDH reverse | This paper | qPCR primer | TTACTCCTTGGAGGCCATGT |
| Sequence-based reagent | Nudt21null forward | This paper | PCR primer | ACAGATTAGCTGTTAGTACAGG |
| Sequence-based reagent | Nudt21null reverse | This paper | PCR primer | GAAGAACCAGAGGAAACGTGAG |
| Sequence-based reagent | Wild-type Nudt21 forward | This paper | PCR primer | AGGAGGCTGACATGGATTGTT |
| Sequence-based reagent | Wild-type Nudt21 reverse | This paper | PCR primer | TCTTCTCCTGGGTTAAGTTCCC |
| Sequence-based reagent | Anti-NUDT21 shRNA 1 | Dharmacon | Clone ID: V2LHS_253272 | CAACCTGTACCCTCTTACCAATTATAC |
| Sequence-based reagent | Anti-NUDT21 shRNA 2 | Dharmacon | Clone ID: V2LHS_197948 | ATCCATATATTCCTGCACATATT |
| Sequence-based reagent | Non-silencing shRNA | Dharmacon | Clone ID: RHS4348 | |
| Sequence-based reagent | P7 adapter (Illumina_4N_21T) | Illumina | Reverse transcription primer | GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNTTTTTTTTTTTTTTTTTTTTT |
| Sequence-based reagent | P5 adapter | Interated DNA Technologies | Reverse transcription primer | 5′HexynylNNNNAGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTAGATCTCGGTGGTCGCCGTATCATT |
| Sequence-based reagent | Universal primer | cDNA amplification primer | AATGATACGGCGACCACCGAG | |
| Sequence-based reagent | Example 3′ indexing primer | cDNA amplification primer | CAAGCAGAAGACGGCATACGAGATCGTGATGTGACTGGAGTTCAGACGTGT | |
| Sequence-based reagent | Ddx6 forward | This paper | qPCR primer | TGGATCTCATCAAGAAAGGC |
| Sequence-based reagent | Ddx6 reverse | This paper | qPCR primer | GTGACAACAATTTATCTGCCTC |
| Sequence-based reagent | Ddx6-long forward | This paper | qPCR primer | CACAGCTGACAGACTCCAACA |
| Sequence-based reagent | Ddx6-long reverse | This paper | qPCR primer | AGCTTACTAACCCAGGCCCA |
| Sequence-based reagent | MECP2 forward | This paper | qPCR primer | GATCAATCCCCAGGGAAAAGC |
| Sequence-based reagent | MECP2 reverse | This paper | qPCR primer | CCTCTCCCAGTTACCGTGAAG |
| Sequence-based reagent | MECP2-long forward | This paper | qPCR primer | GCCTGGAAACCTGTCTGAGG |
| Sequence-based reagent | MECP2-long reverse | This paper | qPCR primer | CTCCAGCTAAGTGTGTCCCG |
| Sequence-based reagent | VMA21 forward | This paper | qPCR primer | TACATATTTGAAGGCGCCC |
| Sequence-based reagent | VMA21 reverse | This paper | qPCR primer | CATACACAAAGAGGGCCAG |
| Sequence-based reagent | VMA21-long forward | This paper | qPCR primer | AGGGGGAGGATTTGGATGTG |
| Sequence-based reagent | VMA21-long reverse | This paper | qPCR primer | TAGCTAAAGAACTCAAGCCCCC |
| Sequence-based reagent | PAK1 forward | This paper | qPCR primer | GAATTACTTGGACAGTTACCTCG |
| Sequence-based reagent | PAK1 reverse | This paper | qPCR primer | ACATCTGTCAAGGAGCCTC |
| Sequence-based reagent | PAK1-long forward | This paper | qPCR primer | CCAGCATTGTGGCTTGTCAT |
| Sequence-based reagent | PAK1-long reverse | This paper | qPCR primer | TTGTGCTGCAGAGGCAGTAG |
| Commercial assay or kit | QuantiTect Reverse Transcription Kit | Qiagen | Cat# 205313 | |
| Chemical compound, drug | 5-Fluoro-2′-deoxyuridine | Sigma-Aldrich | Product Number: F 0503 | 1 µM |
| Software, algorithm | LI-COR Image Studio | LI-COR | Image Studio Lite, RRID:SCR_013715 | |
| Software, algorithm | FreezeFrame 3 | Actimetrics | RRID:SCR_014429 | Conditioned fear assay |
| Software, algorithm | Fusion | Accuscan Instruments | RRID:SCR_017972 | Open field assay |
| Software, algorithm | EthoVision XT | Noldus Information Technology | RRID:SCR_000441 | Morris water maze |
| Software, algorithm | Clampfit 10 | Molecular Devices | RRID:SCR_011323 | EEG |
| Software, algorithm | Proteome Discoverer | Thermo Fisher Scientific | RRID:SCR_014477 | Mass spectrometry |
| Software, algorithm | Prism | Graphpad | RRID:SCR_002798 |
Additional files
-
Supplementary file 1
Intellectual disability associations of genes with misregulated APA and differential gene expression following neuronal NUDT21 inhibition.
Alternative polyadenylation (APA), Differentially expressed gene (DEG), probability of loss of function intolerance (pLI), intellectual disability (ID), Online Mendelian Inheritance in Man (OMIM), autosomal recessive (AR), autosomal dominant (AD), X-linked dominant (XLD), X-linked recessive (XLR)
- https://cdn.elifesciences.org/articles/50895/elife-50895-supp1-v1.xlsx
-
Supplementary file 2
Alternative polyadenylation analysis code.
- https://cdn.elifesciences.org/articles/50895/elife-50895-supp2-v1.zip
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/50895/elife-50895-transrepform-v1.pdf