The CHORD protein CHP-1 regulates EGF receptor trafficking and signaling in C. elegans and in human cells
Figures
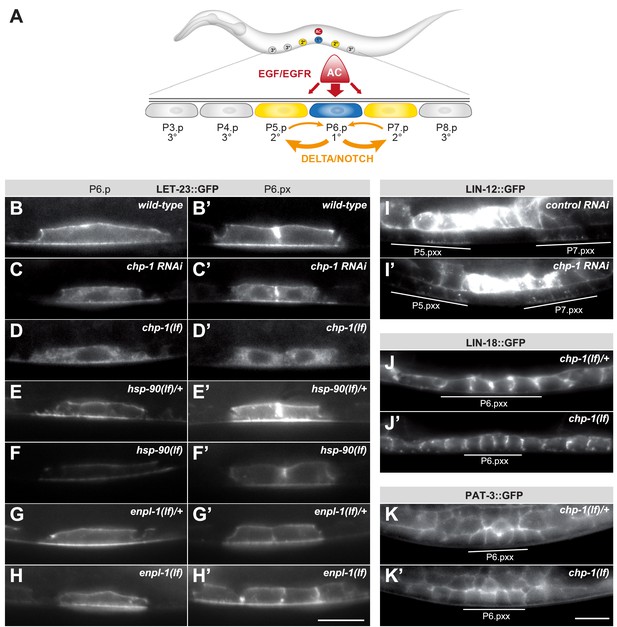
chp-1 is required for the plasma membrane localization of LET-23::GFP.
(A) Overview of the EGFR and NOTCH signaling pathways controlling VPC fate determination. (B) LET-23::GFP localization in P6.p and (B’) the two P6.p daughters (P6.px stage) of a wild-type, (C, C‘) a chp-1 RNAi and (D, D‘) a homozygous chp-1(tm2277lf) mutant larva. (E, E’) LET-23::GFP expression in heterozygous hsp-90(ok1333lf)/+ and (F, F’) and homozygous hsp-90(ok1333lf) larvae at the Pn.p and Pn.px stage. (G, G’) LET-23::GFP expression in heterozygous enpl-1(ok1964lf)/+ and (H, H’) homozygous hsp-90(ok1964lf) larvae at the Pn.p and Pn.px stage. (I) LIN-12::GFP localization in a control RNAi and (I’) a chp-1 RNAi-treated animals at the Pn.pxx stage. Note the unchanged apical localization of LIN-12::GFP in the 2° P5.p and P7.p descendants (underlined). (J) LIN-18::GFP membrane localization in a heterozygous chp-1(tm2277lf)/+ and (J’) a homozygous chp-1(tm2277lf) mutant at the Pn.pxx stage. (K) PAT-3::GFP membrane localization in a heterozygous chp-1(tm2277lf)/+ and (K’) a homozygous chp-1(tm2277lf) mutant at the Pn.pxx stage. The 1° P6.p descendants are underlined. At least 20 animals were analyzed for each genotype. The chp-1, hsp-90 and enpl-1 mutant phenotypes were completely penetrant, and chp-1 RNAi perturbed LET-23 localization in more than 50% of the animals. The scale bars in (H’) and (K’) are 10 µm.
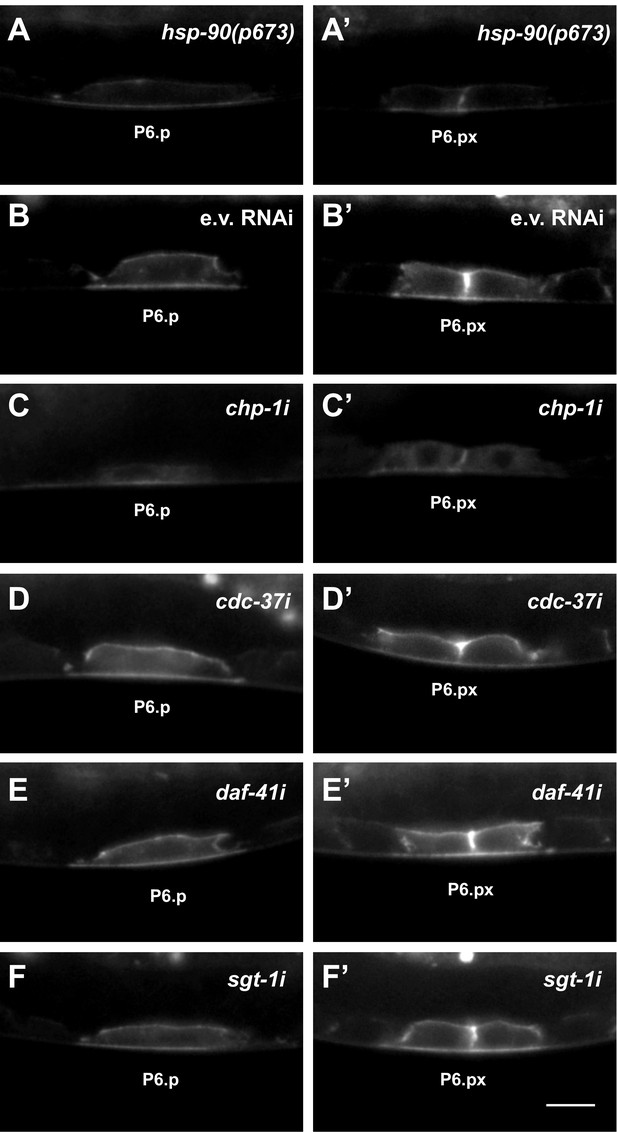
LET-23::GFP localization in hsp-90(p673) mutants and after RNAi knock-down of co-chaperones.
LET-23::GFP localization in P6.p and (left panels) the two P6.p daughters (right panels) in (A, A’) hsp-90(p673) mutants and (B–F’) under the indicated RNAi conditions. E.v. RNAi in (B, B’) are the negative controls treated with empty RNAi vector. At least 20 animals were analyzed for each condition. The scale bar in (F’) is 10 µm.
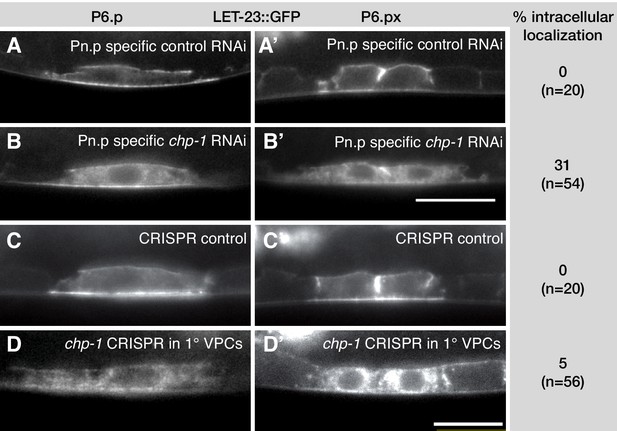
chp-1 acts cell-autonomously in the VPCs.
(A, A’) LET-23::GFP localization in the VPCs of empty vector control and (B, B’) chp-1 RNAi treated animals in the Pn.p cell-specific RNAi background. (C, C’) LET-23::GFP expression in a control siblings without and (D, D’) with the zhEx558[chp-1sg, egl-17p::cas-9] transgene. Note in (D’) the intracellular mislocalization of LET-23::GFP in the two P6.p descendants, while LET-23::GFP remained localized at the plasma membrane in the adjacent P7.p descendant. For each condition, the frequencies of the LET-23::GFP mislocalization phenotype and the numbers of animals analyzed are indicated to the right. The scale bars in (B’) and (D’) are 10 µm.
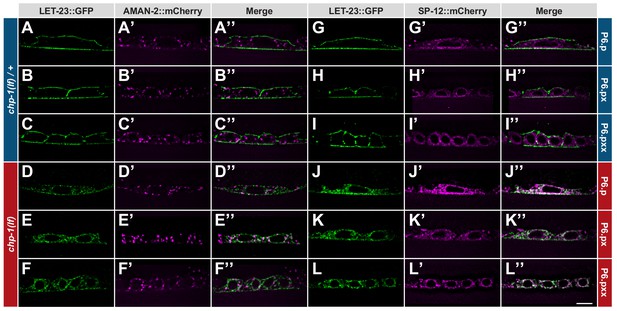
ER mislocalization of LET-23::GFP in chp-1(lf) mutants.
(A–C’’) Localization of LET-23::GFP and the AMAN-2::mCherry Golgi marker in heterozygous chp-1(tm2277lf)/+ control siblings and (D–F’’) homozygous chp-1(tm 2277lf) mutants at the P6.p to P6.pxx stage. (G–I’’) Localization of LET-23::GFP and the SP12::mCherry ER marker in heterozygous chp-1(tm2277lf)/+ control siblings and (J–L’’) homozygous chp-1(tm 2277lf) mutants at the P6.p to P6.pxx stage. The individual panels show the different channels of single mid-sagittal confocal sections through P6.p or its descendants. A voxel by voxel quantification of the co-localization between the AMAN-2::mCherry (Golgi) and SP12::mCherry (ER) markers with the LET-23::GFP signal is shown in Figure 3—figure supplement 1. The scale bar in (L’’) is 10 µm.
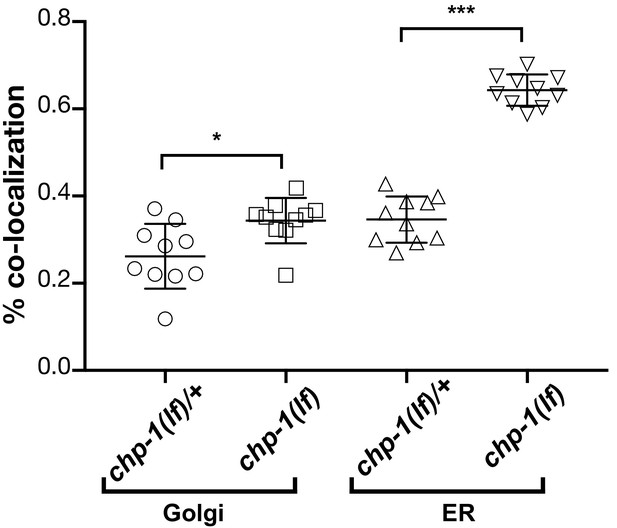
Quantification of the co-localization between the AMAN-2::mCherry (Golgi) and SP12::mCherry (ER) markers with LET-23::GFP in P6.p and its descendants.
Co-localization of the AMAN-2::mCherry Golgi or the SP12::mCherry ER marker with LET-23::GFP was quantified by calculating the thresholded Mander’s coefficient (Manders et al., 1993) as described in Materials and methods. Error bars show the standard error of the mean (SEM). p-Values were calculated by two-tailed t-tests for independent samples (p<0.05 = * and p<0.001 = ***). Ten animals were analyzed for each condition.
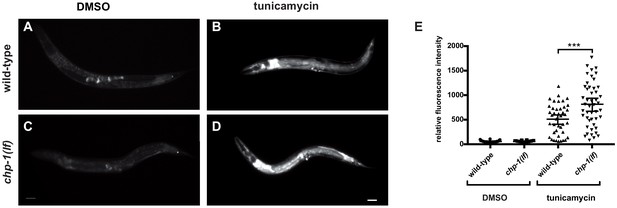
Unfolded protein response in chp-1(lf) mutants after tunicamycin treatment.
(A) HSP-4::GFP expression in untreated (DMSO) controls and (B) tunicamycin-treated wild-type animals, and (C) in untreated and (D) tunicamycin-treated chp-1(lf) mutants. The scale bar is 10 µm. (E) Quantification of HSP-4::GFP signal intensities under the different conditions. Error bars indicate the 95% CI. p-Values were calculated by t-tests for independent samples and are indicated as *** for p<0.01. Between 26 and 47 animals were analyzed for the different conditions.
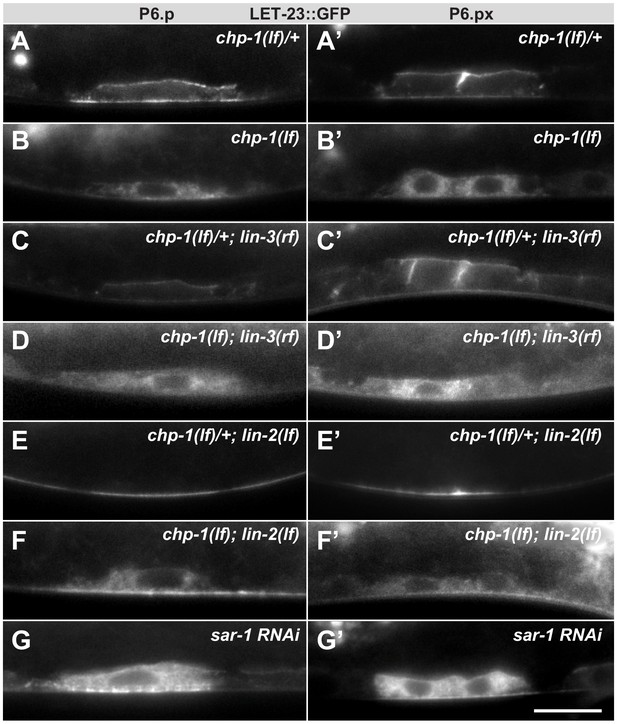
Intracellular mislocalization of LET-23::GFP in chp-1(lf) mutants is ligand independent.
(A, A’) Localization of LET-23::GFP in heterozygous chp-1(tm2277lf)/+ control siblings, (B, B’) homozygous chp-1(tm 2277lf) mutants, (C, C’) chp-1(tm2277lf)/+; lin-3(e1417rf) heterozygous, and (D, D’) in chp-1(tm2277lf); lin-3(e1417rf) homozygous double mutants at the P6.p and P6.px stage. (E, E’) Apical mislocalization of LET-23::GFP in chp-1(tm2277lf)/+; lin-2(n397lf) heterozygous and (F, F’) in chp-1(tm2277lf); lin-2(n397lf) homozygous double mutants. At least 20 animals were analyzed for each condition, and animals of the same genotype all showed the same LET-23::GFP localization pattern. (G, G’) sar-1 RNAi causes the same intracellular accumulation of LET-23::GFP as chp-1(lf). The scale bar in (G’) is 10 µm.
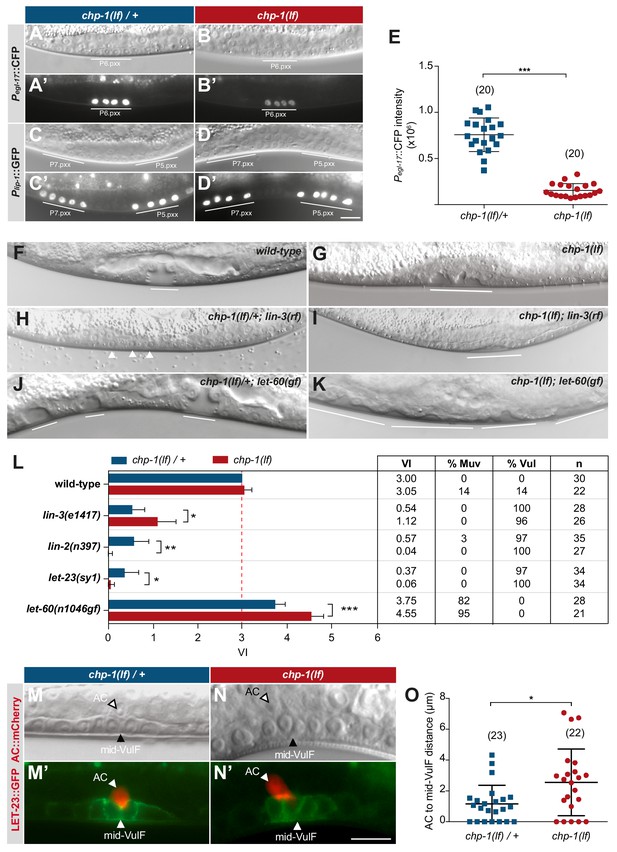
CHP-1 positively regulates EGFR/RAS/MAPK signaling in the VPCs.
(A, A‘) Expression of the 1° cell fate reporter Pegl-17::CFP in heterozygous chp-1(tm2277lf)/+ and (B, B’) homozygous chp-1(lf) mutant at the Pn.pxx stage. The top panels show Nomarski images of the differentiating VPCs and the bottom panel the reporter expression taken with identical exposure settings. (C, C’) Expression of the 2° cell fate reporter Plip-1::GFP in heterozygous chp-1(tm2277lf)/+ and (D, D’) homozygous chp-1(lf) mutant at the Pn.pxx stage. The scale bar in (D’) is 10 µm. (E) Quantification of the Pegl-17::CFP fluorescence intensity in the 1° VPCs at the Pn.pxx stage. The p-value was calculated by a two-tailed t-test for independent samples. The numbers of animals quantified are indicated in brackets. (F) Nomarski images of the vulval morphology in wild-type, (G) homozygous chp-1(tm2277lf), (H) heterozygous chp-1(tm2277lf)/+; lin-3(e1417rf), (I) homozygous chp-1(tm2277lf); lin-3(e1417rf), (J) heterozygous chp-1(tm2277lf)/+; let-60(n1046gf) and (K) homozygous chp-1(tm2277lf); let-60(n1046gf) L4 larvae. The descendants of induced VPCs forming an invagination are underlined and the arrowheads in (H) point at the nuclei of uninduced VPCs. (L) Quantification of the vulval induction index (VI) for the indicated genotypes. The table to the right shows the absolute mean VI, the percentage of animals with a Muv (VI > 3) and a Vul (VI < 3) phenotype and the number of animals scored (n) for each genotype. Error bars indicating the 95% confidence intervals and p-values were calculated by Bootstrapping with a resampling size of 1000, as described in Maxeiner et al. (2019) (p<0.05 = *p<0.01 = ** and p<0.001 = ***). (M, M’) AC to 1° VPC alignment at the Pn.pxx stage in heterozygous chp-1(tm2277lf)/+ and (N, N’) homozygous chp-1(tm2277lf) mutants. The top panels show Nomarski images and the bottom panels the expression of the LET-23::GFP reporter in green and the qyIs23[Pcdh-3:: PLC∂PH::mCherry] reporter labeling the AC in red. The scale bar in (N’) is 10 µm. (O) Quantification of the AC to VulF midline distance. The p-value was calculated by a two-tailed t-test for independent samples, and the numbers of animals scored are indicated in brackets.
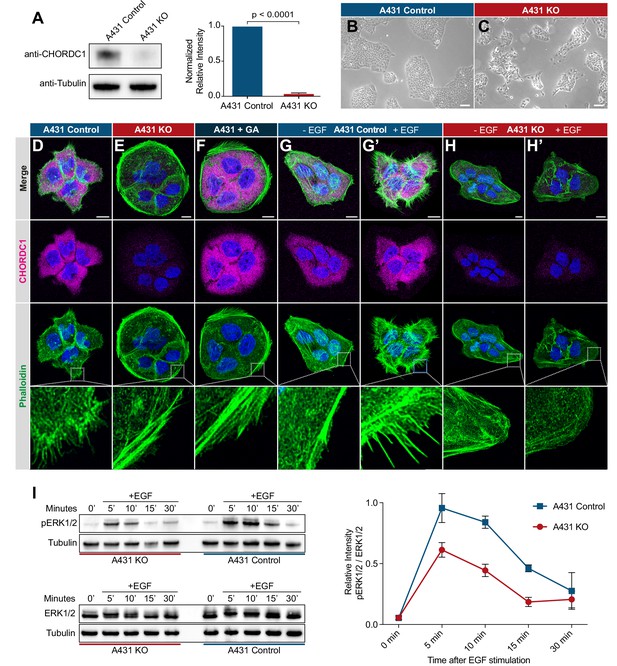
CHORDC1 is required for EGF-induced filopodia formation and sustained ERK activation in A431 cells.
(A) Quantification of CHORDC1 protein levels in A431 control and KO cells by Western blot analysis (average of eleven biological replicates). The bar graph shows the normalized averaged relative intensities ± SEM. The p-value was calculated using a two-tailed t-test for independent samples. (B) Phase contrast images of A431 control and (C) A431 KO cells 12 days post-lentiviral transduction. The scale bars are 100 µm. (D) Immunofluorescence staining of A431 control cells with antibodies recognizing CHORDC1 (magenta), fluorescently labeled phalloidin (green) and DAPI (blue), (E) A431 KO cells, and (F) A431 cells treated for 24 hr with 1 µM geldanamycin (GA). (G, H) Control- and KO cells fixed after 16 hr of serum starvation, and (G’, H’) 10 min after stimulation with 100 ng/ml human EGF. The scale bars are 10 µm. The bottom row shows higher magnifications of the cortical regions outlined by the dashed squares. (I) Total protein lysates of serum-starved cells that had been stimulated with 100 ng/ml human EGF for the indicated times (in minutes) were analyzed on Western blots with antibodies against phospho-ERK1/2 and total ERK1/2. The graph to the right shows the relative phospho-ERK1/2 signals normalized to the total ERK1/2 levels at each time point. The data shown represent the average ratios obtained in three biological replicates. Error bars indicate the SEM.
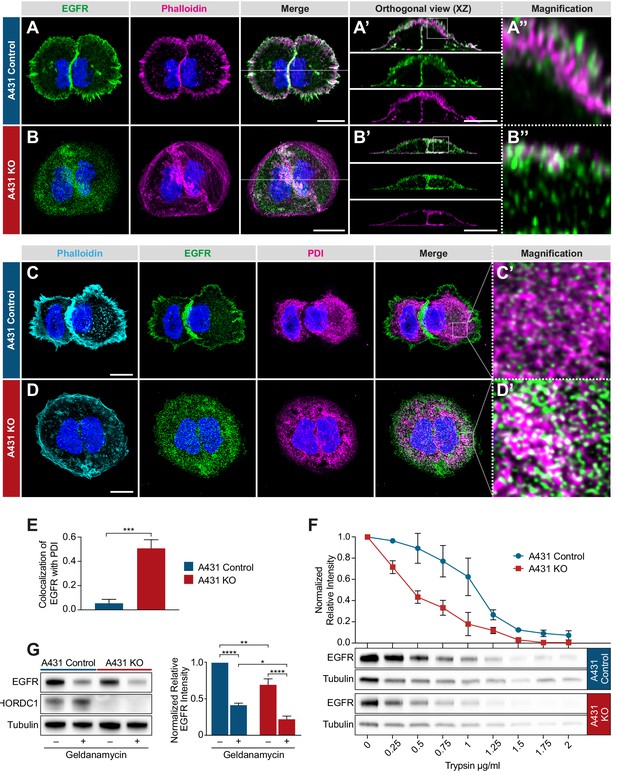
Mislocalization of the EGFR in CHORDC1 mutant A431 cells.
(A) Immunofluorescence staining of A431 control and (B) A431 KO cells with antibodies recognizing EGFR (green), fluorescently labeled phalloidin (magenta) and DAPI (blue). The dotted lines in the merge panels indicate the planes used to create the orthogonal (XZ) views shown in (A’, B’). The doted squares in (A’, B’) indicate the areas shown at higher magnification in (A’’, B’’). (C) Immunofluorescence staining of A431 control- and (D) KO cells with fluorescently labeled phalloidin (light blue), antibodies recognizing EGFR (green), the endoplasmic reticulum marker PDI (magenta), and with DAPI (blue). (C’, D’) show higher magnifications of the regions outlined with dotted squares in the merge panels of (C, D). All images are maximum intensity projection of three confocal sections. The scale bars are 10 µm. (E) Co-localization of EGFR and PDI in A431 control (n = 5) and A431 KO cells (n = 10) was quantified by calculating the Mander’s coefficient as described in Materials and methods (Manders et al., 1993). p-Values were calculated by a two-tailed t-test for independent samples. Error bars show the SEM. (F) Trypsin sensitivity assay. Total protein extracts of A431 control and A431 KO cells were incubated with the indicated trypsin concentrations, and the samples were analyzed by western blotting with antibodies against EGFR and tubulin. The line graph shows a quantification of the EGFR levels double normalized to the tubulin signal in each sample and to the undigested (0 µg/ml) samples. Error bars show the SEM. The average of three biological replicates is shown. (G) Western blot analysis of EGFR and CHORDC1 protein levels in A431 control and A431 KO cells with and without 1 µM geldanamycin treatment. The bar graph shows the normalized averaged relative intensities ± SEM. p-values (p<0.05 = *, p<0.01 = ** and p<0.0001 = ***) were calculated by one-way ANOVA and corrected with a Tukey multiple comparison test. The average of three biological replicates is shown.
Additional files
-
Supplementary file 1
Genotypes of the C. elegans strains used in this study.
- https://cdn.elifesciences.org/articles/50986/elife-50986-supp1-v2.xlsx
-
Supplementary file 2
Oligonucleotide primers used for plasmid constructions.
- https://cdn.elifesciences.org/articles/50986/elife-50986-supp2-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/50986/elife-50986-transrepform-v2.pdf





