Inhibition of ErbB kinase signalling promotes resolution of neutrophilic inflammation
Figures
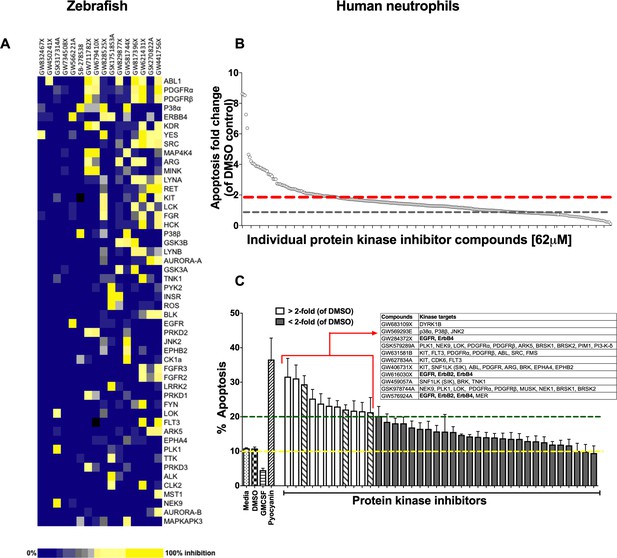
A protein kinase inhibitor compound library screen identifies compounds that promote the resolution of inflammation in vivo and neutrophil apoptosis in vitro.
(A) mpx:GFP zebrafish larvae (three dpf) that had undergone tail fin transection resulting in an inflammatory response at six hpi were incubated with individual PKIS compounds [25 µM] three larvae/well for a further 6 hr. Wells were imaged and manually scored between 0–3 on the basis of GFP at the injury site in the larvae. ‘Hit’ compounds scored ≥1.5 (n = 2, three larvae per compound per experiment). Publicly available kinase profiling information was generated previously by Elkins et al. (2016) and kinase inhibition of each compound [1 µM] is shown as a gradient of blue to yellow. Hit compounds were ranked horizontally (left to right) from the most to least selective. Kinases (listed on the right) were vertically ranked from top to bottom from the most to least commonly targeted by inhibitors in PKIS. (B) PKIS compounds were incubated with primary human neutrophils for 6 hr. The entire library, at [62 µM], was screened on five separate days using five individual donors. Apoptosis was assessed by Annexin-V/TO-PRO-3 staining by flow cytometry and the percentage apoptosis calculated as Annexin-V single plus Annexin-V/TO-PRO-3 dual positive events. Data are expressed as fold change over DMSO control and each circle represents a single compound. Sixty two compounds accelerated apoptosis ≥2 fold as identified by red dotted line (n = 1). Grey dotted line represents level of apoptosis in DMSO control (i.e. no change). (C) Of the 62 compounds identified above, 38 of the most specific inhibitors were incubated with neutrophils at [10 µM] for 6 hr and apoptosis measured as above. Controls included media, DMSO, GMCSF [50 u/mL] and pyocyanin [50 µM]. Eleven compounds (white bars) accelerated apoptosis ≥2 fold over DMSO control (as identified by dotted line). Kinases targeted by the 11 compounds are shown in the inset table. Hatched bars represent data points in which ErbB inhibitors were used. Data are expressed as percentage apoptosis ± SEM, n = 3 neutrophil donors.
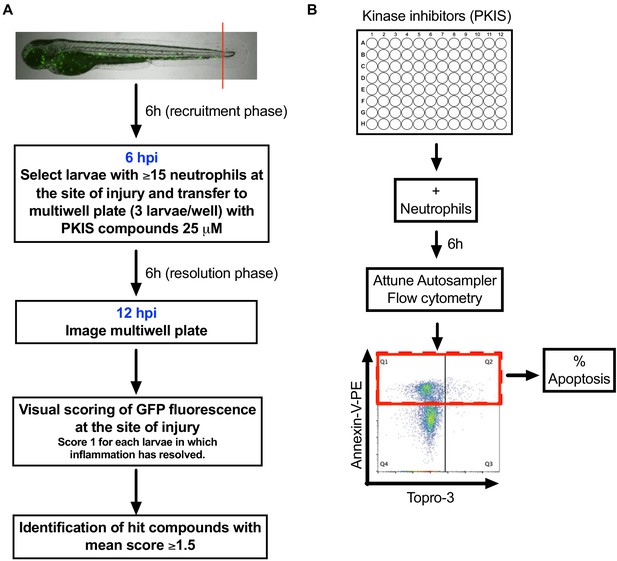
Schematics showing PKIS screen design.
(A) Tail fin transected three dpf Tg(mpx:GFP)i114 zebrafish larvae that had generated an inflammatory response at six hpi were incubated with individual PKIS compounds [25 µM] for a further 6 hr. Larvae were imaged and manually scored between 0–3 on the basis of green fluorescence at the injury site. (B) PKIS compounds were incubated with primary human neutrophils for 6 hr. Apoptosis was assessed by Annexin V/TO-PRO-3 staining by flow cytometry and the percentage apoptosis calculated as Annexin V single plus Annexin V/TO-PRO-3 dual positive events (as indicated by red box).
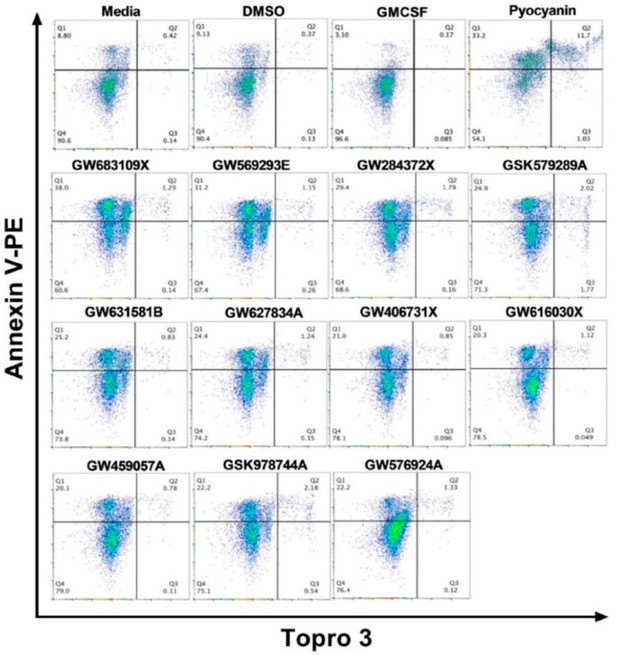
Flow cytometry dot plots for screen validation.
Human neutrophils were incubated for 6 hr with PKIS compounds identified as being pro-apoptotic from the first round of screening (Figure 1B). Neutrophils were also incubated with controls as follows: media, DMSO (vehicle), GM-CSF (anti-apoptotic) or pyocyanin (pro-apoptotic). Figure shows annexin-V (y-axis) and ToPro-3 (x-axis) flow dot plots for all compounds that accelerated apoptosis <2 fold over DMSO control. Dot plots are representative of 3 independent experiments.
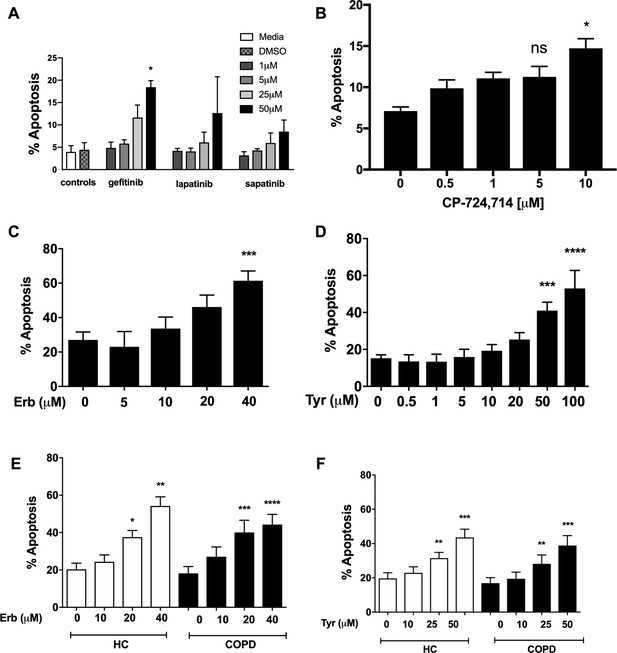
Inhibition of EGFR and ErbB2 drives apoptosis of neutrophils isolated from COPD patients and healthy subjects.
Neutrophils were incubated with media or a concentration range of gefitinib (A), lapatinib (A), sapatinib (A), CP-724714 (B), erbstatin (Erb, C) or tyrphostin AG825 (Tyr, D) for 6 hr. Stars represent significant difference compared to DMSO control (indicated by ‘0’ in B-D). Neutrophils from COPD patients (open bars) and age-matched healthy control subjects (black bars) were incubated with DMSO or a concentration range of erbstatin (E) or tyrphostin AG825 (F) for 6 hr. Apoptosis was assessed by light microscopy. The data are expressed as mean percentage apoptosis ± SEM from 3 (B, D), 4 (A,C), 10 (E,F COPD), or 7 (E,F HC) independent experiments using different neutrophil donors. Statistical significances between control and inhibitor was calculated by one-way ANOVA with Dunnett’s post-test, indicated as *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
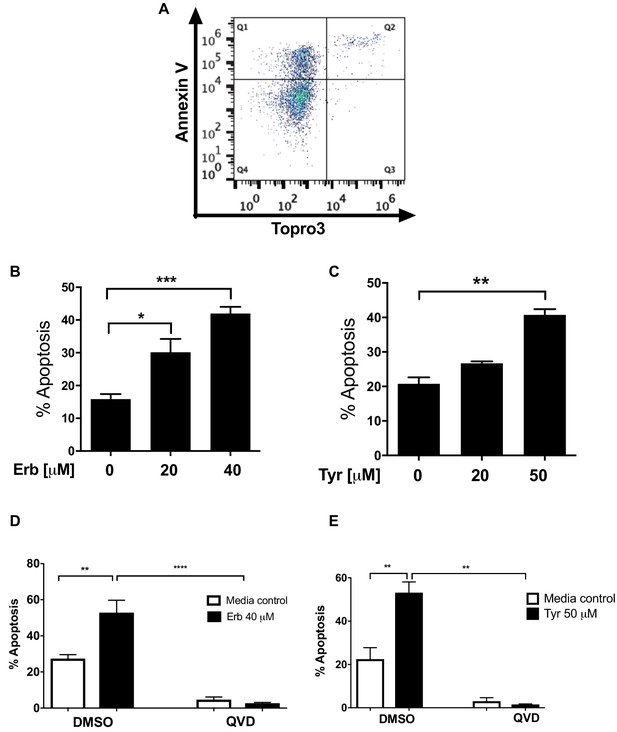
Erbstatin and tyrphostin AG825 induces caspase-dependent neutrophil apoptosis.
Neutrophils were incubated with DMSO or 20 µM or 40 µM erbstatin (Erb, A and B) or tyrphostin AG825 (Tyr, C) for 6 hr. Apoptosis was assessed by Annexin V/TO-PRO-3 staining by flow cytometry and the percentage apoptosis calculated as Annexin V single plus Annexin V/TO-PRO-3 dual positive events. (A) Representative quadrant plot of Erbstatin-treated neutrophils showing distribution of Annexin V and TO-PRO-3 positive events. (D–E) Neutrophils were incubated with DMSO or Erbstatin [40 µM] in the presence or absence of the pan caspase inhibitor, Q-VD-OPh [1 µM] for 6 hr (D) or 20 hr (E). Apoptosis was assessed by light microscopy. The data are expressed as mean percentage apoptosis ± SEM from 3 (B), 4 (C and E), or 5 (D) independent experiments. Statistical differences were calculated by one-way ANOVA (with Dunnett’s post test (B–C) and two-way ANOVA with Sidak (D–E) post-tests) and indicated as *p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001.
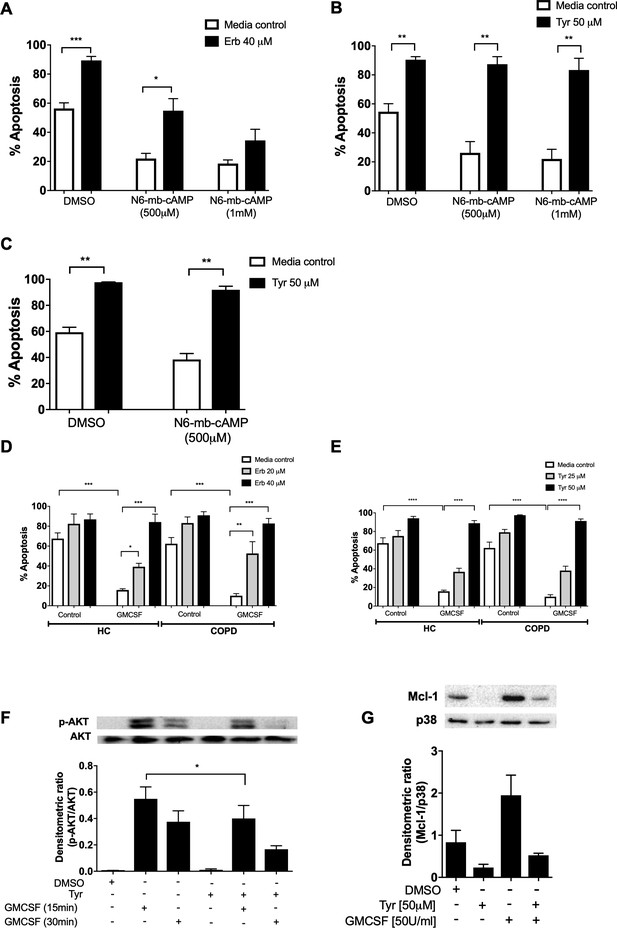
Erbstatin and tyrphostin AG825 overcome pro-survival effects of N6-MB-cAMP and GMCSF.
Neutrophils were incubated with DMSO, Erbstatin [Erb, 40 µM] (A) or tyrphostin AG825 [Tyr, 50 µM] (B) in the presence of DMSO or N6-MB-cAMP [500 µM and 1 mM] for 20 hr. Neutrophils isolated from COPD patients were incubated with DMSO or tyrphostin AG825 [50 µM] in the presence of DMSO or N6-MB-cAMP [500 µM] for 20 hr (C). Neutrophils isolated from COPD patients and age-matched healthy control subjects (HC) were incubated with DMSO, erbstatin (D) [20, 40 µM] or tyrphostin AG825 (E) [25, 50 µM] in the presence or absence of GMCSF [50 u/mL] for 20 hr. Apoptosis was assessed by light microscopy. The data are expressed as mean percentage apoptosis ± SEM from 4 to 6 independent experiments. (F) Neutrophils were incubated with DMSO or tyrphostin AG825 [Tyr, 50 µM] for 60 min before the addition of GMCSF [50 u/mL] for 15 or 30 mins. (G) Neutrophils were incubated with DMSO, tyrphostin AG825 [50 µM] for 60 min before the addition of GMCSF [50 u/mL] for a further 7 hr. Cells were lysed, subjected to SDS-PAGE electrophoresis and membranes probed for p-AKT, Mcl-1 or loading controls, AKT and P38. Images are representative of 3 independent experiments. Charts show densitometric values of 3 individual immunoblots and are expressed as a ratio of target (p-AKT or Mcl-1) over loading control (AKT or P38, respectively). Statistically significant differences were calculated by one-way ANOVA with Sidak post-test (A–C, F–G) or two-way ANOVA with Sidak post-test (D–E) and indicated as *p<0.05, **p<0.01, ***p<0.001.
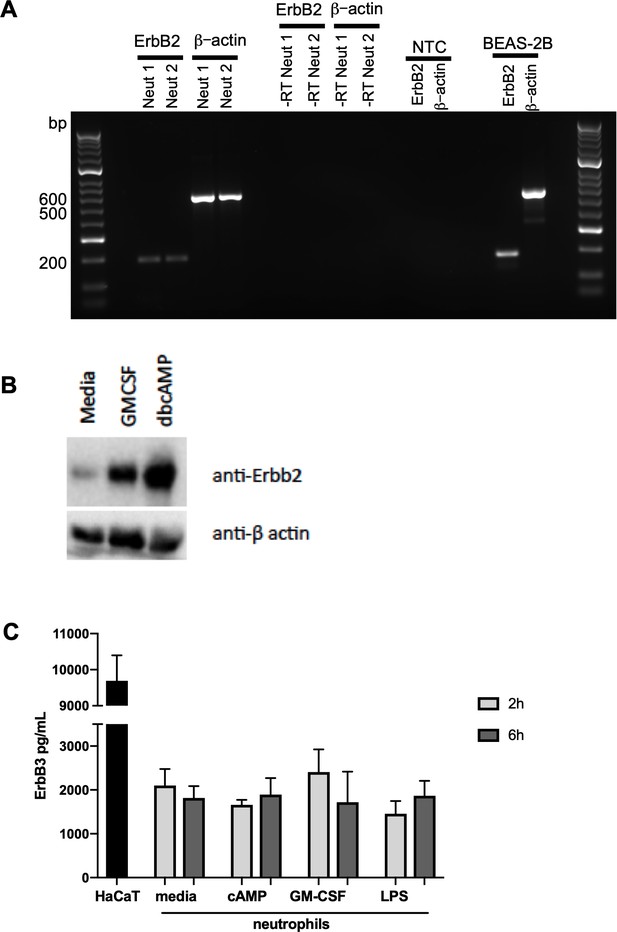
ErbB2 and ErbB3 expression and regulation in human neutrophils.
(A) ErbB2 was detected in neutrophils and the positive control cell line, BEAS-2B, by RT-PCR. Primer sequences are as follows: ErbB2 forward: ACCCAGCTCTTTGAGGACAA, reverse: ATCGTGTCCTGGTAGCAGAG and β-actin forward: ATATCGCCGCGCTCGTCGTC, reverse: TAGCCGCGCTCGGTGAGGAT. NTC – no template control. (B) Neutrophils were treated with GMCSF [50 u/mL] and dbcAMP [10 μM] for 5 hr and lysates subjected to SDS PAGE. Membranes were immunoblotted with antibodies to ErbB2 antibody or β-actin as a loading control. A 60kD band was detected which was upregulated by GMCSF and dbcAMP. The image is representative of three independent experiments. (C) ErbB3 was detected by ELISA in human neutrophils and the positive control cell line, HaCaT. Neutrophils were treated with media, dbcAMP [500 µM], GM-CSF [50 u/mL] or LPS [1 µg/mL] for 2 hr or 6 hr, after which lysates were collected and ELISA detecting total human ErbB3 was carried out (N = 4). Bars indicate mean + SEM and statistical differences between media control and treatments were measured by one-way ANOVA and Sidak post-test (C, ns).
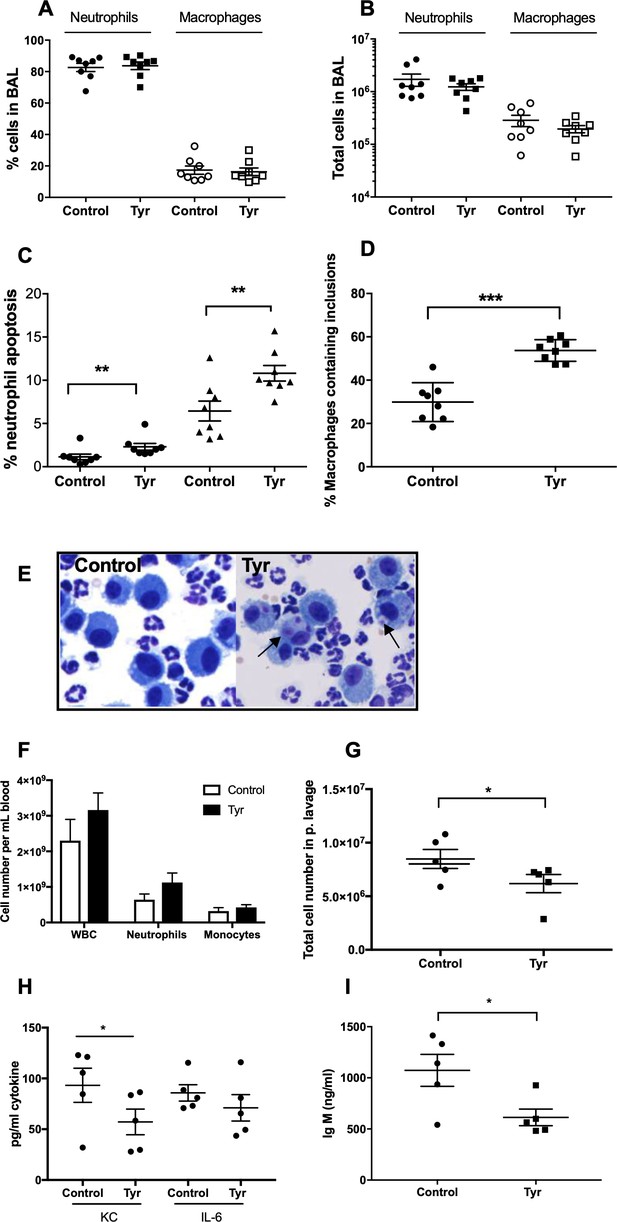
Tyrphostin AG825 increases neutrophil apoptosis and reduces inflammation in murine models of inflammation.
C57BL/6 mice were nebulized with LPS and immediately injected intraperitoneally with either 10% DMSO (control, n = 8) or 20 mg/Kg tyrphostin AG825 (Tyr, n = 8). After 48 hr the mice were sacrificed and subjected to bronchoalveolar lavage. Percentage neutrophils (A, closed icons) and macrophages (A, open icons) and absolute numbers of neutrophils (B, closed icons) and macrophages (B, open icons) in BAL were calculated by haemocytometer and light microscopy. (C) Percentage neutrophil apoptosis (circles) and percentage neutrophil apoptosis calculated by also including numbers of apoptotic inclusions visualised within macrophages (triangles) was assessed by light microscopy. (D) Macrophages containing one or more apoptotic inclusions expressed as a percentage of all macrophages. Light microscopy image showing apoptotic inclusions within macrophages as indicated by black arrows (E). C57BL/6 mice were injected i.p. with 1 mg zymosan and 4 hr later injected i.p. with 20 mg/Kg tyrphostin AG825 (Tyr, n = 5) or 10% DMSO (Control, n = 5). At 20 hr mice were sacrificed and subjected to peritoneal lavage. (F) WBC, neutrophils and macrophages in blood were measured by a Sysmex cell counter. Total cells in peritoneal lavage were counted by flow cytometry (G) and KC, IL-6 (H) and IgM (I) measured in lavage by ELISA. At least two independent experimental replicates each processing 1–3 mice/group were performed. Statistical significance was calculated by Mann–Whitney U test (A–D and G–I) or one-way ANOVA with Sidak post-test (F) and indicated as *p<0.05, **p<0.01, ***p<0.001.
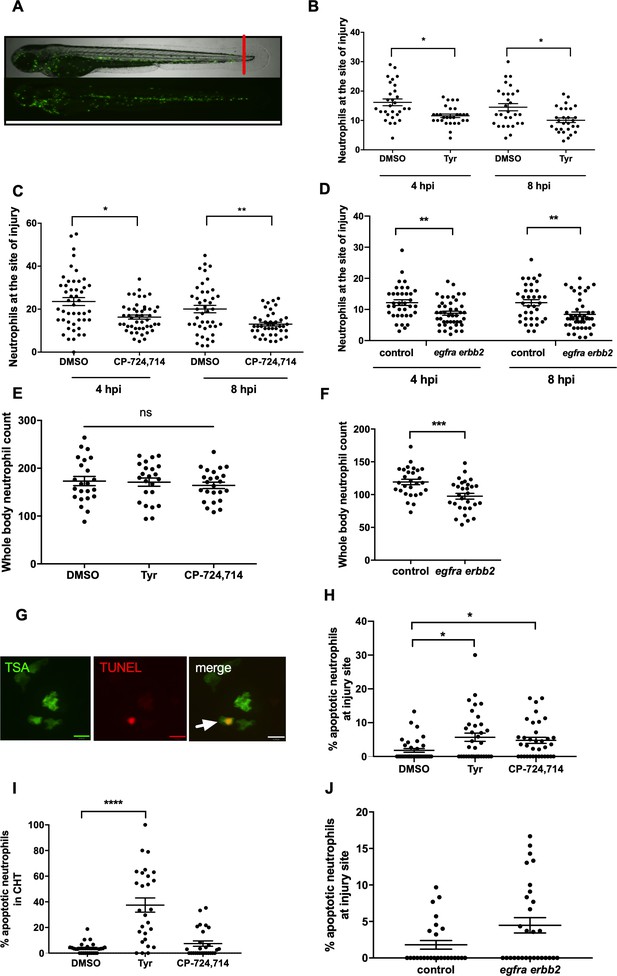
Pharmacological inhibition and genetic knockdown of egfra and erbb2 by CRISPR/Cas9 reduces neutrophil number at the site of injury in a zebrafish model of inflammation.
Tail fin transection was performed as indicated by the red line (A, upper image). Zebrafish larvae (mpx:GFP) were pre-treated at two dpf with DMSO, tyrphostin AG825 [Tyr, 10 µM] (B, minimum n = 28 larvae per condition), or CP-724714 [10 µM] (C, minimum n = 42 larvae per condition) for 16 hr followed by injury. egfra and erbb2 crispants were generated and injured at two dpf (D, minimum n = 36 larvae per condition). The number of neutrophils at the site of injury was determined at 4 and 8 hpi by counting GFP-positive neutrophils. To enumerate neutrophils across the whole body, uninjured inhibitor treated larvae (three dpf) (E, minimum n = 23 larvae per condition) or crispants (two dpf) (F, minimum n = 28 larvae per condition) were imaged by fluorescent microscopy (A, lower image). Apoptosis was measured at the site of injury after 8 hr by TSA and TUNEL double staining (G) (white arrow indicates TUNEL positive neutrophil, scale bar 10 μM) of mpx:GFP tyrphostin AG825 [Tyr, 10 µM] or CP-724714 [10 µM] treated larvae at three dpf (H, minimum n = 35 larvae per condition). Uninjured inhibitor treated larvae were assessed for neutrophil apoptosis in the CHT at three dpf (I, minimum n = 27 larvae per group). Apoptosis at the tail fin injury site of egfra erbb2 crispants at two dpf was also measured at eight hpi (J, minimum n = 26 larvae per group). All data collated from at least three independent experiments, displayed as mean ± SEM. Each icon shows one data point from one individual larvae. Statistically significant differences were calculated by two-way ANOVA with Sidak post-test (B–D) or one-way ANOVA with Dunnett’s post-test(E), Students’ t-test (F), Kruskal-Wallis test with Dunn’s post-test (H–I) or Mann-Whitney U test (J), and indicated as *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Tables
Kinexus antibody microarray analysis.
Ultrapurified neutrophils were incubated with N6-MB-cAMP [100 µM] for 30 and 60 min or lysed immediately following isolation (0’). Lysates from four donors were pooled prior to Kinex antibody microarray analysis. Table shows all targets for which phospho-antibodies had Z ratios of >1.5 compared to t = 0 baseline control, at each timepoint. ErbB related antibodies are in bold.
| Target protein | Z-ratio (30’ v 0’) | Target protein | Z-ratio (60’ v 0’) | |
|---|---|---|---|---|
| PDK1 | 5.69 | PDK1 | 4.79 | |
| ZAP70/Syk | 4.85 | PKCa/b2 | 2.73 | |
| p38a | 3.21 | Zap70/Syk | 2.72 | |
| PLCg1 | 3.16 | p38a | 2.32 | |
| MAP2K1 | 2.70 | S6Ka | 2.05 | |
| FKHRL1 | 2.58 | Rb | 1.96 | |
| GSK3a/b | 2.54 | PKCg | 1.79 | |
| Huntingtin | 2.29 | ErbB2 | 1.53 | |
| BLNK | 2.25 | |||
| Jun | 1.99 | |||
| Rb | 1.99 | |||
| ErbB2 | 1.94 | |||
| Btk | 1.92 | |||
| Bad | 1.81 | |||
| AMPKa1/2 | 1.70 | |||
| Synapsin 1 | 1.69 | |||
| PKBa | 1.64 | |||
Additional files
-
Supplementary file 1
PKIS compounds that accelerated neutrophil apoptosis >2 fold over control.
PKIS compounds were incubated with neutrophils for 6 hr and apoptosis was assessed by Annexin V/TO-PRO-3 staining by flow cytometry. Sixty-two compounds accelerated apoptosis ≥2 fold and compound names are presented here, along with fold change over control. The kinase profiling information for each of these inhibitors is available to download at https://www.nature.com/articles/nbt.3374#supplementary-information (file ‘PKIS Nanosyn Assay Heatmaps’ from Supplementary Data) (Elkins et al., 2016).
- https://doi.org/10.7554/eLife.50990.013
-
Transparent reporting form
- https://doi.org/10.7554/eLife.50990.014





