The Ormdl genes regulate the sphingolipid synthesis pathway to ensure proper myelination and neurologic function in mice
Figures
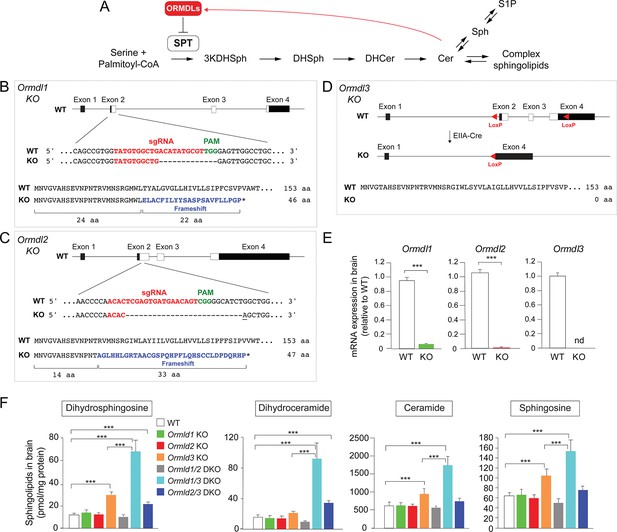
Ormdl3, but not Ormdl1 or Ormdl2, single knockout (KO) mice exhibit significantly increased levels of sphingolipids in the brain.
(A) Schematic of the de novo sphingolipid biosynthetic pathway and its feedback inhibition by ORMDLs through the sensing of ceramide levels. SPT, serine palmitoyltransferase; 3KDHSph, 3-keto-dihydrosphingosine; DHSph, dihydrosphingosine; DHCer, dihydroceramide; Cer, ceramide; Sph, sphingosine; S1P, sphingosine-1-phosphate. (B–D) Generation of Ormdl KO mice. Panels show the intron-exon organizations of the Ormdl genes and the protein coding regions (white). (B, C) Ormdl1 and Ormdl2 KO mice were produced by CRISPR/Cas9-induced mutations, resulting in frameshifts and premature stop codons. The locations of sgRNA sequences (red), PAM sites (green), as well as the changes in DNA and protein are indicated. The base insertion in the CRISPR/Cas9 modified Ormdl2 gene is underlined. (D) Ormdl3 KO mice were generated by germline Cre-LoxP recombination to excise exons 2, 3, and part of exon 4, resulting in the deletion of the entire protein-coding sequence. (E) RT-qPCR of Ormdl WT RNA in brain of Ormdl KO mice relative to that in WT mice. The mice were 8 weeks old. Probes detect the WT Ormdl sequences. Data are expressed as means ± SD. Unpaired Student’s t test; ***p<0.001. nd, not detectable. n = 4 for all genotypes. (F) Levels of dihydrosphingosine, total dihydroceramide, total ceramide, and sphingosine were determined by HPLC-tandem MS on lipid extracts of whole brains harvested from 8-week-old WT, Ormdl1 KO, Ormdl2 KO, Ormdl3 KO, Ormdl1/2 double KO, Ormdl1/3 double KO, and Ormdl2/3 double KO mice (Figure 1—source data 1). Data are expressed as means ± SD. One-way ANOVA with Bonferroni correction; *p<0.05, ***p<0.001. n = 8 for all genotypes. DKO, double knockout.
-
Figure 1—source data 1
Levels of dihydrosphingosine, total dihydroceramide, total ceramide, and sphingosine from brains of WT, Ormdl1 KO, Ormdl2 KO, Ormdl3 KO, Ormdl1/2 double KO, Ormdl1/3 double KO, and Ormdl2/3 double KO mice.
- https://cdn.elifesciences.org/articles/51067/elife-51067-fig1-data1-v1.xlsx
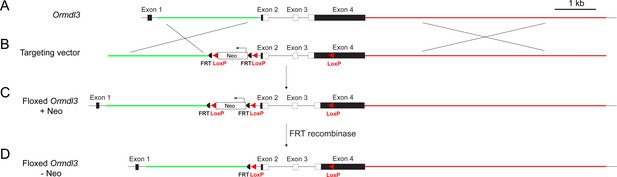
Generation of floxed Ormdl3 mice.
(A) The Ormdl3 gene contains four exons. The protein-coding region (white) extends from exon 2 to exon 4. (B) A gene targeting vector was designed with a 2.4 kb 5′ homology arm (green) and a 5.9 kb 3′ homology arm (red). A LoxP/FRT-flanked neomycin expression cassette was inserted 179 bp upstream of exon 2 in a transcriptional orientation opposite from Ormdl3. A single LoxP site was inserted into the 3′ UTR of exon 4. The genomic region flanked by the LoxP sites was 1.9 kb and included exons 2 and 3, as well as 428 bp of exon 4. (C) Structure of the targeted allele. (D) The neomycin (Neo) cassette was removed by FRT-mediated recombination, resulting in the floxed Ormdl3 -Neo allele.
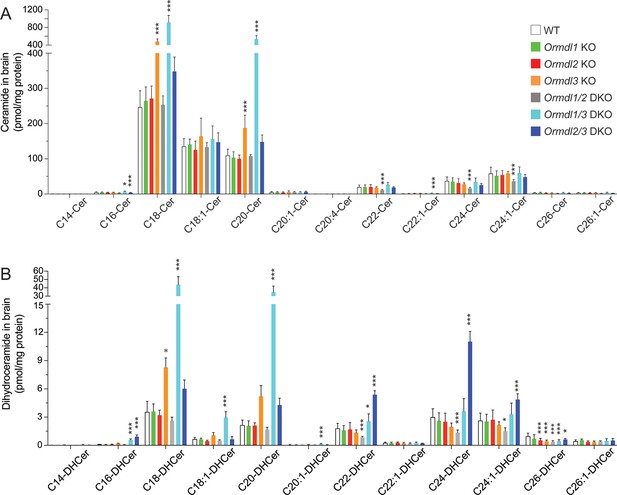
Levels of brain ceramide and dihydroceramide subspecies in Ormdl KO mice.
Sphingolipid concentrations were determined by HPLC-tandem MS on lipid extracts of whole brains harvested from 8-week-old WT, Ormdl1 KO, Ormdl2 KO, Ormdl3 KO, Ormdl1/2 double KO, Ormdl1/3 DKO, and Ormdl2/3 double KO mice (Figure 1—figure supplement 2—source data 1 file 1). (A) Individual ceramide subspecies with different fatty-acid chain lengths and C18 sphingoid bases. (B) Individual dihydroceramide subspecies with different fatty-acid chain lengths. Data are expressed as means ± SD. One-way ANOVA with Bonferroni correction; *p<0.05, ***p<0.001 versus WT mice. n = 8 for all genotypes. DKO, double knockout.
-
Figure 1—figure supplement 2—source data 1
Levels of individual ceramide and dihydroceramide subspecies with different fatty-acid chain lengths from brains of WT, Ormdl1 KO, Ormdl2 KO, Ormdl3 KO, Ormdl1/2 double KO, Ormdl1/3 double KO, and Ormdl2/3 double KO mice.
- https://cdn.elifesciences.org/articles/51067/elife-51067-fig1-figsupp2-data1-v1.xlsx
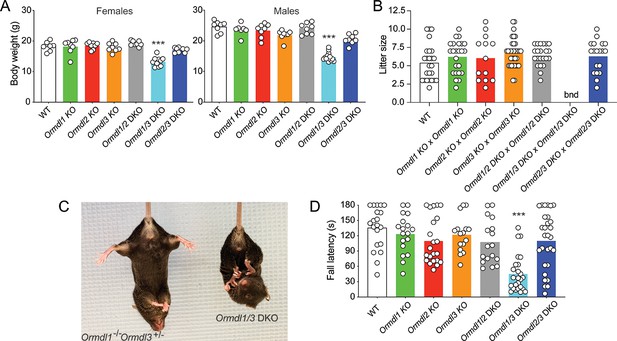
Elevated sphingolipids, neurologic phenotype and reduced viability when multiple Ormdls are deleted.
(A) Body weight of 8-week-old male (n = 7–14) and female mice (n = 7–10) of the indicated genotypes. Each circle represents the weight of an individual mouse. Unpaired Student’s t test; ***p<0.001 versus WT mice. (B) Litter size was determined at weaning for offspring of the indicated matings. Each circle represents the number of weanlings from an individual litter (n = 19–25). bnd, breeding not done. One-way ANOVA with Bonferroni correction. (C) Image of an 8-week-old Ormdl1/3 double KO mouse (right) showing characteristic clasping of the hindlimbs upon tail suspension, a sign of neurodegeneration. An Ormdl1–/– Ormdl3+/– littermate is shown (left). (D) Wire hang behavioral test. Eight-week-old mice were allowed to hang from a suspended wire using the forelimbs and the latency time to fall was recorded. The maximum hanging time was 180 s. Circles represent the mean of three determinations for each mouse (n = 16–29 mice for each genotype). One-way ANOVA with Bonferroni correction; ***p<0.001 versus WT mice. DKO, double knockout.
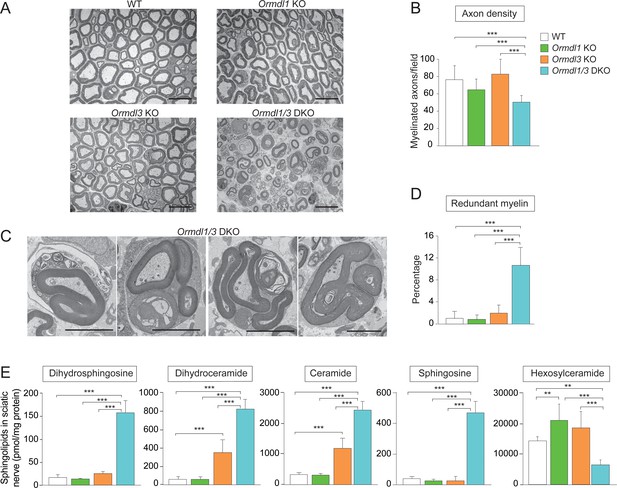
Myelination is disrupted in Ormdl1/3 double KO mice.
(A) Representative transmission EM images of sciatic nerve of 6-week-old WT, Ormdl1 KO, Ormdl3 KO, and Ormdl1/3 double KO mice. Scale bars, 10 μm. (B) Axon density in sciatic nerve of 6-week-old WT, Ormdl1 KO, Ormdl3 KO, and Ormdl1/3 double KO mice was determined by quantifying the number of myelinated axons in 7–12 EM fields per genotype. Data are expressed as means ± SD. One-way ANOVA with Bonferroni correction; ***p<0.001. n = 3 for WT, n = 2 for Ormdl1 KO, n = 2 for Ormdl3 KO, n = 3 for Ormdl1/3 double KO mice. (C) Example images of redundant myelin figures in sciatic nerve axons of 6-week-old Ormdl1/3 double KO mice. Scale bars, 5 μm. (D) Percentage of myelinated axons in sciatic nerve of 6-week-old WT, Ormdl1 KO, Ormdl3 KO, and Ormdl1/3 double KO mice showing redundant myelination was quantified in 7–12 EM fields per genotype. Data are expressed as means ± SD. One-way ANOVA with Bonferroni correction; ***p<0.001. n = 3 for WT, n = 2 for Ormdl1 KO, n = 2 for Ormdl3 KO, n = 3 for Ormdl1/3 double KO mice. (E) Levels of dihydrosphingosine, total dihydroceramide, total ceramide, sphingosine and hexosylceramide were determined by HPLC-tandem MS on lipid extracts of sciatic nerve from 8-week-old WT, Ormdl1 KO, Ormdl3 KO, and Ormdl1/3 double KO mice (Figure 3—source data 1). Data are expressed as means ± SD. One-way ANOVA with Bonferroni correction; ***p<0.001. n = 8 for all genotypes. DKO, double knockout.
-
Figure 3—source data 1
Levels of dihydrosphingosine, total dihydroceramide, total ceramide, sphingosine and total hexosylceramide from sciatic nerves of WT, Ormdl1 KO, Ormdl3 KO, and Ormdl1/3 double KO mice.
- https://cdn.elifesciences.org/articles/51067/elife-51067-fig3-data1-v1.xlsx
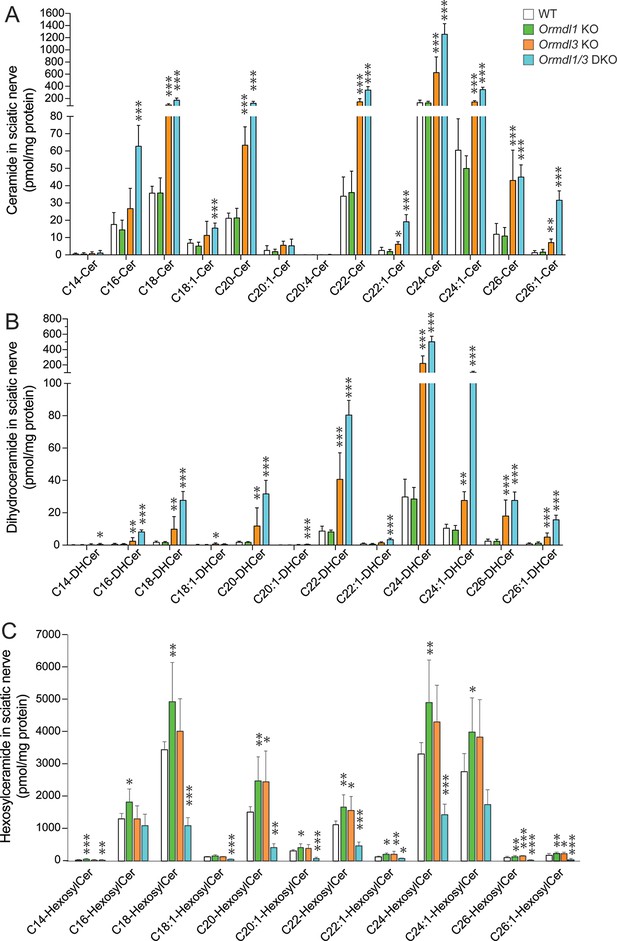
Levels of sciatic nerve ceramide, dihydroceramide and hexosylceramide subspecies in Ormdl1 KO, Ormdl3 KO, and Ormdl1/3 double KO mice.
Concentrations of sphingolipids with C18 sphingoid bases were determined by HPLC-tandem MS on lipid extracts of sciatic nerves harvested from 8-week-old WT, Ormdl1 KO, Ormdl3 KO, and Ormdl1/3 double KO mice (Figure 3—figure supplement 1—source data 1 file 1). (A) Individual ceramide subspecies with different fatty-acid chain lengths. (B) Individual dihydroceramide subspecies with different fatty-acid chain lengths. (C) Individual hexosylceramide subspecies with different fatty-acid chain lengths. Data are expressed as means ± SD. One-way ANOVA with Bonferroni correction; *p<0.05, **p<0.01, ***p<0.001 versus WT mice. n = 8 for all genotypes. DKO, double knockout.
-
Figure 3—figure supplement 1—source data 1
Levels of individual ceramide, dihydroceramide and hexosylceramide subspecies with different fatty-acid chain lengths from sciatic nerves of WT, Ormdl1 KO, Ormdl3 KO, and Ormdl1/3 double KO mice.
- https://cdn.elifesciences.org/articles/51067/elife-51067-fig3-figsupp1-data1-v1.xlsx
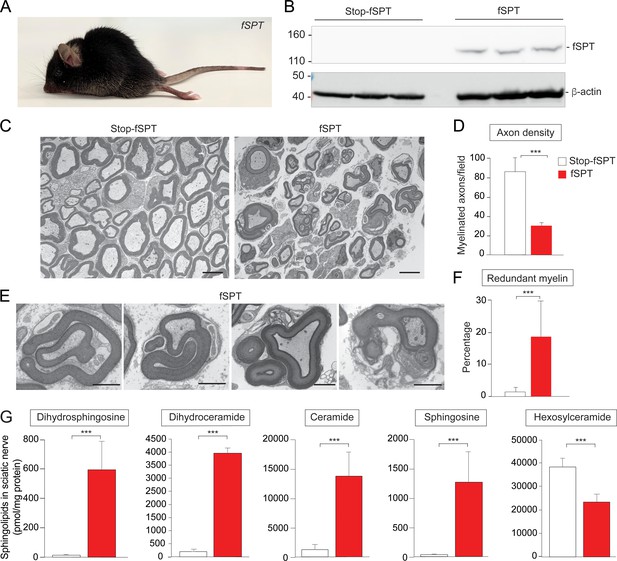
Increased de novo sphingolipid biosynthesis in myelin-producing cells mimicked the phenotype of Ormdl1/3 KO mice.
(A) Six-week-old fSPT/iPlp1-Cre (fSPT) mouse after treatment with tamoxifen for 2 weeks. (B) Western blot of fSPT expression in sciatic nerve of 6-week-old mice carrying Stop-fSPT without and with iPlp1-Cre (fSPT) after treatment with tamoxifen for 2 weeks. Bottom panel represents the same blot reprobed with an antibody against β-actin as a loading control. n = 3 for both genotypes. (C) Representative transmission EM images of sciatic nerve of 6-week-old Stop-fSPT and Stop-fSPT/iPlp1-Cre (fSPT)mice after treatment with tamoxifen for 2 weeks. Scale bars, 5 μm. (D) Axon density in sciatic nerve of 6-week-old Stop-fSPT and fSPT/iPlp1-Cre (fSPT) mice after treatment with tamoxifen for 2 weeks was determined by quantifying the number of myelinated axons in five and six EM fields for Stop-fSPT and fSPT/iPlp1-Cre mice (fSPT), respectively. Data are expressed as means ± SD. One-way ANOVA with Bonferroni correction; ***p<0.001. n = 2 for both genotypes. (E) Example images of redundant myelin figures in sciatic nerve of 6-week-old fSPT/iPlp1-Cre (fSPT) mice after treatment with tamoxifen for 2 weeks. Scale bars, 2 μm. (F) Percentage of myelinated axons showing redundant myelination in sciatic nerve of 6-week-old Stop-fSPT and fSPT/iPlp1-Cre (fSPT) mice after treatment with tamoxifen for 2 weeks, quantified in five and six EM fields for Stop-fSPT and fSPT mice, respectively. Data are expressed as means ± SD. One-way ANOVA with Bonferroni correction; ***p<0.001. n = 2 for both genotypes. (G) Levels of dihydrosphingosine, total dihydroceramide, total ceramide, sphingosine and hexosylceramide were determined by HPLC-tandem MS on lipid extracts of sciatic nerve from 6-week-old Stop-fSPT and fSPT/iPlp1-Cre (fSPT) mice after treatment with tamoxifen for 2 weeks (Figure 4—source data 1). Data are expressed as means ± SD. One-way ANOVA with Bonferroni correction; ***p<0.001. n = 5 for Stop-fSPT, n = 7 for fSPT/iPlp1-Cre (fSPT) mice after treatment with tamoxifen for 2 weeks.
-
Figure 4—source data 1
Levels of dihydrosphingosine, total dihydroceramide, total ceramide, sphingosine and hexosylceramide from sciatic nerves of mice carrying Stop-fSPT, without or with iPlp1-Cre (fSPT), after treatment with tamoxifen.
- https://cdn.elifesciences.org/articles/51067/elife-51067-fig4-data1-v1.xlsx
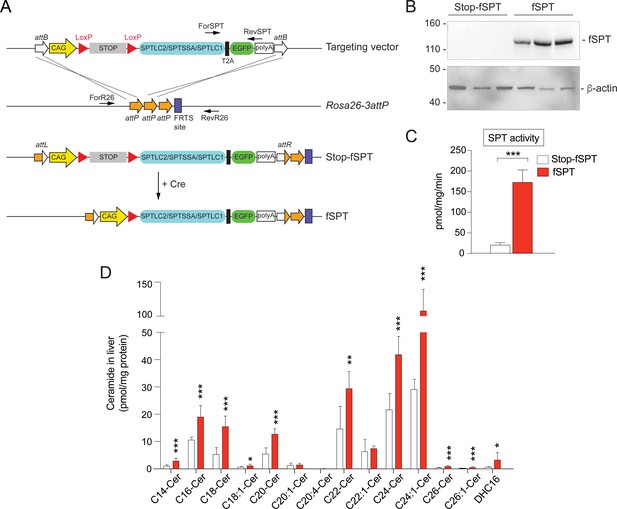
Generation and characterization of fSPT conditional mutant mice.
(A) fSPT was linked to EGFP by a T2A sequence and inserted into a vector containing a CAG promoter, a transcriptional stop sequence flanked by LoxP sites, and a polyA signal. This transcriptional cassette was flanked by attB sites. The plasmid along with phiC31 integrase protein was microinjected into zygotes from FVB Rosa26-3attP mice, which contained three tandem attP sites (3attP) in the Rosa26 locus, resulting in integration into the Rosa26 locus as indicated. The integrase catalyzes recombination between the attP and attB sites, resulting in two new hybrid sites, attL and attR. Expression of Cre excises the Stop sequence, enabling expression of the fSPT. Primers for genotyping (ForSPT, RevSPT, ForR26, RevR26) are shown and described in the 'Materials and methods' section. (B) Western blot of fSPT expression in liver of 12-week-old mice carrying Stop-fSPT, without (left) and with (right) Mx1-Cre (fSPT),after treatment with pIpC for 4 weeks. The bottom panel presents the same blot stripped and reprobed with an antibody against β-actin as a loading control. n = 3 for both genotypes. (C) SPT enzyme activity in liver of 12-week-old mice carrying Stop-fSPT without and with Mx1-Cre (fSPT) after treatment with pIpC for 4 weeks. Data are expressed as mean SPT activity (pmol of [3H] serine incorporated into sphingoid bases/mg protein/min) ± SD. Unpaired Student’s t test; **p<0.01. n = 3 for both genotypes. (D) Levels of individual ceramide subspecies with different fatty-acid chain lengths and C18 sphingoid bases were determined by HPLC-tandem MS on lipid extracts of liver harvested from 12-week-old mice carrying Stop-fSPT, without and with Mx1-Cre (fSPT), after treatment with pIpC for 4 weeks (Figure 4—figure supplement 1—source data 1 file 1). DHC16, C16-dihydroceramide. Data are expressed as means ± SD. Unpaired Student’s t test; *p<0.05, **p<0.01, ***p<0.001. n = 5 for Stop-fSPT, n = 8 for fSPT/Mx1-Cre (fSPT), after treatment with pIpC.
-
Figure 4—figure supplement 1—source data 1
Levels of individual ceramide subspecies with different fatty-acid chain lengths, C16-dihydroceramide, dihydrosphingosine, total ceramide, and sphingosine from liver of mice carrying Stop-fSPT, without and with Mx1-Cre (fSPT), after treatment with pIpC.
DHC16, C16-dihydroceramide.
- https://cdn.elifesciences.org/articles/51067/elife-51067-fig4-figsupp1-data1-v1.xlsx
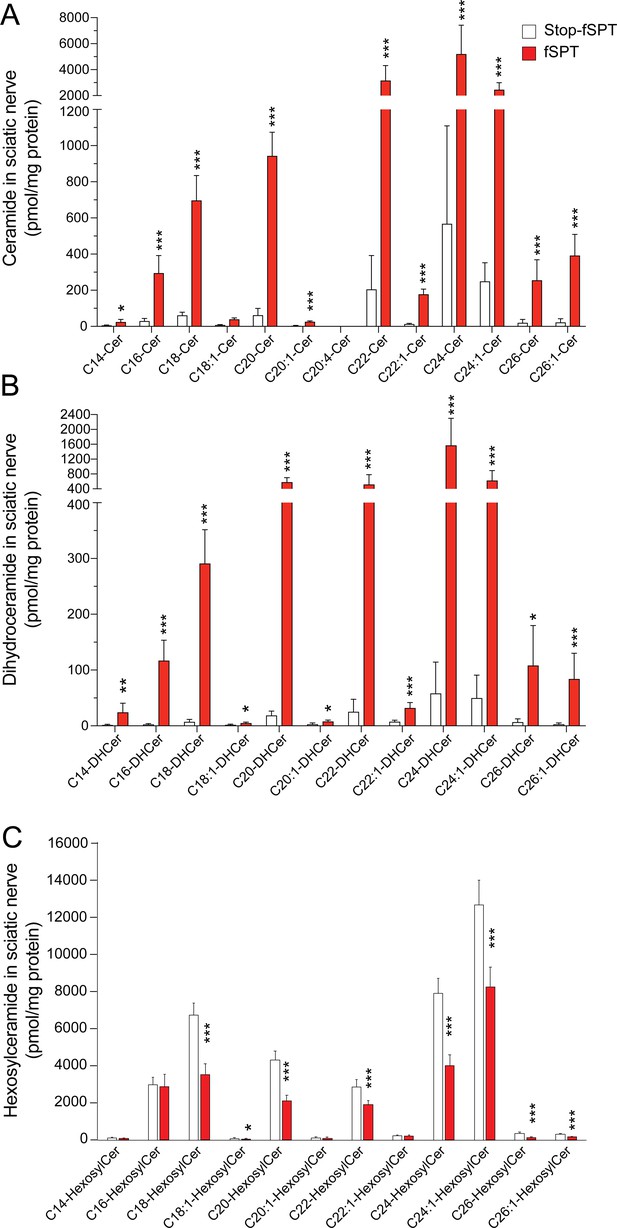
Levels of sciatic nerve ceramide and dihydroceramide subspecies in mice overexpressing fSPT.
Concentrations of sphingolipid with C18 sphingoid bases were determined by HPLC-tandem MS on lipid extracts of sciatic nerve harvested from 6-week-old mice carrying Stop-fSPT, without or with iPlp1-Cre (fSPT), after treatment with tamoxifen for 2 weeks (Figure 4—figure supplement 1—source data 1 file 2). (A) Individual ceramide subspecies with different fatty-acid chain lengths. Data are expressed as means ± SD. One-way ANOVA with Bonferroni correction; *p<0.05, ***p<0.001. n = 8 for both genotypes. (B) Individual dihydroceramide subspecies with different fatty-acid chain lengths. (C) Individual hexosylceramide subspecies with different fatty-acid chain lengths. Data are expressed as means ± SD. One-way ANOVA with Bonferroni correction; *p<0.05, **p<0.01, ***p<0.001. n = 5 for Stop-fSPT, n = 7 for fSPT/iPlp1-Cre (fSPT) mice after treatment with tamoxifen for 2 weeks.
-
Figure 4—figure supplement 2—source data 1
Levels of individual subspecies of ceramide, dihydroceramide and hexosylceramide with different fatty-acid chain lengths from sciatic nerves of mice carrying Stop-fSPT, without or with iPlp1-Cre (fSPT), after treatment with tamoxifen.
- https://cdn.elifesciences.org/articles/51067/elife-51067-fig4-figsupp2-data1-v1.xlsx
Tables
Analysis of offspring from Ormdl1–/– Ormdl2+/– Ormdl3+/– intercrosses.
Mouse genotypes were determined by PCR of tail-snip DNA of 156 pups at weaning from 38 litters derived from Ormdl1–/– Ormdl2+/– Ormdl3+/– intercrosses. The genotype distribution frequency of offspring, predicted by Mendelian considerations and actually obtained, is shown. The Chi-square test was used to determine whether the obtained distribution of genotypes was statistically different from the predicted Mendelian ratios; p<0.0001.
| Ormdl1–/– Ormdl2+/+ Ormdl3+/– | Ormdl1–/– Ormdl2+/+ Ormdl3+/+ | Ormdl1–/– Ormdl2+/+ Ormdl3–/– | Ormdl1–/– Ormdl2+/– Ormdl3+/– | Ormdl1–/– Ormdl2+/– Ormdl3+/+ | Ormdl1–/– Ormdl2+/– Ormdl3–/– | Ormdl1–/– Ormdl2–/– Ormdl3+/+ | Ormdl1–/– Ormdl2–/– Ormdl3+/– | Ormdl1–/– Ormdl2–/– Ormdl3–/– | |
|---|---|---|---|---|---|---|---|---|---|
| Observed # | 16 | 20 | 9 | 33 | 62 | 0 | 11 | 5 | 0 |
| Observed % | 10.26 | 12.82 | 5.77 | 21.15 | 39.74 | 0 | 7.05 | 3.21 | 0 |
| Predicted # | 9.75 | 19.5 | 9.75 | 19.5 | 39 | 19.5 | 9.75 | 19.5 | 9.75 |
| Predicted % | 6.25 | 12.5 | 6.25 | 12.5 | 25 | 12.5 | 6.25 | 12.5 | 6.25 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (M. musculus) | Ormdl1 KO mice | This paper | RRID: IMSR_JAX:034646 | See 'Materials and methods' |
| Genetic reagent (M. musculus) | Ormdl2 KO mice | This paper | RRID: IMSR_JAX:034645 | See 'Materials and methods' |
| Genetic reagent (M. musculus) | Ormdl3 Floxed mice | This paper | RRID: IMSR_JAX:034644 | See 'Materials and methods' |
| Genetic reagent (M. musculus) | EIIA-Cre mice | Jackson Laboratory | RRID:IMSR_JAX:003724 | Mice expressing Cre transgene under the control EIIa promoter |
| Genetic reagent (M. musculus) | STOP-fSPT transgenic mice | This paper | See 'Materials and methods' | |
| Genetic reagent (M. musculus) | PLP/creERT mice | Jackson Laboratory | RRID:IMSR_JAX:005975 | Mice expressing tamoxifen inducible-mouse Plp1 promoter |
| Genetic reagent (M. musculus) | Mx-Cre mice | Jackson Laboratory | RRID:IMSR_JAX:003556 | Mice expressing Cre transgene under the control of the mouse Mx1 promoter |
| Antibody | Monoclonal anti-human SPTLC1 | Santa Cruz Bio-technology | RRID:AB_10917035; clone H-1, cat. # sc-374143 | For western blot (1:1000 dilution) |
| Antibody | Anti-mouse IgG-HRP conjugated | Millipore | RRID:AB_9045; Cat. # AP-124P | For western blot (1:1000 dilution) |
| Antibody | Anti-mouse β-actin-HRP conjugated | Abcam | RRID:AB_8674; AC-15, cat # ab49900 | For western blot (1:100,000 dilution) |
| Sequence-based reagent | Ormdl1For | PCR primer | Genotyping Ormdl1 KO mice: 5′-TGTAATGAACAGCCGTGGTAT | |
| Sequence-based reagent | Ormdl1Rev | PCR primer | Genotyping Ormdl1 KO mice: 5′-GCAGAAGGGGATGCTGAGTAATA | |
| Sequence-based reagent | Ormdl2For | PCR primer | Genotyping Ormdl2 KO mice: 5′-CACATGCAGCAGTCCTACCA | |
| Sequence-based reagent | Ormdl2Rev | PCR primer | Genotyping Ormdl2 KO mice: 5′-GTTGGACTCCTGCCTGATCC | |
| Sequence-based reagent | NDEL1 | PCR primer | Genotyping Ormdl3 KO mice: 5′-GCAGGAGGAAGAGGCCCTCAG | |
| Sequence-based reagent | NDEL2 | PCR primer | Genotyping Ormdl3 KO mice: 5′-CTCTTGACTGCCGCTCTGCAAAAGAG | |
| Sequence-based reagent | SIAE4 | PCR primer | Genotyping Ormdl3 KO mice: 5′-CACGGCGCAGGGTTCTAATACATAC | |
| Sequence-based reagent | ForSPT | PCR primer | Genotyping fSPT mice: 5′-CATCGAGCTGAAGGGCATCG | |
| Sequence-based reagent | RevSPT | PCR primer | Genotyping fSPT mice: 5′-GTTATCTAGAATTATAGACGCGCTAG | |
| Sequence-based reagent | ForR26 | PCR primer | Genotyping fSPT mice: 5′-AGTTCTCTGCTGCCTCCTGGCTTCT | |
| Sequence-based reagent | CreFor | PCR primer | Genotyping fSPT mice: 5′-GCCTGCATTACCGGTCGATGC | |
| Sequence-based reagent | CreRev | PCR primer | Genotyping: 5′-CAGGGTGTTATAAGCAATCCC | |
| Sequence-based reagent | TaqmanFWD | Ormdl1 taqman assay primer | Ormdl1 taqman assay primer: 5′-ATGAATGTTGGAGTTGCCCAC | |
| Sequence-based reagent | TaqmanREV | Ormdl1 taqman assay primer | Ormdl1 taqman assay primer: 5′-GAACACTGCAGAAGGGGATG | |
| Sequence-based reagent | Ormdl1 probe | Applied Biosystems | Custom | Ormdl1 taqman assay probe: 5′-TGTAGATGACCCAAAATGGT |
| Sequence-based reagent | Ormdl2 assay-on-demand | Applied Biosystems | Mm00452481_g1 | |
| Sequence-based reagent | Ormdl3 assay-on-demand | Applied Biosystems | Mm00787910_sH |





