Herpes simplex viral nucleoprotein creates a competitive transcriptional environment facilitating robust viral transcription and host shut off
Figures
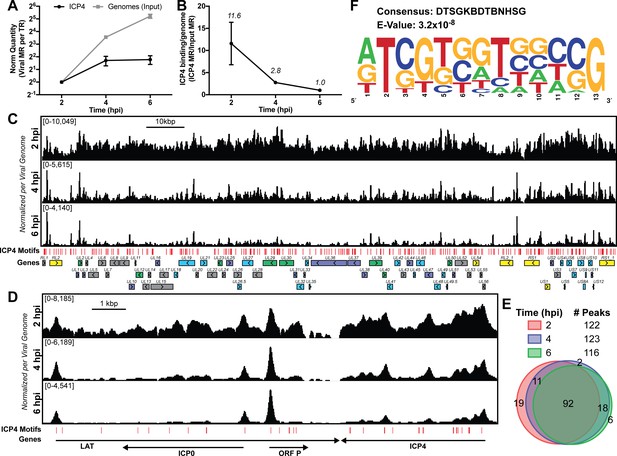
ICP4 binding at key points in the viral life cycle.
MRC5 cells were infected with HSV-1 for 2, 4, or 6 h, and ChIP-Seq for ICP4 was performed. (A) Quantification of viral genomes, measured as Input sample viral mapped reads (MR), normalized for sequencing depth or Total Reads (TR). Samples were normalized to two hpi, which was set as 1. (B) Quantification of ICP4 binding per viral genome measured as viral MR from ICP4 immunoprecipitation (IP) per viral MR from Input. (C–D) All data was normalized for sequencing depth and viral genome number using input ChIP-Seq reads. Viral ORFs are indicated, color coded by gene class with IE as yellow, E as green, leaky late (L1) as blue, and true late (L2) as purple. Find Individual Motif Occurrences (FIMO) identified genome sequences matching the consensus motif in B are indicated in red. (E) Intersection of MACS2 identified ICP4 occupied regions. (F) ICP4 consensus binding motif.
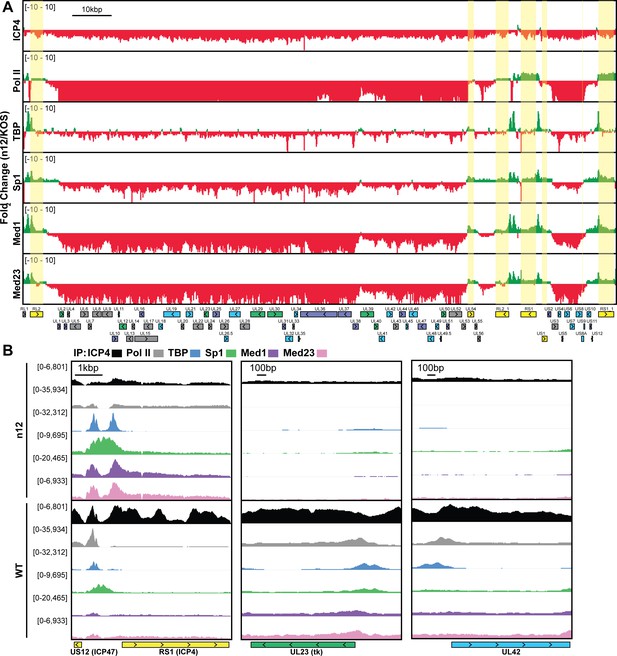
ICP4 recruitment of host Pol II machinery to viral promoters.
MRC5 cells were infected with an ICP4 null mutant (n12) or HSV-1 (WT) for 2.5 h and ChIP-Seq for ICP4, Pol II, TBP, Sp1, Med1, and Med23 was performed. All data was normalized for sequencing depth and viral genome number using input ChIP-Seq reads. Viral ORFs are indicated, color coded by gene class with IE as yellow, E as green, leaky late (L1) as blue, and true late (L2) as purple. (A) Fold change of n12 over WT aligned to the viral genome. Loci with greater binding in n12 or WT are colored in green or red, respectively. (B) ChIP-Seq reads normalized per viral genome and aligned to canonical IE, E, and L1 genes.
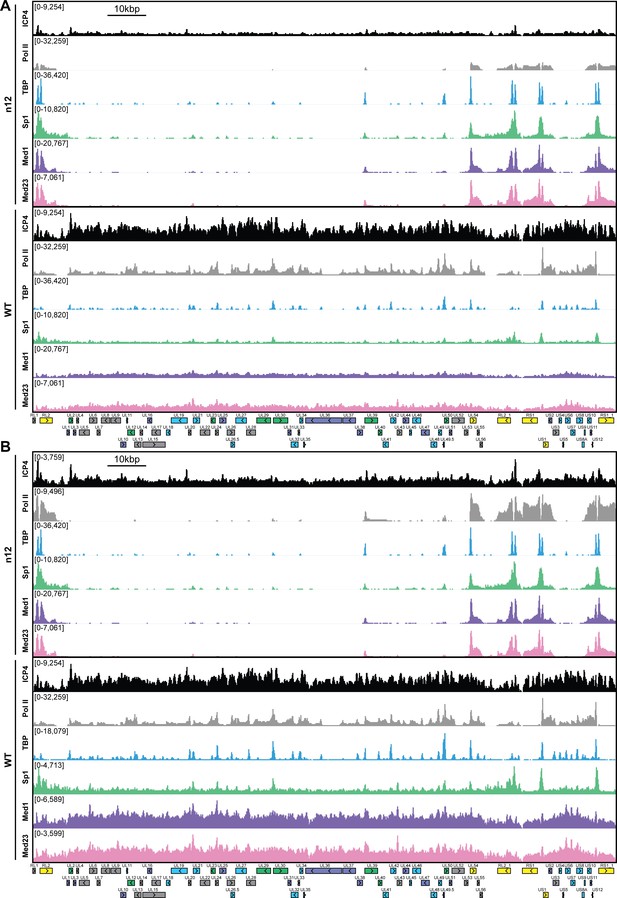
MRC5 cells were infected with an ICP4 null mutant (n12) or HSV-1 (WT) for 2.5 h and ChIP-Seq for ICP4, Pol II, TBP, Sp1, Med1, and Med23 was performed.
All data was normalized for sequencing depth and viral genome number using input ChIP-Seq reads. Viral ORFs are indicated, color coded by gene class with IE as yellow, E as green, L1 as blue, and L2 as purple. y-axes were A) maintained the same between IP’s or B) maximized for each trace to aid in visualization.
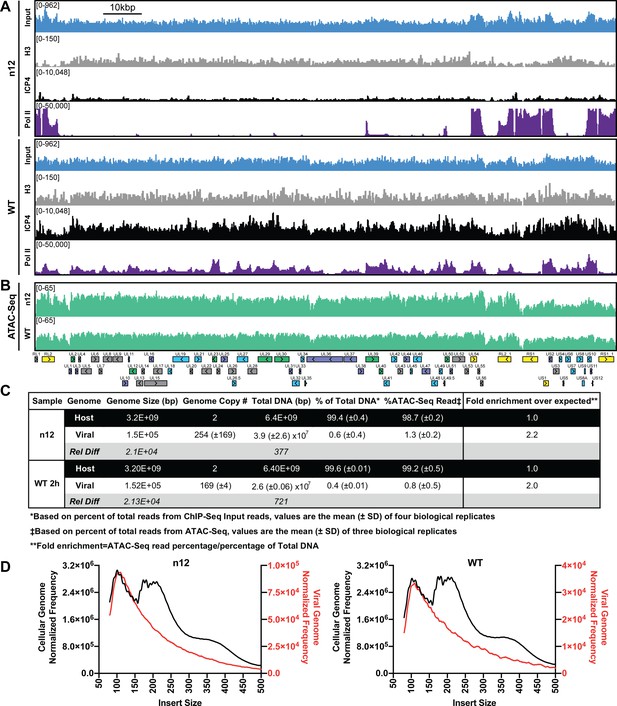
ICP4 dependence of viral genome accessibility and Histone H3 binding.
MRC5 cells were infected with an ICP4 null mutant (n12) or HSV-1 (WT) and harvested prior to genome replication. (A) ChIP-Seq for ICP4, Pol II, H3 and B–D) ATAC-Seq was performed. All data was normalized for sequencing depth and viral genome number using input ChIP-Seq reads. (C) Quantitative analysis of ATAC-Seq data, measuring the relative tagmentation enrichment for the virus or host as compared to expected. (D) Histogram plot of ATAC-Seq fragment size for reads mapped to the viral (red) or cellular (black) genome. Mononucleosome protected fragments are approximately 180–250 bp.
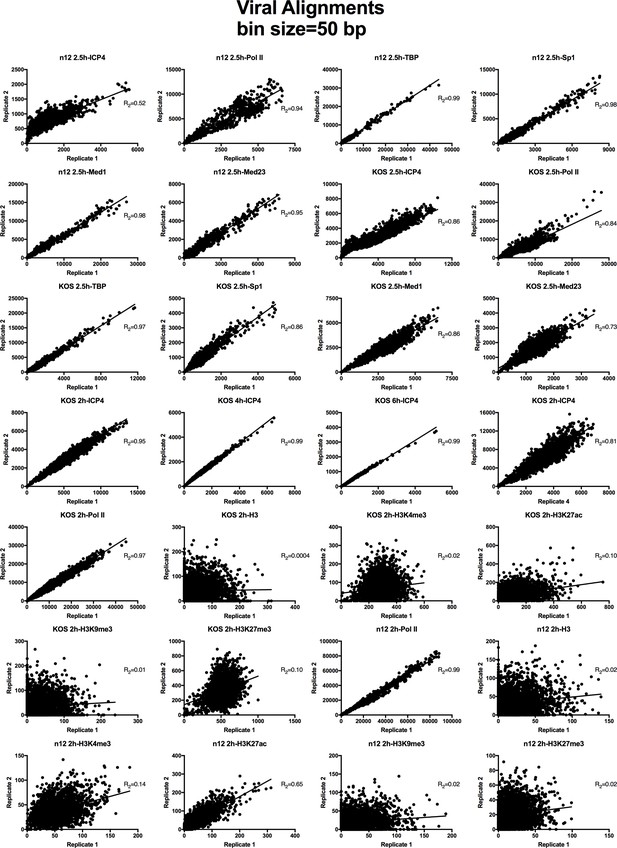
Analysis of ChIP-Seq data quality.
Normalized viral aligned bigwig files were assessed using MultiBigwigSummary in 50 bp bins. The values within these bins were plotted for biological replicates and a linear regression analysis was performed.
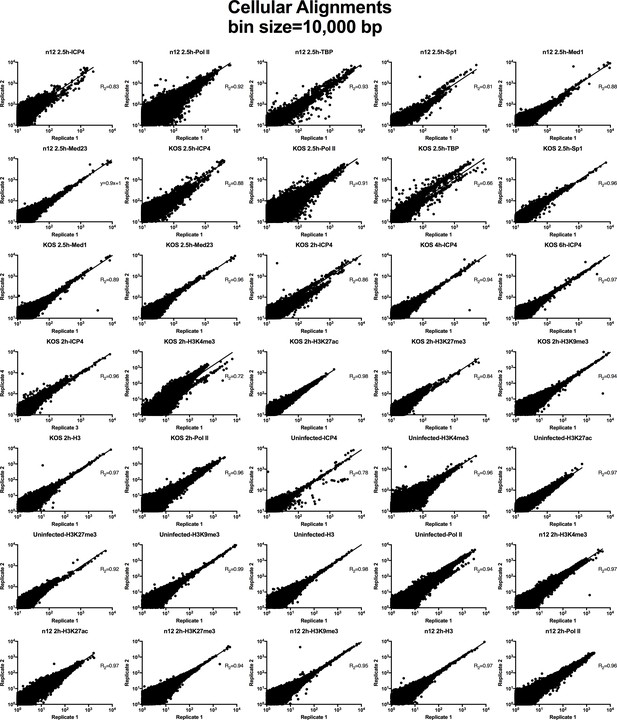
Analysis of ChIP-Seq data quality.
Normalized cellular aligned bigwig files were assessed using MultiBigwigSummary in 10,000 bp bins. The values within these bins were plotted for biological replicates and a linear regression analysis was performed.
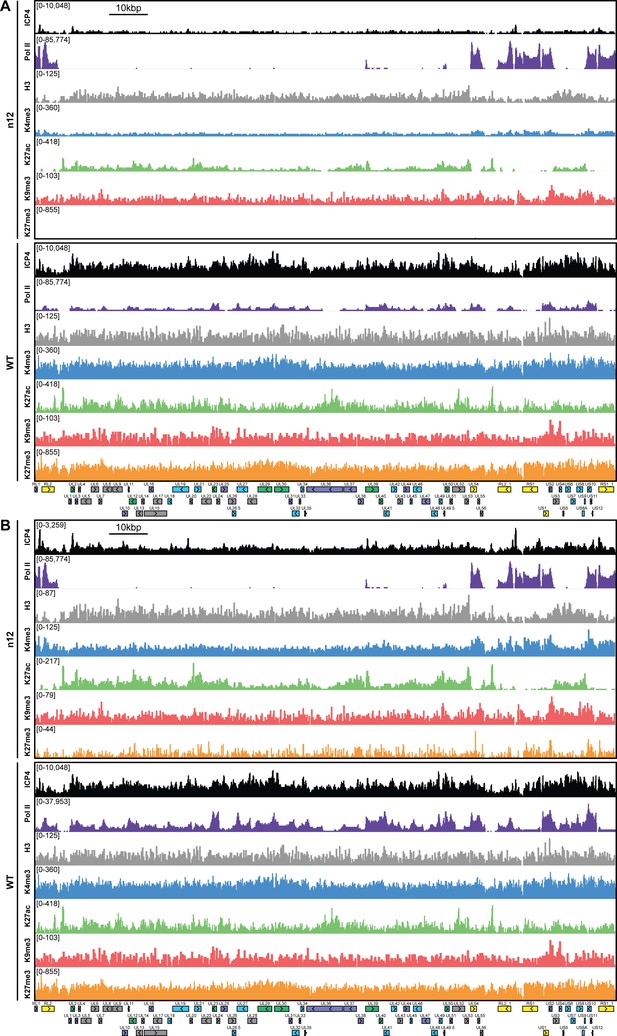
MRC5 cells were infected with an ICP4 null mutant (n12) or HSV-1 (WT) for 2 hr and ChIP-Seq for ICP4, Pol II, H3, H3K4me3, H3K27acetyl, H3K9me3, and H3K27me3 was performed.
All data was normalized for sequencing depth and viral genome number using input ChIP-Seq reads. Viral ORFs are indicated, color coded by gene class with IE as yellow, E as green, L1 as blue, and L2 as purple. y-axes were (A) maintained the same between IP’s or (B) maximized for each trace to aid in visualization.
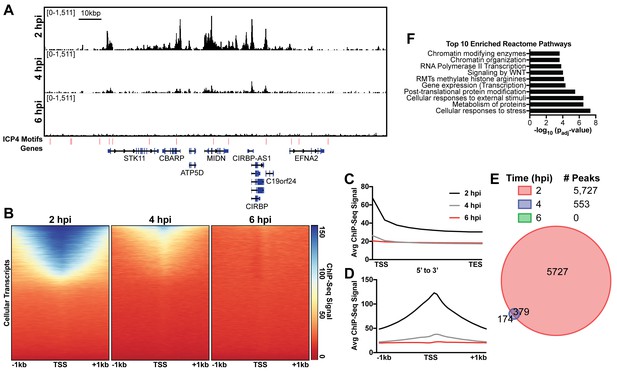
ICP4 binds cellular promoters during early infection.
MRC5 cells were infected with HSV-1 for 2, 4, or 6 h, and ChIP-Seq for ICP4 was performed. Data was aligned to the human genome (hg38) and normalized for cellular genome sampling. (A) Representative region of the cellular genome. ICP4 binding motifs are indicated in red. (B, D) Sequencing data centered + /- 1 kilobase from the transcription start site (TSS) of cellular mRNAs. (C) Sequencing data scaled from mRNA TSS to the transcription end site (TES). (E) Intersection of MACS2 identified ICP4 occupied regions. (F) Top 10 enriched Reactome groups for genes (n = 2190) with ICP4 bound at the promoter at 2 hr.
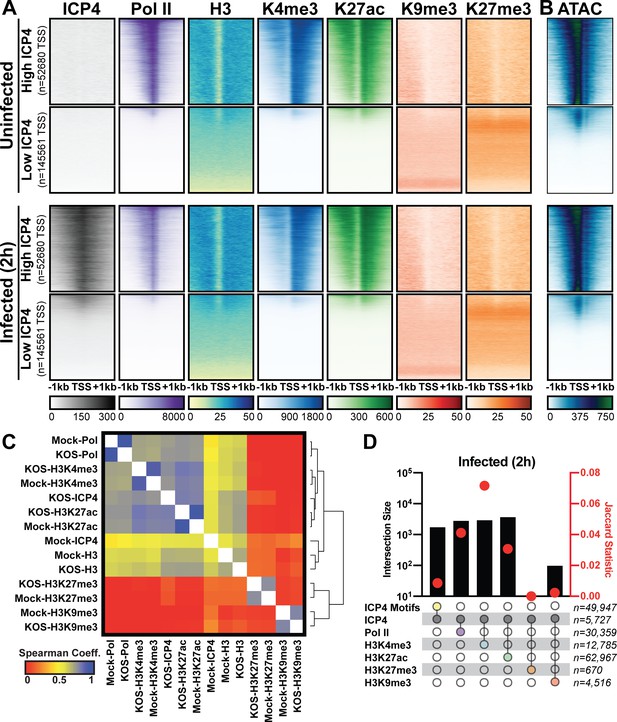
Association between cellular ICP4 binding and chromatin.
MRC5 cells were uninfected or infected with HSV-1 for 2 h. All data was aligned to the human genome (hg38) and normalized for sequencing depth. (A, C–D) ChIP-Seq data for ICP4, Pol II, H3, H3K4me3, H3K27acetyl, H3K9me3, and H3K27me3. (B) ATAC-Seq data. (A–B) Sequencing data centered + /- 1 kilobase from the TSS of cellular mRNAs. Data was stratified for ICP4 binding using K-means clustering. (C) Spearman correlation analysis, limited to cellular transcripts. (D) Intersection of MACS2 peaks, analyzed as number of intersecting peaks or Jaccard statistic.
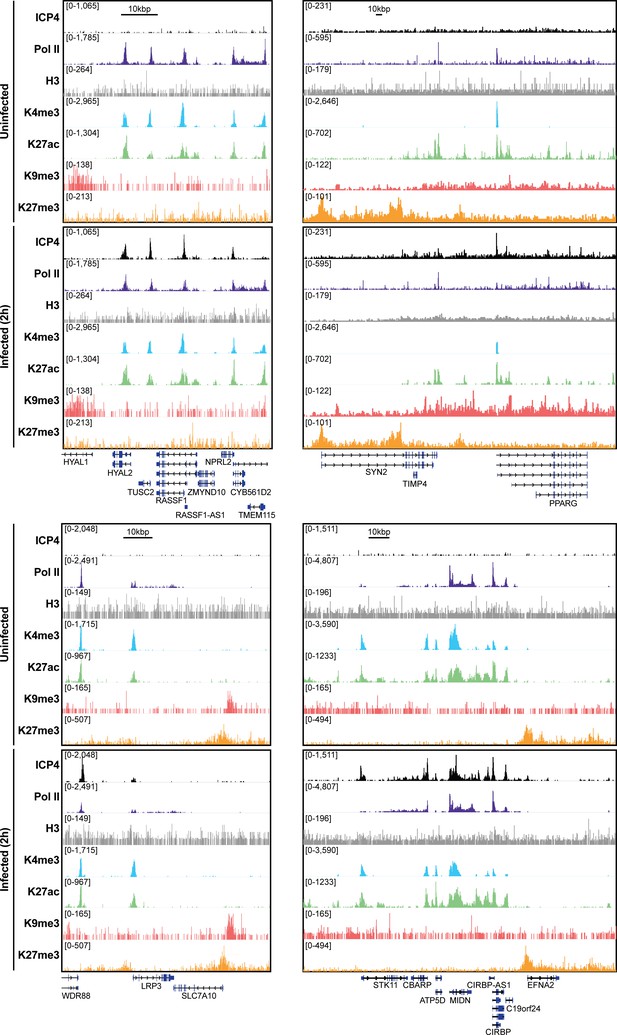
MRC5 cells were uninfected or infected with HSV-1 for 2 h, and ChIP-Seq for ICP4, Pol II, H3, H3K4me3, H3K27acetyl, H3K9me3, and H3K27me3 was performed.
Data was aligned to the human genome (hg38) and normalized for sequencing depth.
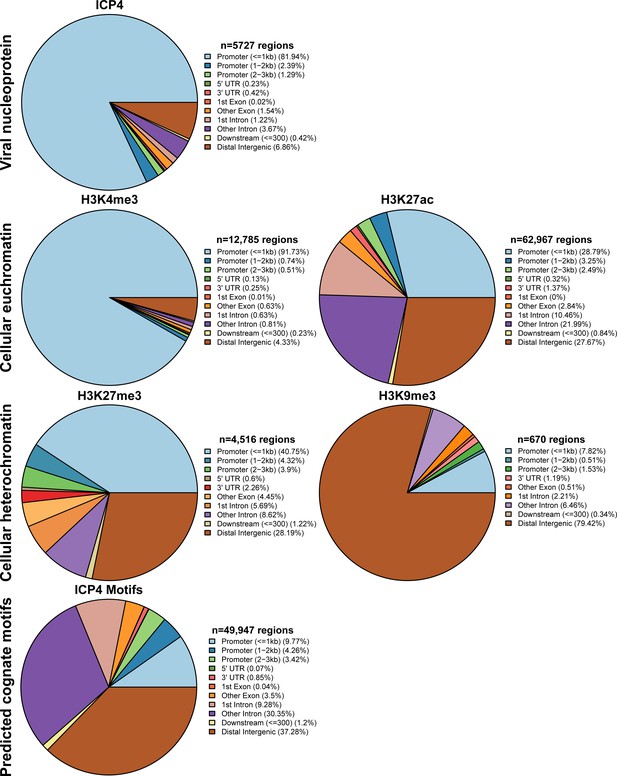
MRC5 cells were infected with HSV-1 for 2 h, and ChIP-Seq for ICP4, Pol II, H3, H3K4me3, H3K27acetyl, H3K9me3, and H3K27me3 was performed.
Data was aligned to the human genome (hg38). IP peaks consistent between biological duplicate experiments were determined using MACS2. ChIPSeeker assessment of bound regions for each set of IP peaks.
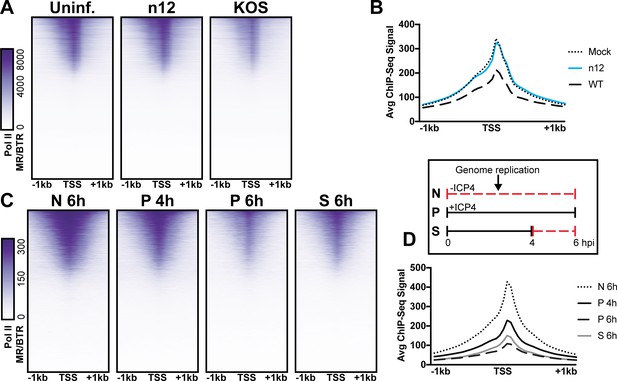
The role of ICP4 in Pol II loss on host promoters.
MRC5 cells were (A-B) uninfected or infected with n12 or WT HSV-1 and harvested prior to genome replication or C-D) infected with tsKos and grown at permissive conditions (P), shifted up from permissive to nonopermissive conditions at four hpi (S), or nonpermissive conditions (N). (A–D) ChIP-Seq for Pol II was performed. Data was aligned to the human genome (hg38) and normalized for cellular genome sampling. ChIP-Seq data was aligned to cellular promoters + /- 1 kilobase from TSS. The average signal for each condition plotted as a line graph.
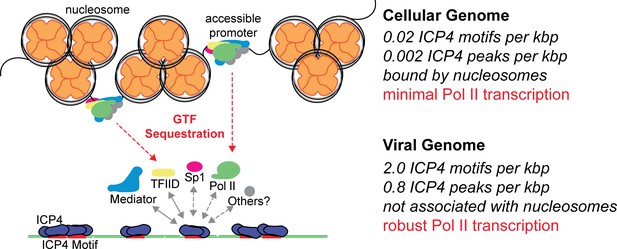
Model for ICP4 function.
ICP4 preferentially binds to the more accessible viral genome recruiting cellular transcription factors preferentially to the viral genome, thus activating the virus and inhibiting cellular transcription.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo-sapiens) | MRC5 | ATCC | ATCC CCL-171 RRID:CVCL_0440 | |
| Cell line (Cercopithecus aethiops) | Vero | ATCC | ATCC CCL-81 RRID:CVCL_0059 | |
| Cell line (Cercopithecus aethiops) | E5 | DeLuca and Schaffer, 1988 | E5 | Vero cells stably expressing ICP4 |
| Strain, strain background (Human herpesvirus 1) | KOS | Smith, 1964 | ||
| Strain, strain background (Human herpesvirus 1) | n12 | DeLuca and Schaffer, 1988 | Strain KOS with nonsense mutation near ICP4 N-terminus | |
| Strain, strain background (Human herpesvirus 1) | tsKos | Dremel and DeLuca, 2019 | Strain KOS with temperature sensitive ICP4 | ICP4 has an A474V mutation |
| Antibody | Anti-RNA polymerase II CTD repeat YSPTSPS (phospho S5) Mouse Monoclonal | Abcam | Cat #ab5408 | |
| Antibody | Anti-TATA binding protein Mouse Monoclonal | Abcam | Cat #ab51841 | |
| Antibody | Anti-Sp1 Mouse Monoclonal | Santa Cruz | Cat #sc-17824 | |
| Antibody | Anti-Human Sur-2 (Med23) Mouse Monoclonal | BD Pharmingen | Cat #550429 | |
| Antibody | Anti-Med1 Rabbit Polyclonal | Bethyl Laboratories | Cat #A300-793A | |
| Antibody | Anti-Histone H3 (tri methyl K4) Mouse Monoclonal | Abcam | Cat #ab12209 | |
| Antibody | Anti-Histone H3 (tri methyl K27) antibody Mouse Monoclonal | Abcam | Cat #ab6002 | |
| Antibody | Anti-Histone H3 (acetyl K27) Rabbit Polyclonal | Abcam | Cat #ab4729 | |
| Antibody | Recombinant Anti-Histone H3 (tri methyl K9) Rabbit Monoclonal | Abcam | Cat #ab176916 | |
| Antibody | Anti-Histone H3 antibody Rabbit Polyclonal | Abcam | Cat #ab1791 | |
| Antibody | Anti-ICP4 (58S) Mouse Monoclonal | Neal DeLuca | Hybridomas available from ATCC HB-8183 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.51109.015





