TORC2-Gad8-dependent myosin phosphorylation modulates regulation by calcium
Figures
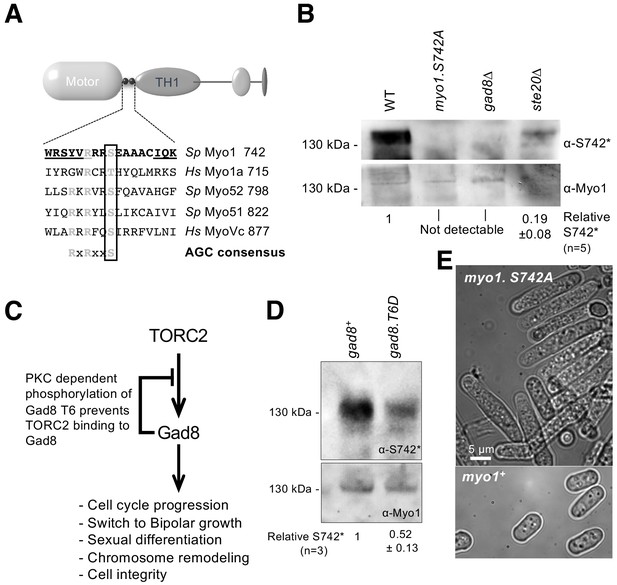
Myo1 serine 742 phosphorylation is TORC2 dependent.
(A) The sequence alignment of myosin IQ regions highlights an AGC kinase consensus sequence that is conserved in class I and V myosins. Underlined residues are those within IQ motifs. (B) Western blots of extracts from myo1+, myo1-S742A, gad8∆ and ste20∆ cells probed with phospho-specific anti-Myo1S742 (upper panel) and anti-Myo1 (lower panel) antibodies demonstrate antigen specificity and a Myo1S742 phosphorylation-state dependence upon the TORC2–Gad8 pathway. Ponceau staining was used to monitor equal loading. Relative Myo1S742 phosphorylation levels were calculated from five independent equivalent experiments (mean ± sd). (C) A schematic of the TORC2–Gad8 signaling pathway. (D) Myo1S742 phosphorylation is reduced in gad8.T6D cells, which have reduced Gad8 kinase activity. Relative Myo1S742 phosphorylation levels were calculated from three independent equivalent experiments (mean ± sd). (E) Nitrogen-starved wildtype (WT) and myo1.S742A cells. In contrast to WT cells, in which growth arrests, myo1.S742A cells continue to grow upon nitrogen-starvation-induced G1 arrest. Scale bar: 5 µm.
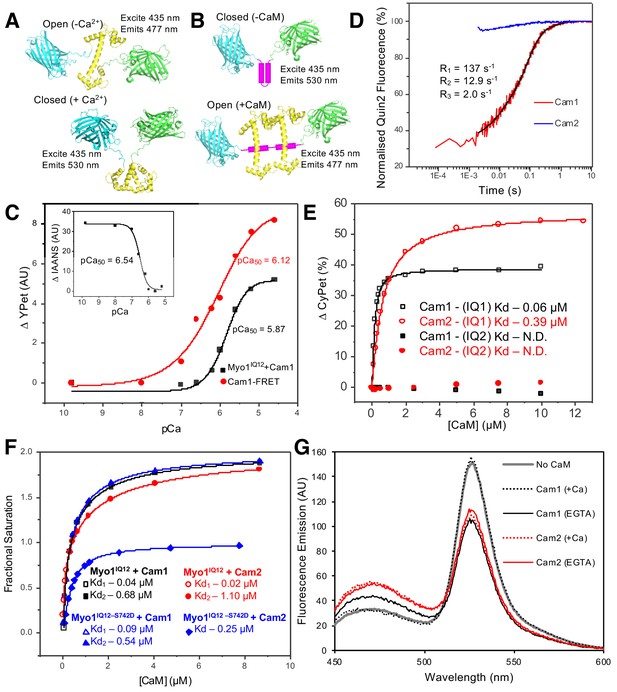
In vitro characterization of interactions between Myo1 and Cam1.
(A) Predicted models of the CyPet–Cam1–YPet FRET reporter protein (Cam1–FRET) in the absence (upper panel) and presence (lower panel) of Ca2+. (B) Predicted models of the CyPet–Myo1IQ12–YPet FRET reporter protein (Myo1IQ12–FRET) in the absence (upper panel) or presence (lower panel) of calmodulin binding (Cypet, cyan; Cam1, yellow; YPet, green; IQ domain, magenta). (C) pCa curve plotting Ca2+-dependent changes in the acceptor fluorescence (plotted as ∆YPet signal) of the Cam1–FRET protein (red), Cam1 association with Myo1IQ12-FRET (black) and change in the fluorescence of IAANS-labelled Cam1–T6C (inset). (D) Transient curves of changes in Quin2 fluorescence induced by Ca2+ release from Cam1 (red) (with three exponential fit best fit (black)) and from Cam2 (blue) illustrate that only Cam1 associates with Ca2+. (E) Curves plotting Cam1- (black) and Cam2-dependent (red) changes of the FRET donor signal of Myo1–FRET proteins containing single IQ domains (IQ1, empty shapes; IQ2, filled shapes) each show that CaM associates with IQ1 but not with an equivalent single IQ2 motif region. (F) Curves plotting Cam1- (squares and triangles) and Cam2-dependent (circles and diamonds) changes in the FRET donor signal of either 0.5 µM wild type (black and red) or S742D phosphomimetic (blue) Myo1IQ12–FRET proteins show that although phosphorylation does not significantly impact Cam1 binding, it results in a drop of about 50% in Cam2 interaction. (G) Spectra of 0.5 µM Myo1IQ12–FRET reporter alone (grey line) or of 0.5 µM Myo1IQ12–FRET reporter mixed with 10 µM saturating concentrations of: Cam1 + Ca2+ (black dotted line), Cam1 – Ca2+ (black solid line), Cam2 + Ca2+ (red dotted line), or Cam1 – Ca2+ (red solid line).
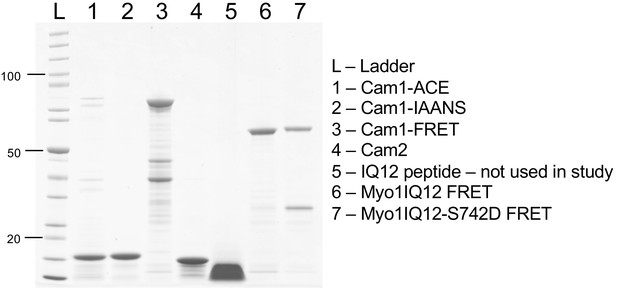
Purified proteins used during in vitro studies.
Coomassie-stained SDS-PAGE gel of recombinant proteins expressed and purified during this study. From left to right, the lanes contain (L) protein standard; (1) Nt-acetylated Cam1; (2) Nt-acetylated Cam1-T6C; (3) Cam1-FRET; (4) Cam2; (5) IQ12 peptide (not used during this study); (6) Myo1IQ12-FRET; and (7) Myo1IQ12S742D-FRET.
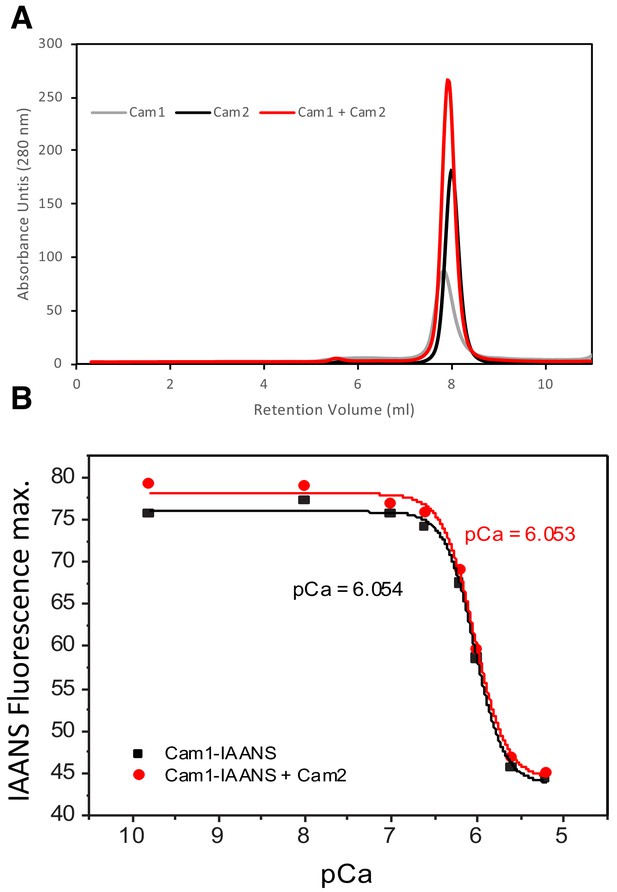
Cam1 and Cam2 do not interact directly.
(A) Overlaid OD280 spectra of eluate from a Superdex 75 gel filtration column that had been loaded withCam1 (gray line), Cam2 (black line) or both Cam1 and Cam2 (red line) under identical 4 mM EGTA buffer conditions. (B) Maximum IAANS fluorescence values (440 nm) of 0.5 μM Cam1-IAANS over a range of pCa values. Black symbols show values for Cam1-IAANS, red symbols show values for Cam1-IAANS with 5 μM Cam2 protein. 2 mM Ca- EGTA buffers were used to give the indicated pCa values. pCa50 values were calculated from Origin fitting analysis - Hill equation.
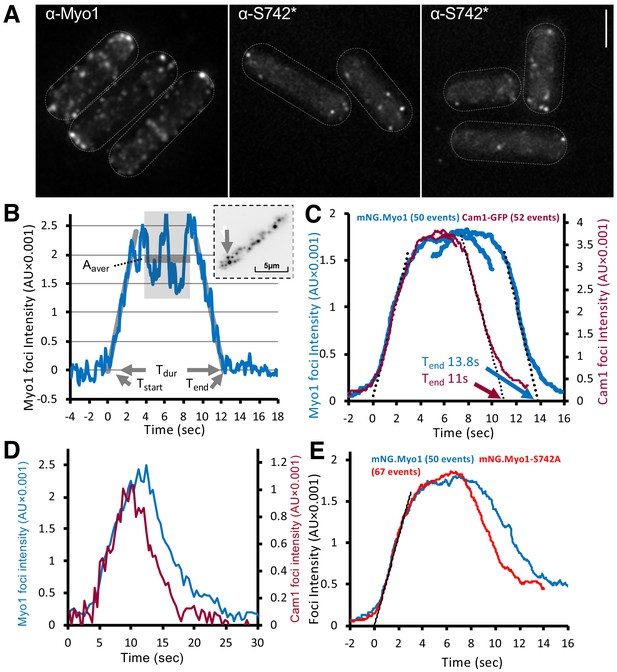
Myo1 and Cam1 dynamics in wild-type and myo1.S742A cells.
(A) Maximum projections from 31-z stack widefield immunofluorescence images of wild-type cells, probed with anti-Myo1 (left panel) or anti-Myo1S742* phosphospecific (right panels) antibodies, illustrate that S742 phosphorylated Myo1 localizes to cortical foci (scale bar, 5 µm). (B) An example relative-intensity trace of a single mNeongreen.Myo1 endocytic event. Linear fitting (grey lines, 60 points) was used to find the maximum gradient for both the rising and the falling slope. The intercept with zero intensity level was used to calculate Tstart, Tend, and subsequently, the duration of the event Tdur. See detailed description in the Materials and methods section. Insert: an arrow highlights the analyzed endocytotic event (5 × 5 pixels area). (C) Averaged profile for individual Myo1 (blue) and Cam1 (red) membrane-association events, synchronized relative to Tstart and Tend. Dotted lines show fitted rising (Myo1, 537 AU/sec; Cam1, 1073 AU/sec) and falling (Myo1, 567 AU/sec; Cam1, 1028 AU/sec) gradients. (D) An example fluorescence trace from simultaneous two-color imaging of a Myo1 (blue line) and Cam1 (red line) membrane-association event observed in mNeongreen.myo1 cam1.mCherry cells is consistent with the relative intensities and timings observed using single-fluorophore strains. (E) Averaged intensity trajectories of individual Myo1 (blue line) and Myo1.S742A (red line) endocytosis events from TIRFM imaging of mNeongreen.myo1 and mNeongreen.myo1.S742A cells, respectively.
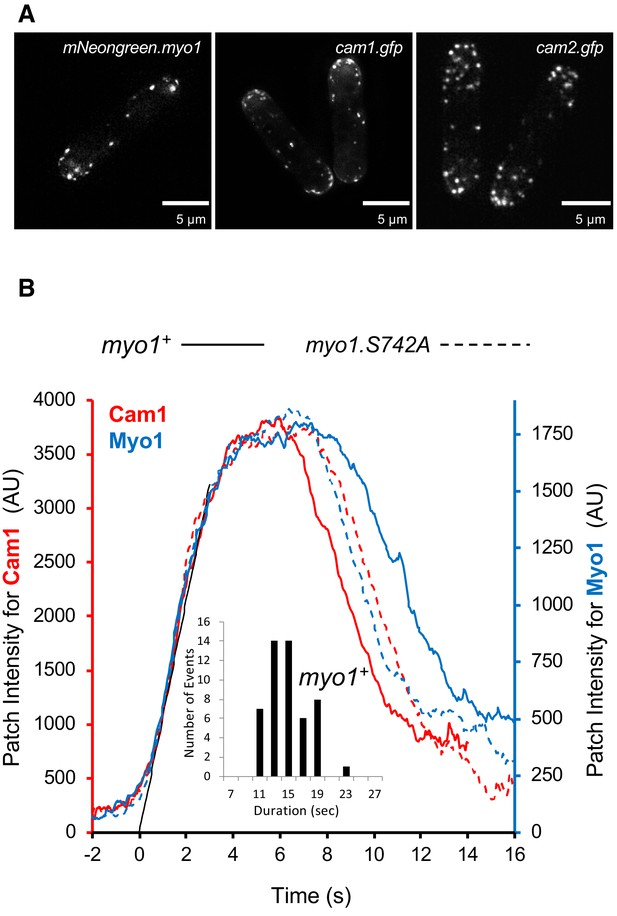
Relative TIRF profiles.
(A) Maximum projections of 31-z stack widefield images of mNG.myo1, cam1.gfp and cam2.gfp cells (scales bars, 5 µm). (B) Combined profiles of averaged intensity trajectories of Myo1 (blue) and Cam1 (red) endocytosis events in wild-type (solid lines) or myo1.S742A strains (dashed lines). See Materials and methods section for the description of TIRF imaging. Insert: the distribution of the durations of individual wild-type Myo1 events.
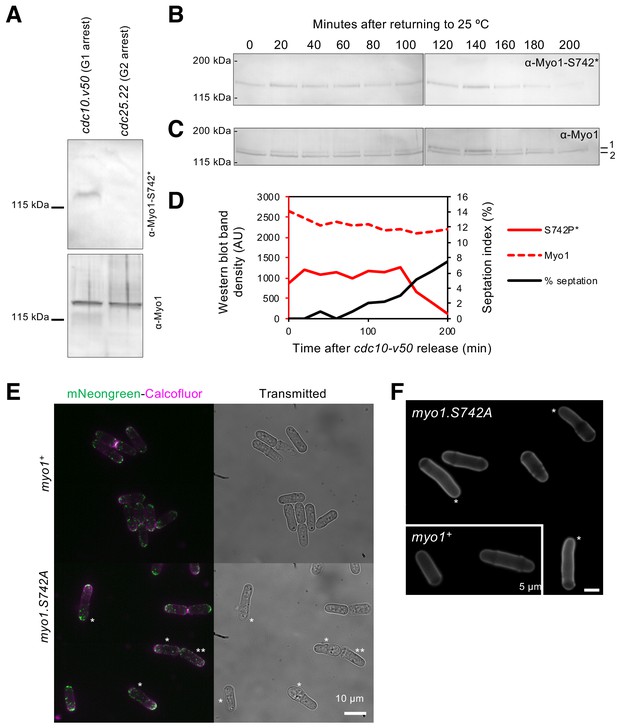
Myo1 S742 is phosphorylated in a cell-cycle-dependent manner to affect polarized growth.
(A) Western blots of extracts from G1-arrested cdc10.v50 cells and pre-mitotic G2-arrested cdc25.22 cells probed with phospho-specific anti-Myo1S742 (upper panel) and anti-Myo1 (lower panel) antibodies demonstrate that Myo1 S742 phosphorylation occurs before the Cdc10 execution point in monopolar G1 cells, and is not detectable by the Cdc25 execution point at the end of G2 (n = 3). (B–D) A cdc10.v50 culture was synchronized in G1 by shifting to 36°C for 240 min before returning to 25°C at time 0. Samples of cells were taken every 20 min from the release and processed for western blotting to monitor Myo1S742 phosphorylation. The membrane was first probed with phosphospecific anti-Myo1S742* antibodies (B), and subsequently probed with anti-Myo1 antibodies to monitor total Myo1 (C). Both phosphorylated (1) and non-phosphorylated (2) Myo1 bands can be observed in panel (C). Equal loading was monitored by Ponceau staining of the membrane. (D) Densitometry measurements of phosphorylated Myo1S742 (from panel (B)) and total Myo1 (both bands from panel (C)) are plotted along with the % of cells in the culture with septa. (E) Myosin-1 distribution (green), calcofluor-stained regions of cell growth (magenta), and cell outline (transmitted image) of prototroph mNeongreen.myo1+ and mNeongreen.myo1.S742A cells cultured in EMMG medium at 34°C. Asterisks highlight cells that have morphology defects. Scale bar, 10 µm. (F) Calcofluor-stained WT and myo1.S742A cells. Asterisks highlight long bent cells displaying monopolar growth. Scale bar, 5 µm.
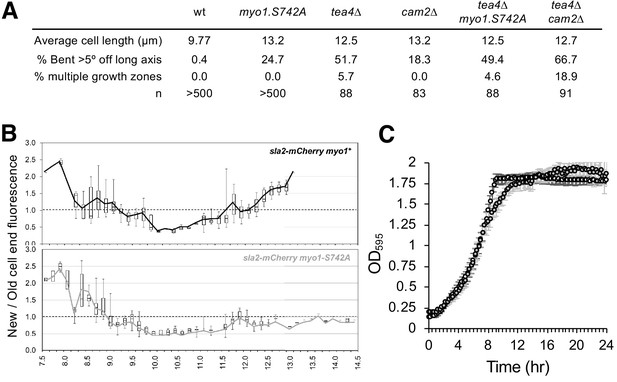
Myo1S742 phosphorylation impacts polarized cell growth.
(A) Average length and frequency of growth defects in WT, myo1.S742A, tea4∆, cam2∆, tea4∆ myo1.S742A, and tea4∆ cam2∆ cells. (B) Ratio of Sla2-mCherry fluorescence at ‘new’:‘old’ cell ends, averaged from >30 growing mid-log sla2-mCherry myo1+ (upper panel) and sla2-mCherry myo1.S742A (lower panel) cells. Boxes plots show the median and quartile for each length measured, whereas the lines are plots of the mean values at each length measured. (C) Averaged growth curves from three independent experiments of prototroph wild-type (empty circles) and myo1.S742A (gray-filled circles) cells cultured in EMMG medium at 34°C. Slower growth is apparent at the end of log phase in myo1.S742A cells, which grow more until reaching the stationary phase. Error bars denote the s.d. of the mean.
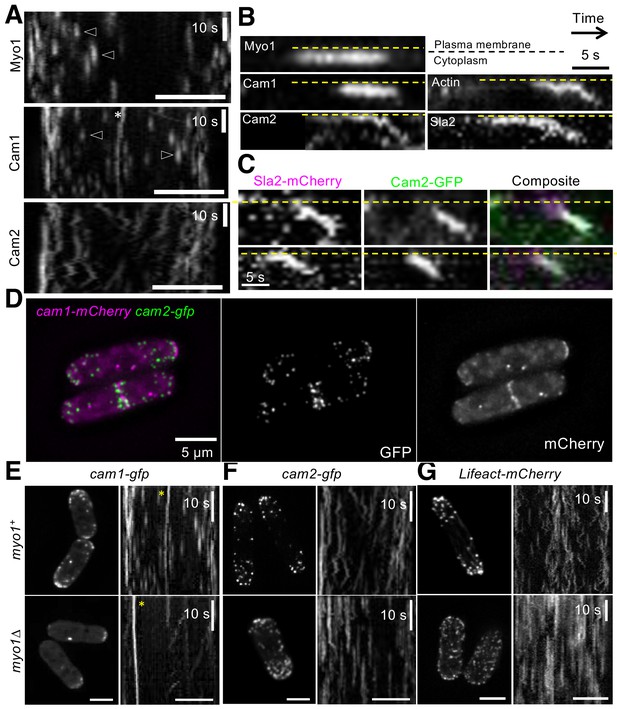
Cam2 associates with internalized endocytic vesicles.
(A) Kymographs of GFP-labelled foci from maximum projections of 13-z plane time-lapse images of mNeongreen.myo1 (upper panel), cam1.gfp (middle panel) and cam2.gfp (bottom panel) cells illustrate the static nature of Myo1 and Cam1 endocytic foci when associated with the plasma membrane (arrowheads). Cam1 foci that are associated with a spindle pole body (SPB) are highlighted. By contrast, Cam2 foci displayed extensive lateral movements. (B) Kymographs generated from single z-plane time-lapse images of single endocytic foci surfaces during vesicle formation and subsequent internalization. Myo1 and Cam1 only associate with the plasma membrane, whereas Cam2, Sla2 and actin are internalized on the vesicle after scission. These kymographs are not aligned temporally. (C) Kymographs of Cam2 and Sla2 co-internalization in sla2.mCherry cam2.gfp cells. (D) Maximum projection of a 31-z slice image of cam1.mCherry cam2.gfp cells reveals that Cam1 (magenta) and Cam2 (green) colocalize in a subset of endocytic foci. (E–G) Single frames (left panels) and kymographs (right panels) from maximum projections of 13-z plane time-lapse images of cam1.gfp (E), cam2.gfp (F) and LifeACT.mCherry (G) in either myo1+ (upper panels) or myo1∆ (lower panels) cells. These images show that although only Cam1 recruitment to endocytic foci is dependent upon Myo1, the myosin is required for the internalization of Cam2-GFP and LifeACT.mCherry foci. Scale bar, 5 µm.
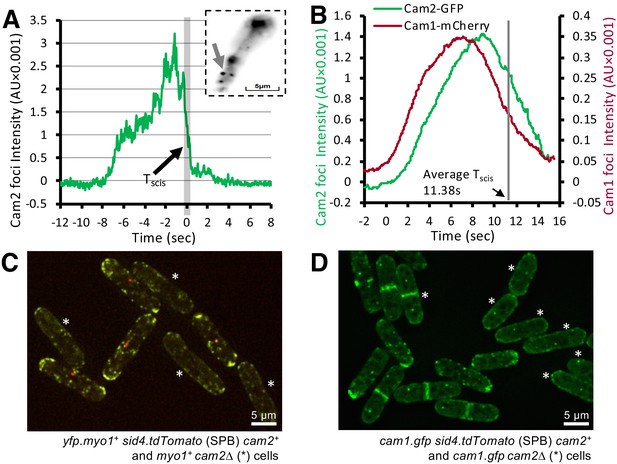
Cam2 does not impact Myo1 or Cam1 dynamics in vegetative cells.
(A) An example fluorescence trace of a Cam2 membrane binding and vesicle internalization event from TIRFM imaging of cam2.gfp cells. An abrupt drop in the fluorescence was marked as ‘scission time’ (Tscis, gray vertical line). Insert: an arrow shows the location of the monitored endocytic event (5 × 5 pixels area). (B) Averaged profile from 65 individual Cam2 membrane association events (green line), together with the averaged Cam1-mCherry profile (red) from two-color TIRFM imaging of cam1.mCherry cam2.gfp cells. The events were synchronized relative to the Cam1 Tstart. The gray line denotes the mean time of vesicle scission (Tscis). See detailed description in the Materials and methods section. (C) Maximum projection of a 31-z slice widefield image of a mixture of yfp.myo1 sid4.tdTomato (WT, with a red labeled SPB marker) and yfp.myo1 cam2∆ (asterisks) cells. Red-labeled SPBs allow differentiation between cam2+ and cam2∆ cells in the same field. (D) Maximum projection of a 31-z slice widefield image of a mixture of prototroph cam1.gfp sid4.tdTomato (WT, with a red-labeled SPB marker) and cam1.gfp cam2∆ cells (asterisks). Red-labeled SPBs allow differentiation between cam2+ and cam2∆ cells in the same field. Scale bars, 5 µm.
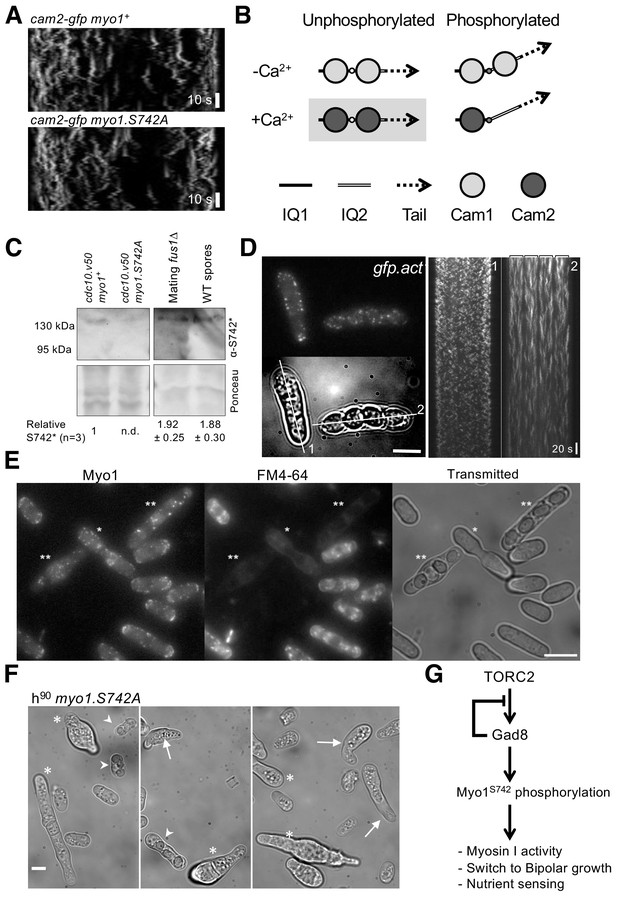
Myo1 S742 phosphorylation regulated Cam1 and Cam2 dynamics during meiosis.
(A) Kymographs of Cam2.GFP foci dynamics in myo1+ (upper panel) and myo1.S742A (lower panel) cells. (B) Scheme of the consequences of phosphorylation of Myo1Ser742 (small empty circle) and Ca2+ levels upon the binding of Cam1 (light gray filled circle) and Cam2 (dark gray filled circle) to the IQ1 (solid thick black line) and IQ2 (double line) motifs of Myo1, and the impact on the relative orientation of the myosin lever arm (dashed arrow). The highlighted combination of unphosphorylated Myo1S742 and Ca2+ does not normally occur in cells. (C) Western blots of extracts from G1-arrested cdc10.v50 myo1+, cdc10.v50 myo1-S742A, and conjugation arrested (starved, premeiotic cells) fus1∆ cells, and from meiotic spores, probed with phospho-specific anti-Myo1S742 antibodies (upper panel). Ponceau staining (lower panel) confirms that Myo1S742 remains phosphorylated from the end of G1, through conjugation until the end of meiosis (n = 5). Relative Myo1S742 phosphorylation levels were calculated from three independent equivalent experiments (mean ± sd). (D) Left panel: maximum projection of a 13-z slice GFP fluorescence image (top) and a transmitted light image (bottom) from a time-lapse of vegetative (cell 1) and meiotic (cell 2) gfp-act1 cells. Image from a GFP-act signal. The kymographs in the right panels were generated along the two dotted axes. (E) Maximum projection of mNeongreen-Myo1 fluorescence (left), FM4-64 fluoresence (middle) and transmitted light images of a mixed population of vegetative, fusing (*) and sporulating (**) mNG-myo1+ cells, illustrating that endocytosis is reduced in meiotic cells. Scale bar, 10 µm. (F) Micrographs illustrating myo1.S742A cell morphology on solid starvation medium. Asterisks highlight cells with unregulated growth and polarity defects; arrows highlight cells with elongated or abnormally bent shmooing (conjugation) tips; arrow heads highlight meiotic cells with defective spore formation. Scale bar, 5 µm. (G) A schematic of the TORC2–Gad8–Myo1S742 signaling pathway.
Videos
Single-molecule Myo1 membrane-association events.
TIRFM imaging of mNeongreen.myo1 cell showing rapid single-molecule interactions of Myo1 with the plasma membrane, which are apparent as small bright green spots of a diffraction limited size that are visible over background from the camera noise (highlighted by tracking white lines). These single myosin1 molecules have limited residency time (off rate 7.8 s−1) and mobility (0.03 µm2.s−1) at the plasma membrane. 63 frames per second (fps) @ 23°C. The plot at the end of the Video was constructed from the analysis of a 50 s full-length video.
Endocytic Myo1 events.
TIRFM imaging of mNeongreen.myo1 cells showing endocytosis-associated interactions of Myo1 at the plasma membrane. The myosin1 quickly accumulates at the site of endocytosis, and within 5–8 s of reaching a maximum, rapidly leaves the endocytic site. The accumulated myosin remains immobile on the membrane for the duration of the event. 20 fps @ 23°C.
Endocytic Cam2 events.
TIRFM imaging of a cam2.gfp cell showing Cam2 recruitment to endocytic vesicles, to which it remains associated after scission and internalization of the endosome. At the start of each event, each spot is immobile, but at the end of the endocytic event, the vesicle oscillates as it is internalized into the cytoplasm. 20 fps @ 23°C.
Comparison of Cam1 and Cam2 dynamics.
TIRFM imaging of a cam1.mCherry cam2.gfp cell showing early recruitment of Cam1 (red) and subsequent recruitment of Cam2 (green) to the sites of endocytosis. Cam1 disassociates prior to vesicle scission, while Cam2 remains associated with the internalized endosome, so that at the beginning of the endocytic event, the spots are red. During the event, they become yellow as Cam2 (green) is recruited to the vesicle. At the end of the event, the vesicle becomes green as Cam1 dissociates from the endocytotic site. 20 fps @ 23°C.
Myo1 dynamics in interphase and meiotic cells.
Time-lapse of maximum projections from 13-z slice widefield images of mNeongreen.myo1 cells showing typical examples of Myo1 dynamics in vegetative and meiotic (highlighted with arrow) cells. Frame rate: 650 msec/frame.
Cam1 dynamics in interphase and meiotic cells.
Time-lapse of maximum projections from 13-z slice widefield images of cam1.gfp cells showing typical examples of Cam1 dynamics in vegetative and meiotic (highlighted with arrow) cells. Frame rate: 650 msec/frame.
Cam2 dynamics in interphase and meiotic cells.
Time-lapse of maximum projections from 13-z slice widefield images of cam2.gfp cells showing typical examples of Cam2 dynamics in vegetative and meiotic (highlighted with arrow) cells. Frame rate: 650 msec/frame.
Act1 dynamics in interphase and meiotic cells.
Time-lapse of maximum projections from 13-z slice widefield images of gfp.act1 cells showing typical examples of Act1 dynamics in vegetative and meiotic (highlighted with arrow) cells. Frame rate: 650 msec/frame.
Additional files
-
Supplementary file 1
Strains used during this study.
- https://doi.org/10.7554/eLife.51150.022
-
Supplementary file 2
Oligonucleotides used during this study.
- https://doi.org/10.7554/eLife.51150.023
-
Transparent reporting form
- https://doi.org/10.7554/eLife.51150.024





