Structure of a mitochondrial ATP synthase with bound native cardiolipin
Figures
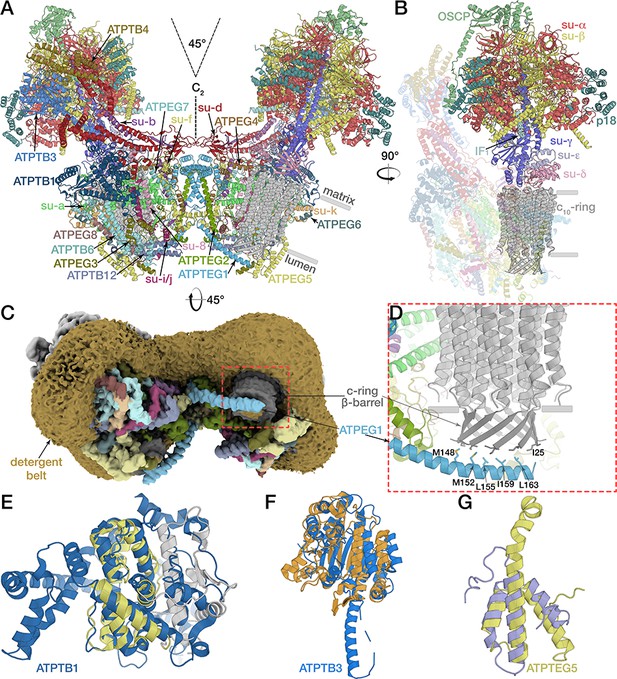
Structure of the E. gracilis ATP synthase dimer.
(A) Atomic model of the complete E. gracilis ATP synthase dimer with both subcomplexes in rotational state-1. The 2-MDa dimeric F1Fo-complex contains 29 different proteins. Dashed lines indicate C2-symmetry axis and 45° dimer angle. (B) OSCP/F1/c-ring subcomplex in rotational state-1, bound to its natural inhibitor protein IF1 (cyan), remaining Fo transparent. (C) Density map showing the lumen-exposed Fo region. Detergent belt shown in yellow; c-ring β-barrel in dark grey, Fo subunits as in (A). (D) Close-up of the lumenal interface of ATPEG1 (blue) with the c-ring (grey). The interaction occurs mostly via hydrophobic interactions (blue and grey sticks). (E–G) Euglenozoa-specific Fo-subunits with known folds. (E) ATPTB1 in blue superposed with Mdm38 (PDB ID: 3SKQ) (Lupo et al., 2011), six conserved helices coloured yellow, rest grey. (F) ATPTB3 in blue superposed with a bacterial homoisocitrate dehydrogenase in orange (PDB ID: 4YB4)(Takahashi et al., 2016), adopts a Rossman-fold. (G) ATPEG5 in yellow is a structurally conserved ortholog of the cytochrome c oxidase subunit VIb superfamily; bovine subunit VIb in purple (PDB ID: 2Y69) (Kaila et al., 2011).
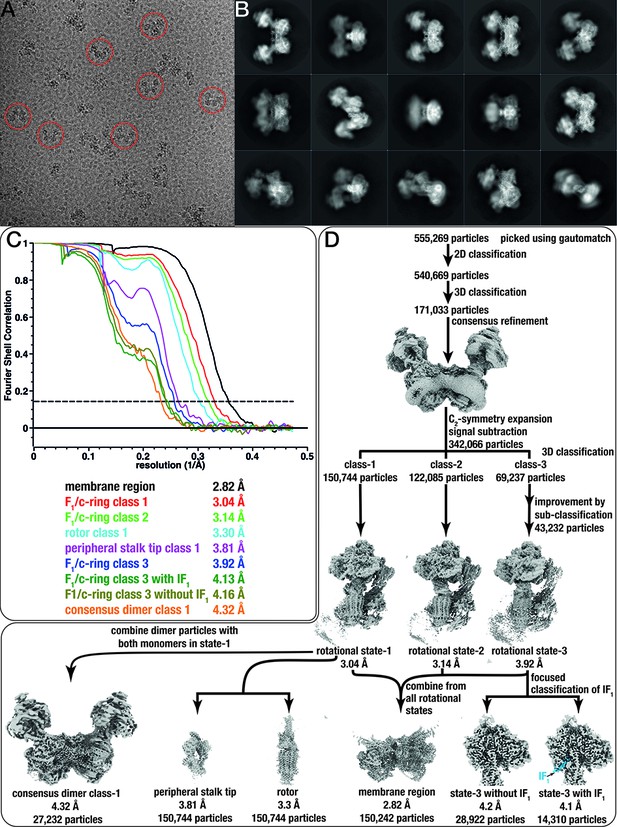
Cryo-EM data processing and classification scheme.
(A) Representative micrograph. Scale bar 500 nm. (B) Representative 2D classes. (C) Fourier Shell Correlation (FSC) plots of all seven maps shown in D, with resolutions calculated according to the 0.143-cutoff criterion. (D) Cryo-EM data processing scheme. Final maps include the membrane region (from all rotational states), F1/c-ring maps from rotational states 1–3, the rotor and peripheral stalk tip (both from rotational state-1), as well as a consensus map of the ATP synthase dimer with both F1/c-ring subcomplexes in rotational state-1. Rotational state-3 displayed partial occupancy of IF1 and was separated into two classes with or without bound inhibitor using focused classification.
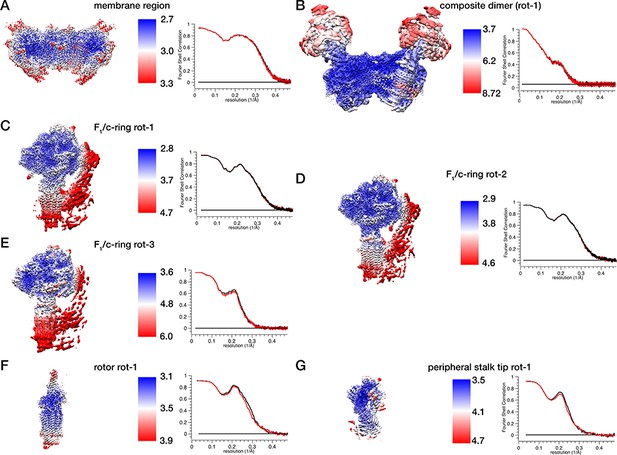
Local resolution estimation and model-map-correlations.
(A–G) Density maps of masked regions of the ATP synthase coloured according to local resolution (left panels, local resolution as highlighted in color bars in Å) with Model-Map-FSC curves of respective atomic model refined into one half map to give FSCwork (black) and FSCtest (red)(right panels). (A) Membrane region, (B) consensus map of complete ATPS synthase dimer with both monomers in rotational state-1. (C–E) F1/c-ring maps of rotational states 1–3, (F) rotor in rotational state-1, (G) peripheral stalk tip of rotational state-1.
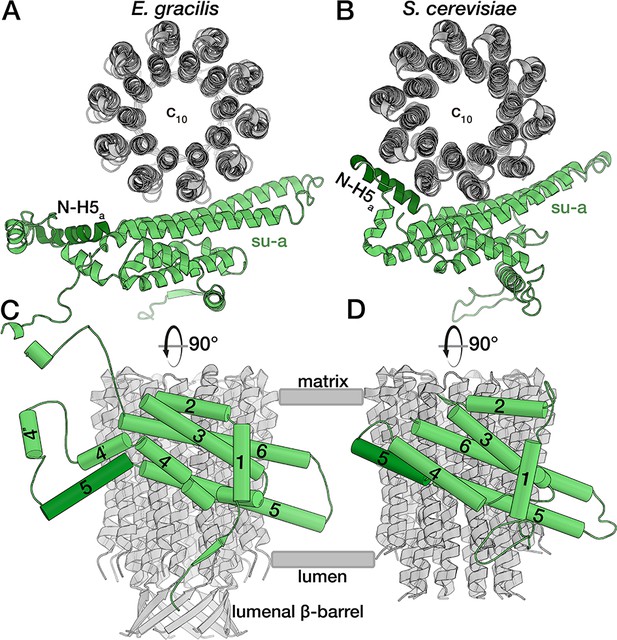
E. gracilis subunit a fold comparison.
Top view (upper panel) and side view (lower panel) of the E. gracilis (left) and S. cerevisiae (right) (Guo et al., 2017) subunit a (green) and c-ring (grey). Both structures contain the conserved H1-6a, with E. gracilis displaying two helices (H4’a and H4’’a) in an extension segment and a C-terminal extension. Whereas the N-terminal region of H5a (dark green) is kinked towards the c-ring in the yeast complex, it extends towards the lumen in the E. gracilis structure, thereby diminishing its interface with the c-ring. Unlike its yeast homolog, the N-terminus of E. gracilis subunit a is not involved in dimerisation, but contributes a strand to a β-sheet along the lumenal side of the detergent micelle.
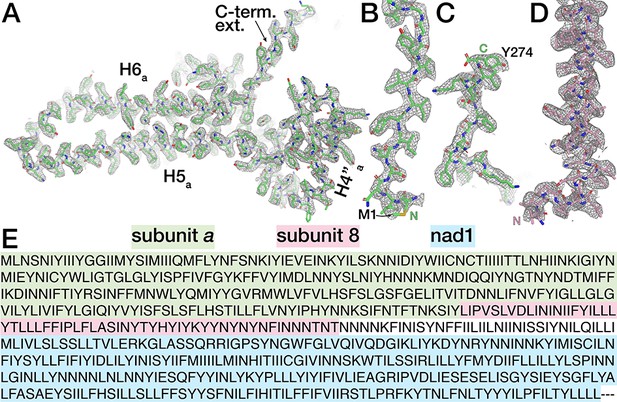
Identification of subunits a and 8.
(A-D) Map quality and model fit of E.gracilis subunit a. H4’’a, H5 a and H6 a (A), subunit a N-terminus (B), C-terminus (C), and subunit 8 N-terminus and transmembrane helix (D) are shown with map density as mesh. (E) Sequence of the translated E. gracilis genome contig containing the newly identified subunit a (green), subunit 8 (pink), as well as NADH dehydrogenase subunit 1 (nad1, blue) in a single predicted open reading frame (contig_684056 of PRJEB27422, E. gracilis Genome Project). An identical consensus contig was obtained by assembling sequence reads of the E. gracilis mitochondrial genome (PRJNA294935)(Dobáková et al., 2015), indicating that subunit a and subunit 8 are mitochondrially encoded.
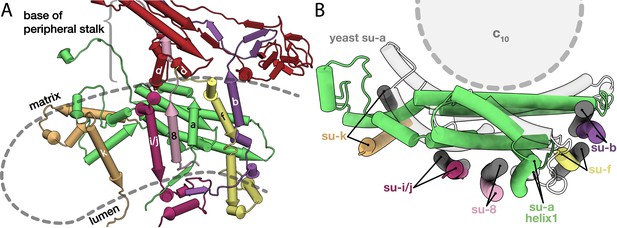
Conserved subunits of the Fo region.
(A) Side view of the conserved E. gracilis Fo subunits. Transmembrane helices with structural equivalents in yeast are labelled. (B) Top view of the superimposed conserved Fo subcomplexes from E. gracilis (coloured) and yeast (grey) PDB ID: 6B2Z (Guo et al., 2017). Although subunit k does not superimpose well, it occupies the same position relative to the H5a.
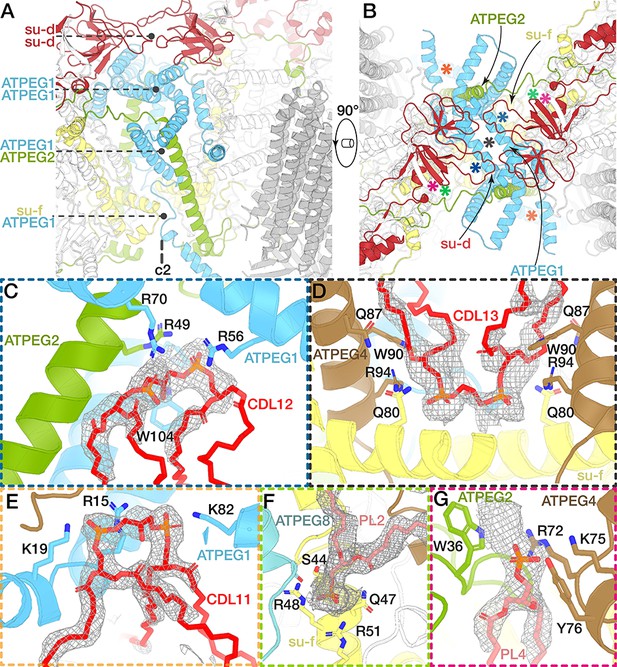
The dimer interface and associated lipids.
(A and B) Views of the dimer interface along (A) and perpendicular (B) to the membrane plane. The dimerisation motifs (interacting subunits coloured) are stacked along the C2-symmetry axis and formed by two copies of subunit d (red) and ATPEG1 (blue), which interacts with its symmetry-related copy, as well as ATPEG2 (green) and subunit f (yellow). Asterisks in (B) indicate positions of lipid-binding sites. (C to G) Close-ups of the lipid-binding sites indicated in (B). Bound lipids at the dimer interface identified as cardiolipin (CDL; C to E) or phospholipids modelled as phosphatidic acid (PL; F and G). Interacting residues (subunits coloured) include at least one arginine residue. Density shown as grey mesh.
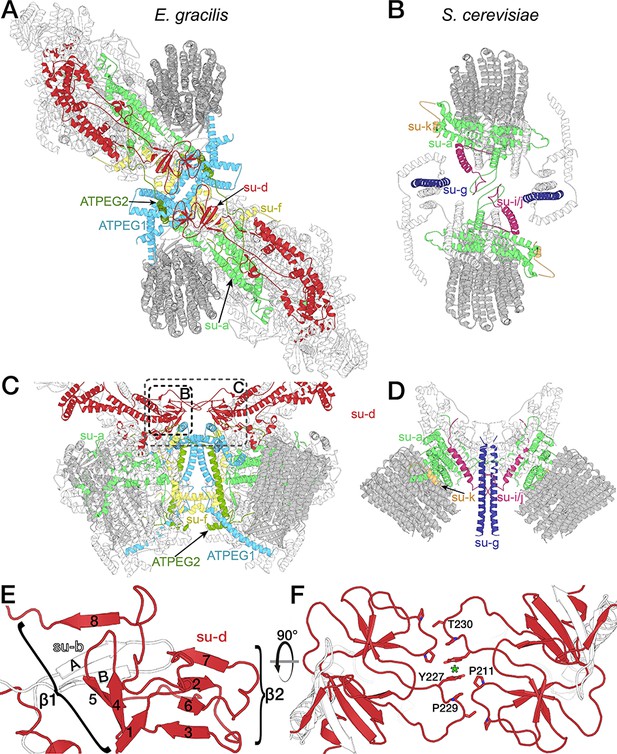
Architecture and dimer interface comparison between the yeast and E. gracilis ATP synthase.
The E. gracilis (A,C) and yeast (B,D) Fo subcomplexes display different architectures. (A,C) The E. gracilis complex is held together by subunit d (red), subunit f (yellow), ATPEG-1 (light blue) and ATPEG2 (dark green). The subunit a/c-ring subcomplexes (light green and dark grey respectively) are offset along the along the Fo long axis. (B,D) The yeast ATP synthase dimer (PDB ID: 6B2Z) (Guo et al., 2017) is held together by subunit a (light green), subunit i/j, as well as subunit k (orange) and subunit g (dark blue, interaction not modelled). (E-F) Subunit d extension contributes to the dimer interface and peripheral stalk. (E) Subunit d (red) and subunit b (white) together form a ten-stranded all-β-fold at the dimer interface on the matrix side that contributes to the dimer interface. The smaller β-sheet (β2) is formed by subunit d, which adopts a ferredoxin-like fold containing a β-hairpin insertion (strands 4 an 5), which is part of the larger β-sheet (β1) that contains two β-strands (labelled A and B) of subunit b. (F) Top view of the dimer interface formed by subunit d, with interacting residues shown; green asterisk marks C2-symmetry axis.
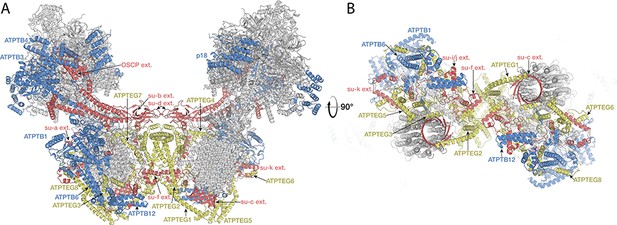
Species-specific subunits and extensions form the dimer interface.
The E. gracilis ATP synthase dimer side view (A) and bottom view (B). Phylum-specific subunits previously identified in T. brucei (ATPTB1, ATPTB3, ATPTB4, ATPTB6, ATPTB12, p18) shown in blue. Species-specific subunits with no detectable homologs outside euglenoids (ATPEG1-8) are shown in yellow. Conserved subunits with homologs or close structural equivalents in yeast are shown in grey, with species-specific extensions highlighted in red.
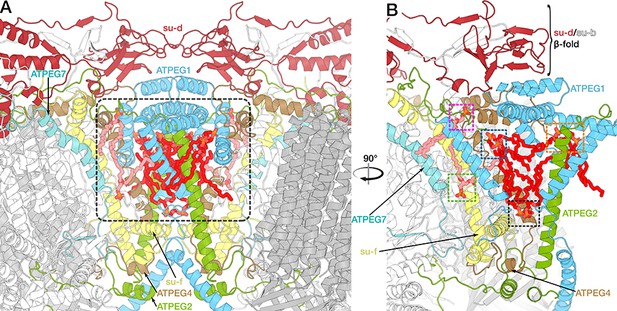
Bound lipids of the dimer interface.
Side view (A) and cut view (B) of the dimer interface with central cavity (grey rectangle in (A)) containing bound lipids (cardiolipin red, unidentified phospholipids light pink). Subunits involved in dimerization or lipid binding are highlighted. (B) View of ATP synthase monomer cut along a plane though the C2-symmetry axis. Coloured rectangles highlight lipid binding sites at the dimer interface as shown in Figure 4C to G.
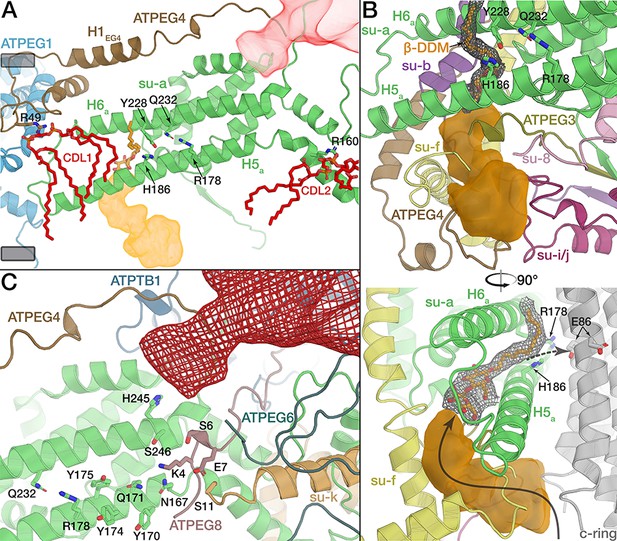
The rotor-stator interface is flanked by bound cardiolipin.
(A) View from the c-ring towards the membrane-embedded stator subunits. H5a and H6a are augmented by the tilted, amphipathic H1EG4 (brown). Cardiolipin molecules flanking subunit a are shown in red (tails of acyl chains are mostly disordered and shown only for illustration). Proton half-channels on the lumen and matrix side are shown in orange and red respectively. Remaining subunits not shown for clarity. The conserved R178 and the H186 at the lumen channel exit are shown with interacting residues. (B) Entrance of the lumenal channel (orange) is lined by the termini of subunit f, ATPEG4, subunit 8, ATPEG3, as well as a lumenal segment of subunit i/j. Inside the Fo, the lumen channel is confined by transmembrane helices of subunits f and b. β-DDM occupying the exit of the lumen channel shown in orange with density map as mesh, c-ring in grey. Arrows indicate proposed path of proton flow. (C) Polar and protonatable residues between R178 and the matrix-side half channel (red mesh). Subunit k contributes a horizontal helix (H1k) to the rotor-stator interface.
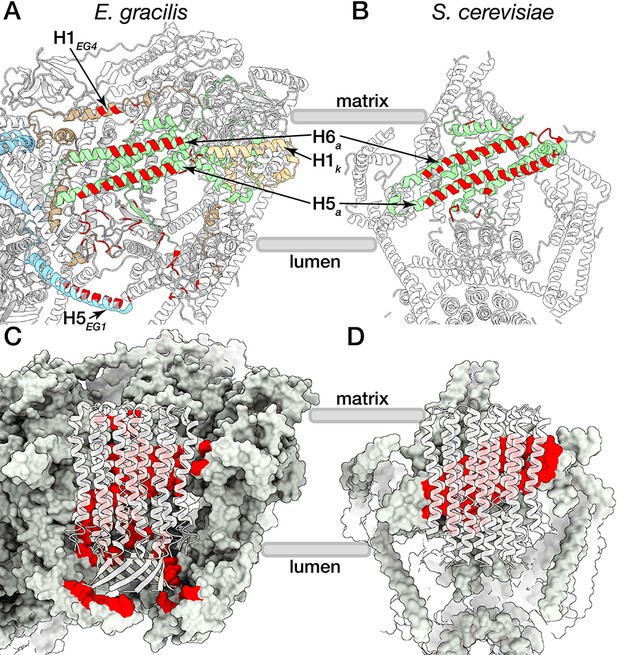
Comparison between E. gracilis and S. cerevisiae rotor-stator interfaces.
The E. gracilis ATP synthase (A,C) displays a larger rotor-stator interface than S. cerevisiae (B,D). (B) In yeast, the interactions (red) with the c-ring are mainly formed by the horizontal H5a and H6a in the membrane (green). (A) In E. gracilis, four horizontal helices (H5a, H6a, H1EG4, H1k) are found at the rotor-stator interface, with the H5EG1 forming a lumenal c-ring interaction. (C,D) Interactions between the Fo stator (grey) and the c-ring (white transparent) are highlighted in red.
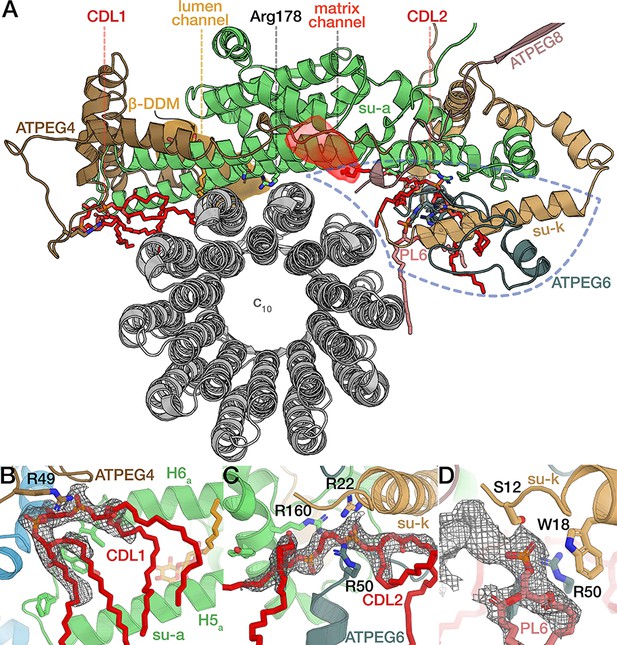
Native phospholipids of the rotor-stator interface.
Associated native phospholipids of the E. gracilis rotor-stator interface. (A) Top view of the rotor-stator interface. The offset half-channels are separated by the central R178 (lumenal channel with β-DDM shown in orange; internal end of matrix channel shown in red). The half channels are flanked on either side by a cardiolipin (shown in red). Species-specific structural elements constituting the rotor-stator interface near the matrix channel enclosed by dashed blue line. (F-H) Cardiolipins (B,C) and unidentified phospholipid (D) of the rotor-stator interface (map density shown in grey mesh).
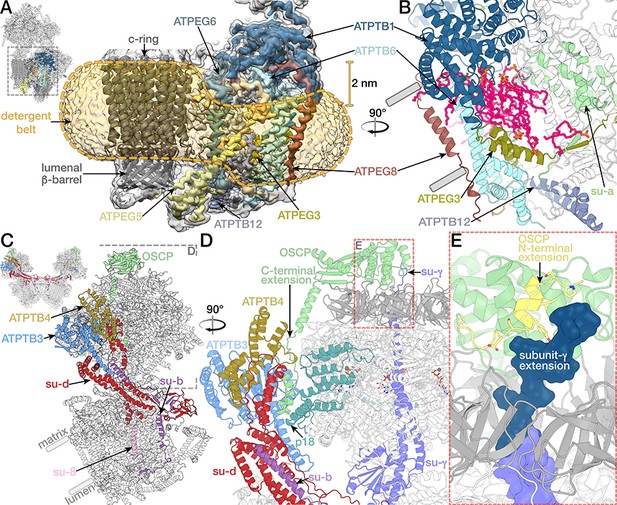
Peripheral Fo subcomplex and the peripheral stalk.
(A) Euglenozoa-specific subunits form a peripheral Fo subcomplex. Density of the Fo with proteins of the peripheral region coloured, c-ring model shown in grey, outline of the detergent belt (yellow dashed lines) with 2 nm offset towards the lumen indicated as determined by the density (transparent gold) (B) Atomic model of the Fo periphery, cavity lipids are shown in magenta. (C to E) Attachment of the peripheral stalk to F1. (C) E. gracilis ATP synthase with proteins constituting the peripheral stalk coloured. (D) Side view of the peripheral stalk tip and F1 (white, crown domain light grey). C-terminal helix of OSCP (light green) extending towards the membrane attaches OSCP to the rest of the peripheral stalk via subunit d (red). (E) The N-terminal extension of OSCP (yellow) interacts with the C-terminal extension of the rotor subunit γ (conserved region purple, extension dark blue).
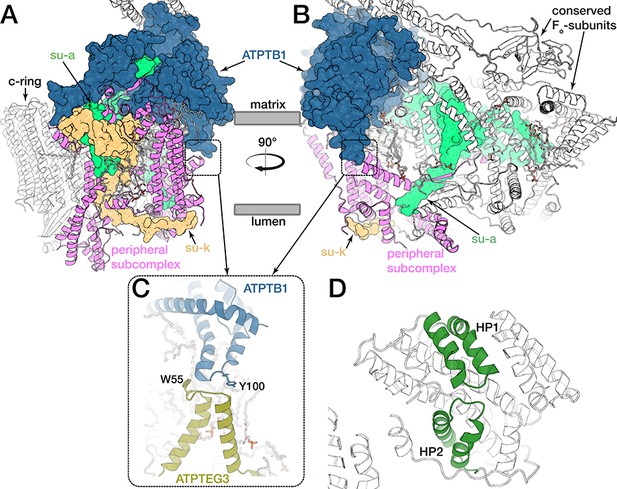
The peripheral Fo subcomplex and inverted topology structural motif.
Side view (A) and front view (B) of the Fo region. The peripheral subcomplex (pink) and the conserved Fo-subunits (white) are joined together by ATPTB1 on the matrix side and by subunits a and k in the lumen and separated by the lipid-filled Fo cavity (B). (C) Hairpin helices of ATPTB1 and ATPEG3 extend into the membrane region from opposite sides in the membrane, where they interact, forming an inverted topology motif. (D) Inverted topology motif of the prokaryotic aspartate transporter GltPh consisting of two hairpin loops (HP1 and HP2, PDB ID: 1XFH) (Yernool et al., 2004).
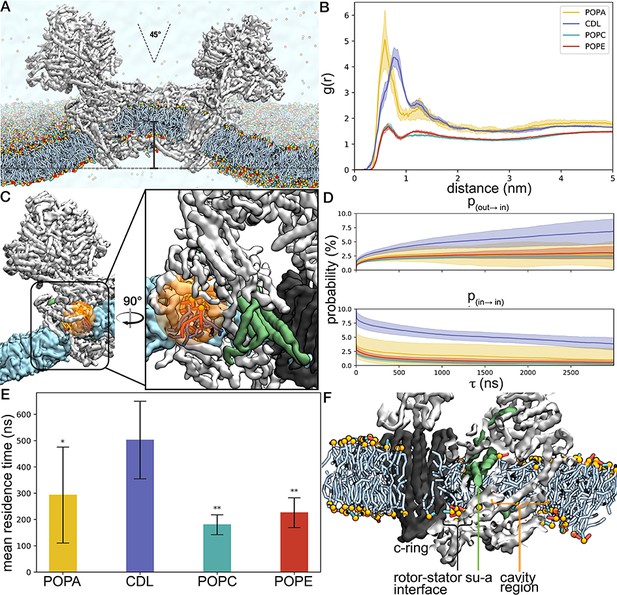
Coarse-grained molecular dynamics simulations of the E. gracilis ATP synthase dimer.
(A) The 45°-angle of the E. gracilis ATP synthase dimer (grey) generates significant membrane curvature (acyl chains in light blue; phosphate groups in orange, ethanolamine groups in red, choline groups in green), resulting in a 7 nm local displacement of the membrane from the bilayer plane (marked with bar). (B) Radial distribution function g (r) of the center of mass of head groups from different lipid types. The first two maxima indicate well-defined coordination shells around the transmembrane region. Phosphatidic acid (PA) and cardiolipin (CDL) have a higher density in the annulus of the transmembrane region than phosphatidyl choline (PC) and phosphatidylethanolamine (PE). (C) View of Fo cavity with orange sphere indicating region used for lipid binding analysis. (D) Probabilities of entering and staying bound in the cavity shown for each lipid type. (E) Mean residence time of each lipid type in the cavity. Error bars indicating the 90% confidence interval obtained from bootstrapping. Nonparametric significance tests were performed for each lipid type’s residence time against cardiolipin: * p-value<0.05, ** p-value<1×10−5. (F) Side view of Fo cavity with lipids forming a bilayer-like array.
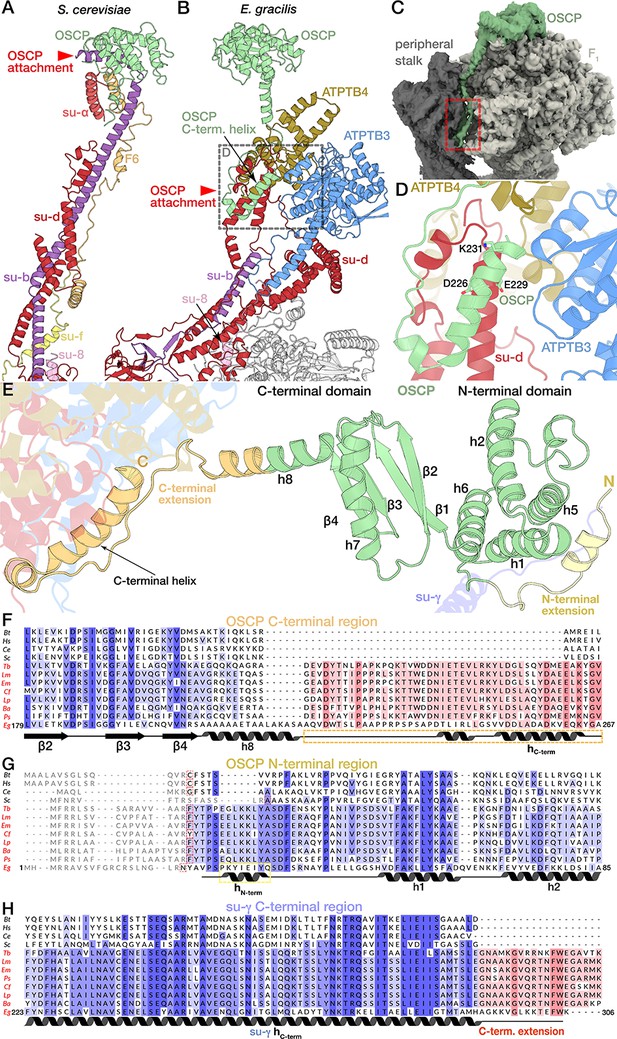
Interactions of OSCP extension with F1 and the peripheral stalk.
(A, B) Peripheral stalk architectures of S. cerevisiae (A) (Guo et al., 2017) and E. gracilis (B) viewed from the F1 (not shown) towards the peripheral stalk. The two structures have subunits b, d, 8 and OSCP in common. Unlike in S. cerevisiae, subunit f does not contribute to the peripheral stalk in E. gracilis. Subunit F6 is not found in E. gracilis. Red arrowheads indicate different heights of OSCP attachment to subunit b and subunit d respectively. (C) The C-terminal extension of E. gracilis OSCP (red rectangle) extends in between the peripheral stalk (dark grey) and F1 (light grey). (D) Close-up of OSCP/subunit d interaction in the E. gracilis peripheral stalk. The indicated residues are conserved in Euglenozoa. (E) The E. gracilis OSCP contains conserved structural elements (light green)(Srivastava et al., 2018) with the N-terminal domain consisting of helices 1,2,5 and 6 and the C-terminal domain consisting of a four-stranded β-sheet and helices 7 and 8 (dihydrolipicolinate reductase domain 2-like fold). The N-terminal extension (light yellow) interacts with subunit γ (blue) of the rotor, whereas the C-terminal extension (orange) is anchored to the rest of the peripheral stalk. (F-H) Multiple sequence alignments of the OSCP C-terminal region (F), with the Euglenozoa-specific extension highlighted red; OSCP N-terminal region (G) with the N-terminal helix found in the E. gracilis OSCP highlighted yellow; red rectangles indicate predicted cleavage sites of mitochondrial processing peptidase. (H) Subunit γ C-terminal region, with euglenozoa-specific extension highlighted red; Bos tarus (Bt), Homo sapiens (Hs), Caenorhabditis elegans (Ce), Saccharomyces cerevisiae (Sc), the trypanosomatids Trypanosoma brucei (Tb), Leishmania major (Lm), Endotrypanum monterogeii (Em), Crithidia fasciculata (Cf), Leptomonas pyrrhocoris (Lp), Blechomonas ayalai (Ba), Phytomonas sp. (Ps) and the euglenoid Euglena gracilis (Eg).
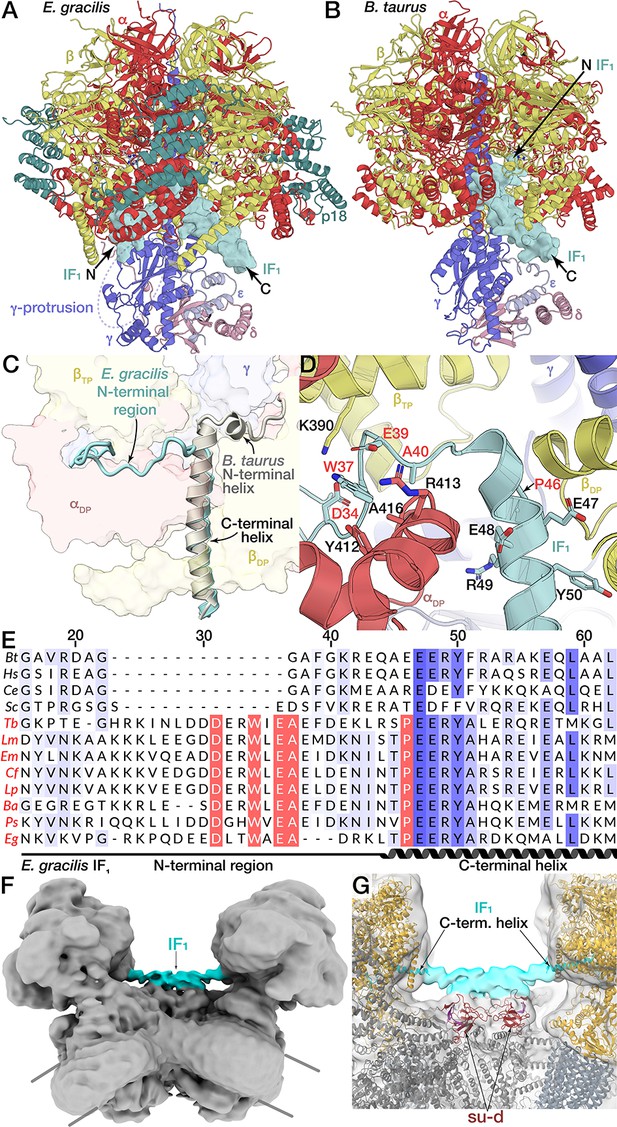
Unusual binding mode of IF1.
(A and B) Comparison of IF1 binding mode between E. gracilis (rotational state-1) and Bos taurus (PDB ID: 2V7Q) (Gledhill et al., 2007). In the E. gracilis structure, both termini are located outside the F1. (C) Superposition of the IF1-inhibited E. gracilis F1 and the bovine counterpart. Both structures share the conserved C-terminal helix but differ in the structure of the N-termini. (D) Close-up of the IF1 binding site. The C-terminal helix of the E. gracilis IF1 contains the conserved EERY, followed by the helix-breaking P46. In the Euglenozoa-specific N-terminal region W37 and E39 interact with αDP, whereas D34 interacts with βTP. (E) Multiple sequence alignment of IF1 from different species; Bos tarus (Bt), Homo sapiens (Hs), Caenorhabditis elegans (Ce), Saccharomyces cerevisiae (Sc), the trypanosomatids Trypanosoma brucei (Tb), Leishmania major (Lm), Endotrypanum monterogeii (Em), Crithidia fasciculata (Cf), Leptomonas pyrrhocoris (Lp), Blechomonas ayalai (Ba), Phytomonas sp. (Ps) and the euglenoid Euglena gracilis (Eg). Euglenozoan species names shown in red. Residues are shaded in blue according to conservation. Residues conserved within Euglenozoa are highlighted red. (F,G) Bridging density in ATP synthase dimer. (F) E. gracilis ATP synthase dimer with both monomers in rotational state-1; map shown at low threshold. (G) From the C-terminal helix of IF1 a continuous density (cyan) extends towards the C2-symmetry axis, contacting the all-β fold formed by subunits d and b, thereby bridging the two F1 subcomplexes.
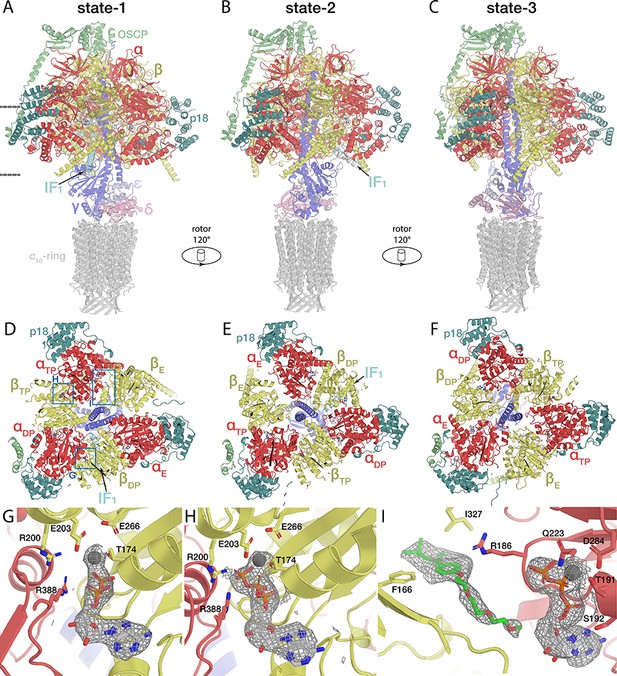
Three main rotational states with bound nucleotides and IF1.
(A-C) F1/c-ring/OSCP subcomplexes in the rotational states 1 (A), 2 (B), and 3 (C), related by a ~ 120°-rotation of the rotor. (G-F) Nucleotide binding sites of the βDP containing ADP (G), the βTP containing ATP (H) and the αTP containing ATP and Triton-X100 (green, I). In both the βDP and βTP, the arginine finger (αR388) extends towards the nucleotide, the former being in contrast to the IF1-inhibited bovine F1, in which the arginine finger points away from the nucleotide (Gledhill et al., 2007).
Videos
Density map of E. gracilis ATP synthase dimer with regions corresponding to protein shown in grey and the detergent belt coloured gold.
Atomic model of the E. gracilis ATP synthase dimer with bound lipids.
The cryo-EM map of the membrane region is shown as mesh. Closeup view of cardiolipin (CDL11) linking H1EG1 and H3EG1 of the two ATPEG1 copies at the matrix side of the membrane region.
Coarse-grained molecular dynamics simulation showing the diffusion of lipids into and out of the peripheral Fo cavity (orange sphere, as described in Figure 6—figure supplement 2C) within the lipid bilayer over a period of 4 µs.
A sliced view of the membrane bilayer is initially shown for reference, but later removed to allow viewing of the binding and unbinding phospholipids. Cardiolipin is indicated with purple, phosphatidic acid with yellow, phophatidylethanolamine with red, phosphatidylcholine with cyan acyl chains respectively. Lipids considered to be bound in the beginning or end of the simulation are visualized, demonstrating that cardiolipin replaces other lipids in the cavity during the simulation.
Additional files
-
Supplementary file 1
Cryo-EM data collection.
- https://cdn.elifesciences.org/articles/51179/elife-51179-supp1-v2.docx
-
Supplementary file 2
E. gracilis ATP synthase dimer atomic model statistics *FSC corrected for the effect of the mask according to 0.143-cutoff criterion ** FSC (masked) according to 0.5-cutoff criterion.
- https://cdn.elifesciences.org/articles/51179/elife-51179-supp2-v2.docx
-
Supplementary file 3
Subunits of the E. gracilis ATP synthase dimer identified in this study.
- https://cdn.elifesciences.org/articles/51179/elife-51179-supp3-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/51179/elife-51179-transrepform-v2.pdf





