RNA polymerase mutations cause cephalosporin resistance in clinical Neisseria gonorrhoeae isolates
Figures
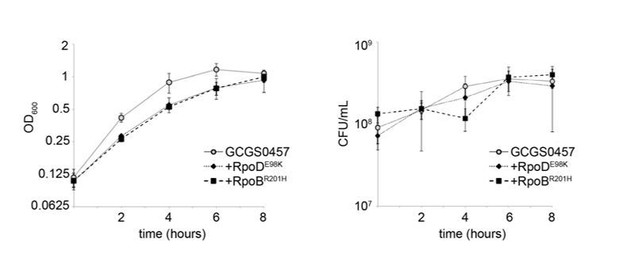
RNA polymerase-mediated reduced cephalosporin susceptibility among gonococcal isolates.
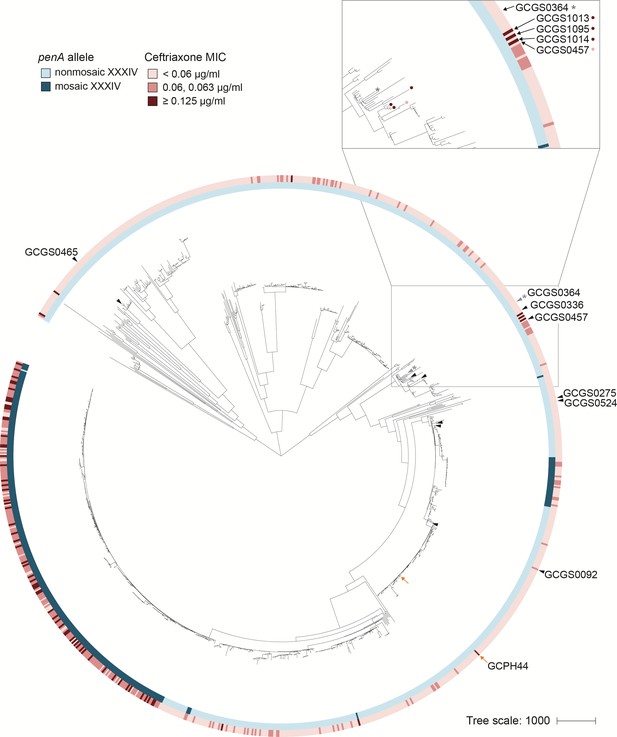
Maximum likelihood phylogeny of the 1102 isolates of the GISP dataset and one United Kingdom isolate (GCPH44). Most high-level reduced cephalosporin susceptibility (MIC ≥0.125 μg/mL) in this dataset is associated with the mosaic penA XXXIV allele, but some isolates with high MICs lack this allele. In four of these isolates – GCGS1095, GCGS1014, GCGS1013 (inset, red-marked leaves) and the U.K. isolate GCPH44 (orange arrow) – CRORS is caused by mutations in the RNA polymerase holoenzyme. Transformation of the CRORS allele rpoB1 from GCGS1095 confers phenotypic reduced susceptibility to the phylogenetic neighbor GCGS0457 (inset, pink-marked leaf) and to other susceptible clinical isolates (black arrowheads). The isolate GCGS0364 (gray arrowhead, denoted with *) spontaneously develops CRORS via rpoB mutation in vitro.
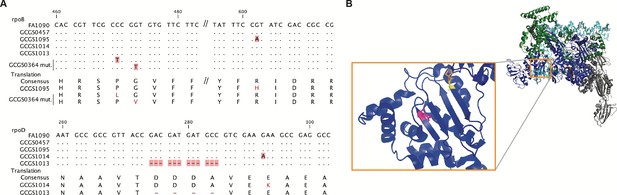
Location of CRORS-associated RNA polymerase mutations.
(A) Alignment of mutant RNA polymerase alleles associated with reduced ceftriaxone susceptibility. (B) Crystal structure of the RNA polymerase holoenzyme from E. coli by Zuo et al. (PDB 4YLO) (Zuo and Steitz, 2015), showing the location of the residues homologous to N. gonorrhoeae RpoB R201 (magenta), G158 (yellow), and P157 (orange). The flexible region of σ70 1.1 that includes the variant positions of rpoD1 and rpoD1 is not included in this structure, but is predicted to interact with this region of RpoB (Murakami, 2013).
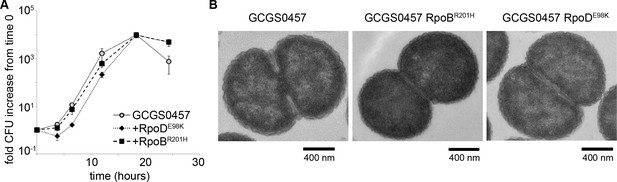
RNA polymerase mutations do not change growth phenotypes.
(A) Growth of GCGS0457 and RNAP mutant transformants on solid GCB agar (mean and standard deviation of three technical replicates shown, representative of two independent experiments). RNAP mutations do not result in a growth rate defect. (B) Transmission electron micrographs of GCGS0457 and RNAP mutant transformants. CRORS transformants are slightly smaller than the parental GCGS0457 strain, but are otherwise morphologically similar.
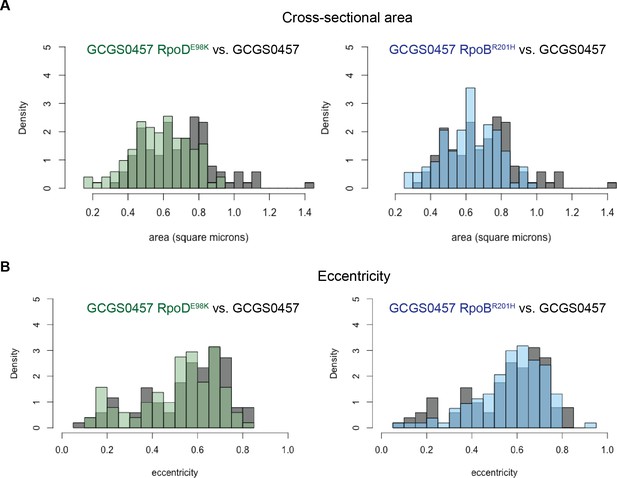
Size and eccentricity of GCGS0457 and CRORS transformants.
100–105 cellular cross sections from TEM images were manually measured in ImageJ to calculate the (A) cross-sectional area and (B) eccentricity of cells from the CROS strain GCGS0457 and its CRORS transformants GCGS0457 RpoDE98K and GCGS0457 RpoBR201H. The cross-sectional area of CRORS transformant cells is slightly smaller (mean area: 0.585 μm2 for GCGS0457 RpoDE98K; 0.621 μm2 for GCGS0457 RpoBR201H) than cells of the parental strain GCGS0457 (mean area 0.693 μm2; p=3.977×10−5 compared to GCGS0457 RpoDE98K and p=0.00436 compared to GCGS0457 RpoBR201H by Welch’s t-test). The degree of eccentricity in the CRO cells is not significantly different between these populations.
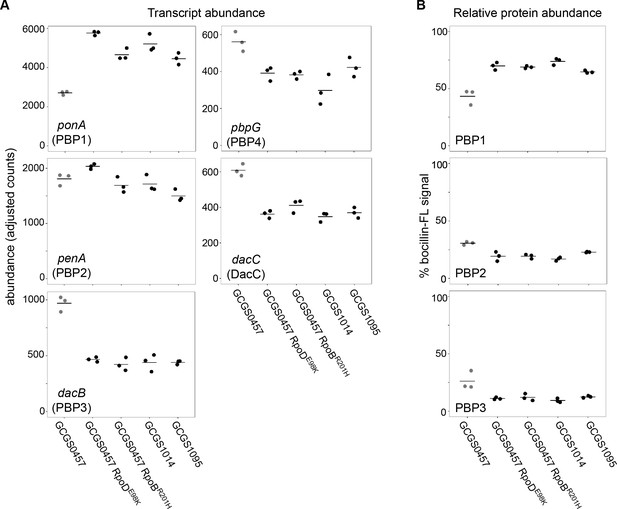
Effect of CRORS-associated RNA polymerase mutations on PBP abundance.
(A) Normalized abundance of transcripts encoding penicillin binding proteins (PBPs) in parental isolates (GCGS0457, GCGS1095, GCGS1014) and RNAP mutant transformants in the GCGS0457 background, measured by RNAseq. (B) Relative protein abundance of PBP1, PBP2, and PBP3, as measured by bocillin-FL labeling. Total bocillin-FL signal for each strain was set at 100%. The relative contribution of each PBP to that signal is shown here. PBP4 protein was not observed on these gels, in agreement with previous reports (Stefanova et al., 2003).
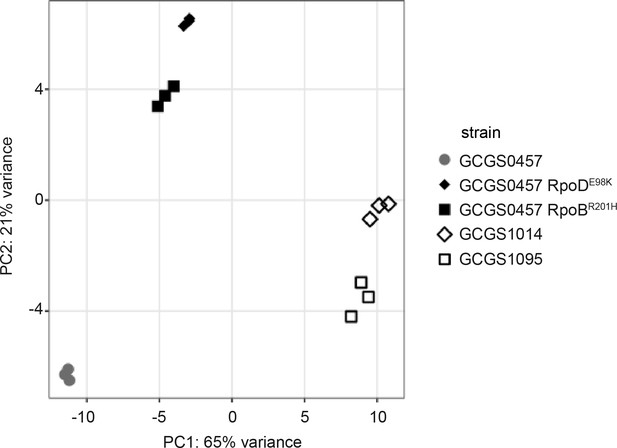
Principle components analysis of RNA-seq data.
The first two principle components are shown for transcriptomic data collected from the CROS isolate GCGS0457, the CRORS isolates GCGS1095 and GCGS1014, and the CRORS transformants GCGS0457 RpoDE98K and GCGS0457 RpoBR201H (three biological replicates/strain). Replicates from both CRORS transformants cluster tightly, despite having different RNAP mutations.
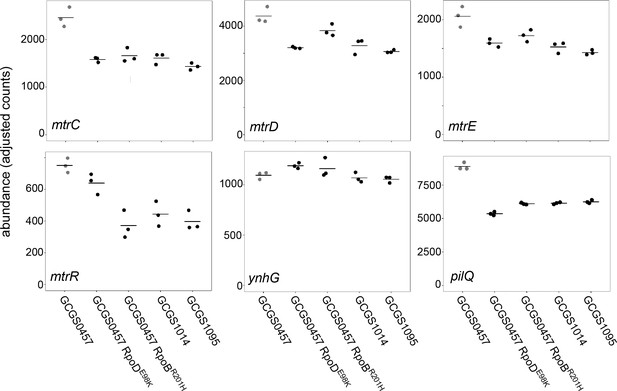
Transcript abundance of various genes in CRORS strains with RNA polymerase mutations.
Normalized abundance of various transcripts in parental isolates (GCGS0457, GCGS1095, GCGS1014) and RNAP mutant transformants in the GCGS0457 background. Shown: mtrC, mtrD, and mtrE, which encode the components of the Mtr efflux pump; mtrR, which encodes the transcriptional repressor of the mtrCDE operon; ynhG, a putative L,D-transpeptidase; and pilQ, which encodes the pilus pore subunit PilQ.
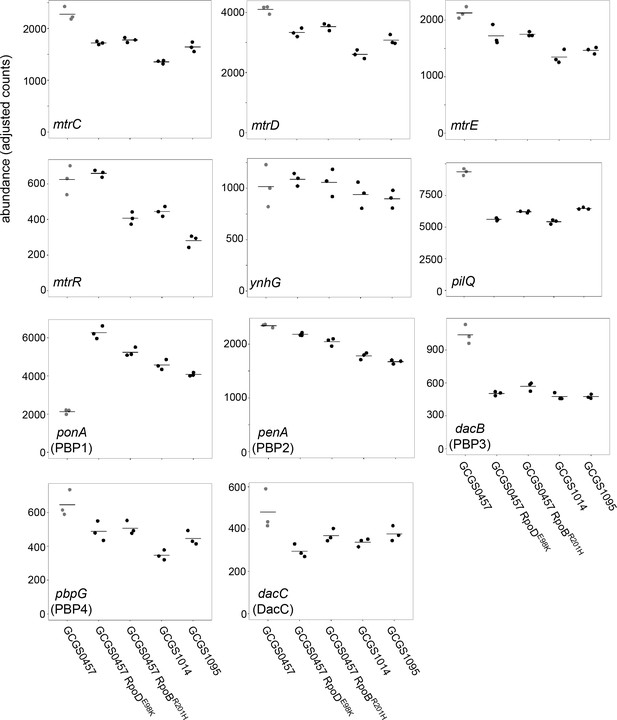
Transcript abundance of various genes in CRORS strains with RNAP mutations, exposed to sub-inhibitory ceftriaxone.
Normalized abundance of various transcripts in parental isolates (GCGS0457, GCGS1095, GCGS1014) and RNAP mutant transformants, exposed to 0.008 μg/mL CRO for 90 min. Shown: mtrC, mtrD, and mtrE, which encode the components of the Mtr efflux pump; mtrR, which encodes the transcriptional repressor of the mtrCDE operon; ynhG, a putative L,D-transpeptidase; pilQ, which encodes the pilus pore subunit PilQ; and transcripts for each of the penicillin-binding proteins.
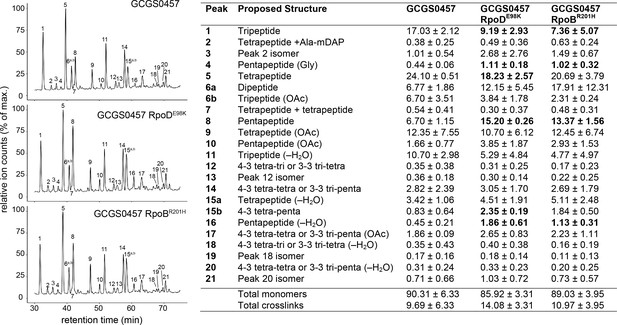
Effect of CRORS-associated RNA polymerase mutations on cell wall structure.
Relative abundance of muropeptide peaks in cell wall digests of GCGS0457 and its CRORS derivatives, GCGS0457 RpoDE98K and GCGS0457 RpoBR201H. Transformant cell walls contain a higher proportion of peptidoglycan with pentapeptide stems (peaks 4, 8, 15b, and 16). Relative abundance values for each muropeptide species (right) were calculated by extracting the peak mass from the total ion chromatogram, integrating the resulting peak area, and dividing by the sum of all of the peak areas within each sample. Shown: mean and standard deviation from 3 biological replicates (data from individual replicates can be found in Figure 5—source data 1). Muropeptide species that are significantly altered in abundance each transformant compared to the parental strain GCGS0457 are indicated in bold (p<0.05 by two-tailed t test). See Supplementary file 3 for a list of all muropeptide masses detected.
-
Figure 5—source data 1
Relative abundance of muropeptide species in individual biological replicates.
- https://cdn.elifesciences.org/articles/51407/elife-51407-fig5-data1-v2.xlsx
Tables
Selected strains, phenotypic ceftriaxone susceptibility, and relevant genotypes.
| strain | CRO MIC (μg/mL) | penA (PBP2) | ponA (PBP1) | PorB 120/121 | RpoB | RpoD |
|---|---|---|---|---|---|---|
| GCGS0457 | 0.012 | Type IX non-mosaic (NG STAR 9.001) | 421P | 120G/121V | wildtype | wildtype |
| GCGS1095 | 0.19 | Type IX non-mosaic (NG STAR 9.001) | 421P | 120N/121A | R201H (rpoB1) | wildtype |
| GCGS1014 | 0.125 | Type IX non-mosaic (NG STAR 9.001) | 421P | 120D/121A | wildtype | E98K (rpoD1) |
| GCGS1013 | 0.19 | Type IX non-mosaic (NG STAR 9.001) | 421P | 120G/121A | wildtype | Δ92–95 (rpoD2) |
| GCPH44 | 0.125 | Type II non-mosaic (NG STAR 2.001) | 421L | 120G/121A | H553N | Δ92–95 (rpoD2) |
| GCGS0457 rpoB1 | 0.19 | Type IX non-mosaic (NG STAR 9.001) | 421P | 120G/121V | R201H (rpoB1) | wildtype |
| GCGS0457 rpoD1 | 0.125 | Type IX non-mosaic (NG STAR 9.001) | 421P | 120G/121V | wildtype | E98K (rpoD1) |
| GCGS0457 rpoD2 | 0.19 | Type IX non-mosaic (NG STAR 9.001) | 421P | 120G/121V | wildtype | Δ92–95 (rpoD2) |
| GCGS0364 | 0.023 | Type IX non-mosaic (NG STAR 9.001) | 421P | 120K/121D | wildtype | wildtype |
| GCGS0364 rpoB2 | 0.5 | Type IX non-mosaic (NG STAR 9.001) | 421P | 120K/121D | G158V (rpoB2) | wildtype |
| GCGS0364 rpoB3 | 0.75 | Type IX non-mosaic (NG STAR 9.001) | 421P | 120K/121D | P157L (rpoB3) | wildtype |
Antibiotic susceptibility profiles of CRORS strains with RNAP mutations.
| Strain | MIC (μg/ml) | |||||||
|---|---|---|---|---|---|---|---|---|
| CRO | CFX | PEN | EPM | AZI | TET | CIP | RIF | |
| GCGS0457 | 0.012 | 0.016 | 2 | 0.016 | 0.25 | 1 | ≤0.015 | 0.125 |
| GCGS1095 | 0.19 | 1 | 2 | 0.032 | 0.5 | 1 | ≤0.015 | ≤0.0625 |
| GCGS1014 | 0.125 | 0.5 | 1.5 | 0.032 | 0.25 | 1 | ≤0.015 | ≤0.0625 |
| GCGS1013 | 0.19 | 0.5 | 1.5 | 0.023 | 0.5 | 1 | ≤0.015 | ≤0.0625 |
| GCGS0457 RpoBR201H | 0.19 | >0.5 | 2 | 0.047 | 0.25 | 1 | ≤0.015 | ≤0.0625 |
| GCGS0457 RpoDE98K | 0.125 | 0.5 | 1.5 | 0.023 | 0.25 | 1 | ≤0.015 | ≤0.0625 |
| GCGS0457 RpoDΔ92-95 | 0.19 | 0.5 | 2 | 0.023 | 0.25 | 1 | ≤0.015 | ≤0.0625 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Neisseria gonorrhoeae) | penA | Neisseria gonorrhoeae PubMLST | NEIS1753 | |
| Gene (Neisseria gonorrhoeae) | ponA | Neisseria gonorrhoeae PubMLST | NEIS0414 | |
| Gene (Neisseria gonorrhoeae) | rpoB | Neisseria gonorrhoeae PubMLST | NEIS0123 | |
| Gene (Neisseria gonorrhoeae) | rpoD | Neisseria gonorrhoeae PubMLST | NEIS1466 | |
| Strain, strain background (Neisseria gonorrhoeae) | GCGS0457 | (Grad et al., 2016) | ||
| Strain, strain background (Neisseria gonorrhoeae) | GCGS1095 | (Grad et al., 2016) | ||
| Strain, strain background (Neisseria gonorrhoeae) | GCGS1014 | (Grad et al., 2016) | ||
| Strain, strain background (Neisseria gonorrhoeae) | GCGS1013 | (Grad et al., 2016) | ||
| Strain, strain background (Neisseria gonorrhoeae) | GCGS0364 | (Grad et al., 2016) | ||
| Strain, strain background (Neisseria gonorrhoeae) | GCGS0092 | (Grad et al., 2016) | ||
| Strain, strain background (Neisseria gonorrhoeae) | GCGS0126 | (Grad et al., 2016) | ||
| Strain, strain background (Neisseria gonorrhoeae) | GCGS0161 | (Grad et al., 2016) | ||
| Strain, strain background (Neisseria gonorrhoeae) | GCGS0249 | (Grad et al., 2016) | ||
| Strain, strain background (Neisseria gonorrhoeae) | GCGS0275 | (Grad et al., 2016) | ||
| Strain, strain background (Neisseria gonorrhoeae) | GCGS0336 | (Grad et al., 2016) | ||
| Strain, strain background (Neisseria gonorrhoeae) | GCGS0353 | (Grad et al., 2016) | ||
| Strain, strain background (Neisseria gonorrhoeae) | GCGS0374 | (Grad et al., 2016) | ||
| Strain, strain background (Neisseria gonorrhoeae) | GCGS0465 | (Grad et al., 2016) | ||
| Strain, strain background (Neisseria gonorrhoeae) | GCGS0489 | (Grad et al., 2016) | ||
| Strain, strain background (Neisseria gonorrhoeae) | GCGS0497 | (Grad et al., 2016) | ||
| Strain, strain background (Neisseria gonorrhoeae) | GCGS0501 | (Grad et al., 2016) | ||
| Strain, strain background (Neisseria gonorrhoeae) | GCGS0524 | (Grad et al., 2016) | ||
| Strain, strain background (Neisseria gonorrhoeae) | GCGS0560 | (Grad et al., 2016) | ||
| Strain, strain background (Neisseria gonorrhoeae) | GCGS0576 | (Grad et al., 2016) | ||
| Strain, strain background (Neisseria gonorrhoeae) | GCGS0698 | (Grad et al., 2016) | ||
| Strain, strain background (Neisseria gonorrhoeae) | GCGS0769 | (Grad et al., 2016) | ||
| Strain, strain background (Neisseria gonorrhoeae) | GCPH44 | (De Silva et al., 2016) | ||
| Strain, strain background (Neisseria gonorrhoeae) | 28BL | Gift of S. Johnson | ||
| Chemical compound, drug | Ceftriaxone disodium salt hemi(heptahydrate) | Sigma Aldrich | Cat. # C5793 | |
| Chemical compound, drug | Ceftriaxone Etest | bioMérieux | SKU # 412302 and 412300 | |
| Chemical compound, drug | Penicillin G Etest | bioMérieux | SKU # 412262 | |
| Chemical compound, drug | Ertapenem Etest | bioMérieux | SKU # 531640 | |
| Chemical compound, drug | Cefixime trihydrate | Sigma Aldrich | Cat. # 18588 | |
| Chemical compound, drug | Azithromycin | Sigma Aldrich | Cat. # PHR1088 | |
| Chemical compound, drug | Tetracycline hydrochloride | Sigma Aldrich | Cat. # T7660 | |
| Chemical compound, drug | Ciprofloxacin | Sigma Aldrich | Cat. # 17850 | |
| Chemical compound, drug | Rifampicin | Chem-Impex | Cat. # 00260 | |
| Chemical compound, drug | Bocillin FL penicillin, sodium salt | Thermo Fisher | Cat. # B13233 | |
| Sequence-based reagent | RpoB-US | This paper | PCR primers | ATGCCGTCTGAATATCAGATTGATGCGTACCGTT |
| Sequenced-based reagent | RpoB-DS | This paper | PCR primers | CGTACTCGACGGTTGCCCAAG |
| Sequenced-based reagent | RpoD-US | This paper | PCR primers | AACTGCTCGGACAGGAAGCG |
| Sequenced-based reagent | RpoD-DS | This paper | PCR primers | CGCGTTCGAGTTTGCGGATGTT |
Additional files
-
Supplementary file 1
Allelic diversity of RNA polymerase holoenzyme components and sigma factors.
- https://cdn.elifesciences.org/articles/51407/elife-51407-supp1-v2.xlsx
-
Supplementary file 2
Ceftriaxone susceptibility and genotype of isolates that have or can acquire CRORS-associated RNA polymerase mutation.
- https://cdn.elifesciences.org/articles/51407/elife-51407-supp2-v2.docx
-
Supplementary file 3
Muropeptide masses detected in cell wall digests of GCGS0457 and RNAP mutants.
- https://cdn.elifesciences.org/articles/51407/elife-51407-supp3-v2.docx
-
Supplementary file 4
N. gonorrhoeae strains used in this study.
- https://cdn.elifesciences.org/articles/51407/elife-51407-supp4-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/51407/elife-51407-transrepform-v2.docx