Developmentally regulated Tcf7l2 splice variants mediate transcriptional repressor functions during eye formation
Figures
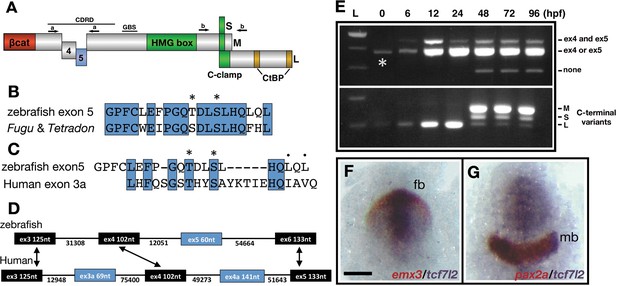
Description and expression of a new alternatively spliced exon in zebrafish tcf7l2.
(A) Schematic representation of variants of Tcf7l2 arising from different splice forms (not to scale). Labels 4 and 5 represent the region of Tcf7l2 coded by alternative exons 4 and 5. Short (S) Medium (M) and Long (L) C-terminal variants coded by alternative splice variants in the 5’ end of exon 15 are indicated. Red box, β-catenin (βcat) binding domain. Green boxes, High-Mobility Group (HMG) Box, which is the primary DNA interacting domain, and C-clamp DNA-helper binding domain. Yellow boxes, CtBP interaction domains. CDRD labelled line over exons 4 and 5 indicates the Context Dependent Regulatory Domain and Groucho Binding Site (GBS) marks the region of interaction with Groucho/Tle transcriptional co-repressors. Arrows indicate the position of primer sets ‘a’ and ‘b’ used for RT-PCR experiments in (E). (B–C) Alignment of the amino acid sequences coded by zebrafish, Takifugu rubripens and Tetradon tcf7l2 exon 5 (B) or human exon 3a (C). Identical amino acids marked by blue boxes. Asterisks over sequence mark putative phosphorylated amino acids. Dots over sequence indicate similar amino acids. (D) Schematic of the genomic region of zebrafish and human tcf7l2. Introns depicted as lines and exons as boxes. Blue exon boxes depict human tcf7l2 alternative exons 3a and 4a, and zebrafish alternative exon 5. Black exon boxes indicate equivalent exons in both species emphasised by arrows. Numbers under introns and within exons represent their nucleotide size (not to scale). (E) RT-PCR experiments performed on cDNA from embryos at stages indicated in hours post fertilisation (hpf). L, 1 Kb ladder. Top panel shows results of PCRs using primer set ‘a’ (indicated in Figure 1A, Materials and methods) amplifying the region of alternative exons 4 and 5. Middle band contains amplicons including either tcf7l2 exon 4 or exon 5. Bottom panel shows results of PCRs using primer set ‘b’ (indicated in Figure 1A, Materials and methods) amplifying the region of alternative exon 15. Asterisk shows maternal expression of tcf7l2. (F–G) Double in situ hybridisation of tcf7l2, in blue, and emx3 (F) or pax2a (G) in red. 10hpf flat mounted embryos, dorsal view, anterior up, posterior down; fb, prospective forebrain; mb, prospective midbrain. Scale Bar in (F) is 200 µm.
-
Figure 1—source data 1
Zebrafish exon five nucleotide and coded amino acid sequences.
(A) Nucleotide sequence of zebrafish tcf7l2 exon 5 (bold and highlighted) and neighbouring exons. (B) Amino acid sequence of the translated sequence of exons in (A). Amino acids Y128, V161, T172, S175 and L181 numbered.
- https://cdn.elifesciences.org/articles/51447/elife-51447-fig1-data1-v1.docx

Alignment of the amino acid sequence coded by tcf7l2 exon five in zebrafish and other fish species.
(A) Alignment of the zebrafish and other fish species amino acid sequence coded by tcf7l2 exon 5. (B) Table showing the nucleotide (nt) homology between human and zebrafish exons surrounding zebrafish new exon five and the amino acid (aa) identity of the protein regions they code. Hs, Homo sapiens; Dr, Danio rerio.
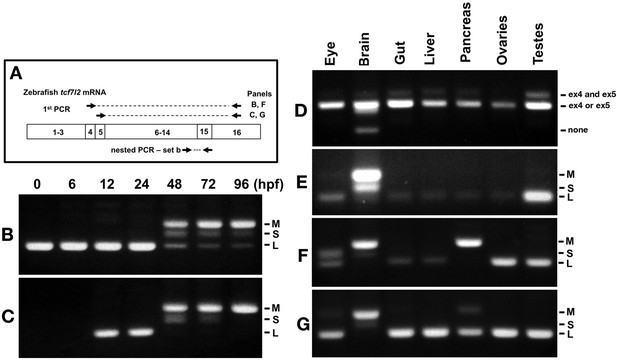
Expression of tcf7l2 alternative exons 4, 5 and 15 varies across development and in adult organs.
RT-PCR analysis of alternative exons 4, 5 and 15 of zebrafish tcf7l2 across development and in various adult organs. (A) Schematic representation of nested RT-PCR strategy used for panels B, C, F and G. (B–C) cDNA from embryos of ages indicated (hpf) was PCR amplified using a forward primer that anneals over exon 4 (B) or exon 5 (C) and a reverse primer that anneals to exon 16 common to all tcf7l2 mRNA variants. The product from this first PCR was then used as a template for a nested PCR using primer set ‘b’ (as in Figure 1. A) that amplifies exon 15 and reveals the different possible Ct ends of Tcf7l2. This last PCR product is shown in these panels. M (Medium), S (short) and L (Long) C-terminal Tcf7l2 variant end. (D–E) RT-PCR experiments performed on cDNA of the indicated adult organs using primer set ‘a’ (Materials and methods) amplifying the region of alternative exons 4 and 5 (D) or using primer set ‘b’ (Materials and methods) amplifying the region of alternative exon 15 (E). (F–G) Same PCR amplification strategy used in panels (B–C) to detect the C-terminal Tcf7l2 variants associated with exons 4 or 5, but using the indicated adult organ cDNA as template in the 1st PCR reaction.
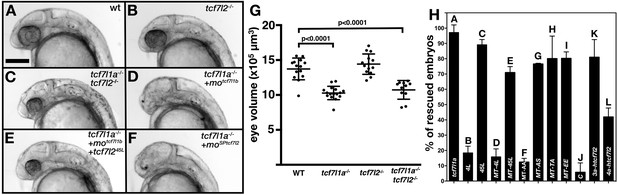
Alternative exon 5 of tcf7l2 impacts eye formation.
(A–F) Lateral views (anterior to left, dorsal up) of 28hpf live wildtype (A), tcf7l2 zf55/ zf55 (B), double tcf7l1a-/-/tcf7l2 zf55/ zf55 (C) and tcf7l1a-/- (D–F) zebrafish embryos with injected reagents indicated top right showing representative phenotypes. (D) 0.12 pmol motcf7l1b (E), 0.12 pmol motcf7l1b and 20 pg of 45L-tcf72 splice variant mRNA, (F) 1.25 pmol moSPtcf7l2. Scale bar in (A) is 200 µm. (G) Plot showing the volume of eyes (µm3) of 30hpf fixed embryos coming from a double heterozygous tcf7l1a/tcf7l2 mutant incross. Error bars are mean ± SD, only P values greater than 0.1 from unpaired t test with Welch's correction are indicated. Data in Supplementary file 1C. (H) Bars represent the percentage of tcf7l1a-/- embryos that develop eyes (with distinguishable lens and pigmented retina) coming from multiple tcf7l1a+/- female to tcf7l1a-/- males crosses, injected with 0.12 pmol of motcf7l1b (all bars) and co-injected with constructs stated on X axis: 10 pg of tcf7l1a mRNA (A) 20 pg of tcf7l2 mRNA splice variants 4L-tcf7l2 (B) 45L-tcf7l2, (C) MT-4L-tcf7l2 (D) MT-45L-tcf7l2 (E) MT-tcf7l2-AA (F) MT-tcf7l2-AS (G) MT-tcf7l2-TA (H) MT-tcf7l2-AA (I) htcf7l2-C (J) htcf7l2-3a (K) and htcf7l2-4a (L). Data for all these plots are included in Supplementary file 1E. Error bars are mean ± SD.
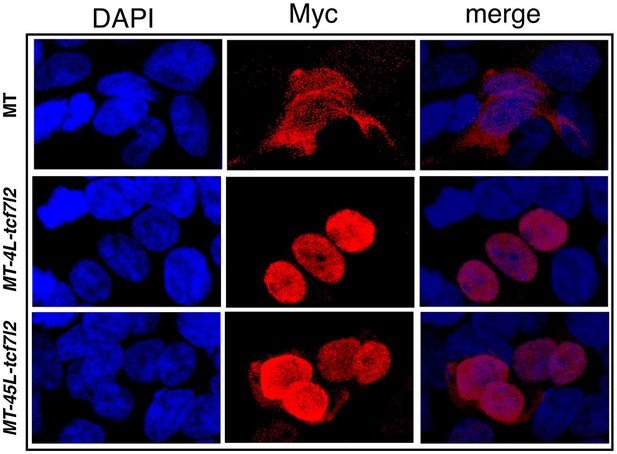
Tcf7l2 variants localise to the nucleus.
Sub-cellular localisation of 4L-Tcf7l2 and 45L-Tcf7l2 myc tagged splice variants. HEK293 cells were transfected with empty myc tag (MT) vector (top row), MT-4L-tcf7l2 (middle row) and MT-45L-tcf7l2 (right row) splice variants and immunostained with anti-myc antibody (2nd column) and stained with DAPI (1 st column). Merged images in 3rd column.
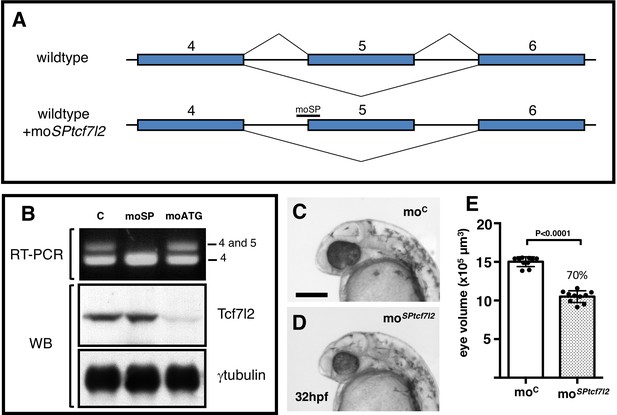
Splicing specific morpholino knockdown of tcf7l2 splice variants that include exon5.
(A) Cartoon showing the rationale of using moSPtcf7l2 that targets intron 4/exon five splice site boundary blocking the splicing machinery and making it skip to the next splicing acceptor site in exon 6. (B) RT-PCR (top panel) and western blot (WB, middle and bottom panels) performed using RNA and proteins extracted from 24hpf zebrafish embryos injected with: control morpholino (first lane), moSPtcf7l2 (moSP, second lane), and moATGtcf7l2 (moATG, third lane). cDNA was amplified using set ‘a’ primers (Figure 1A, 5’F1 vs Splice R1, Materials and methods). Anti human Tcf7l2 and anti-gamma tubulin antibodies were used in Western blot experiments (middle and bottom panel). (C, D) 32hpf embryos injected with 1.25 pmol of (C) control morpholino (moC) or (D) tcf7l2 exon five splicing morpholino (moSPtcf7l2). Scalebar in (C) is 200 µm. (E) Plot showing estimated eye volume of 32hpf live embryos injected with injected with moC or moSPtcf7l2 as in (C, D). Error bars are mean ± SD, P values from unpaired t test. Percentage indicates average eye profile size relative to control.
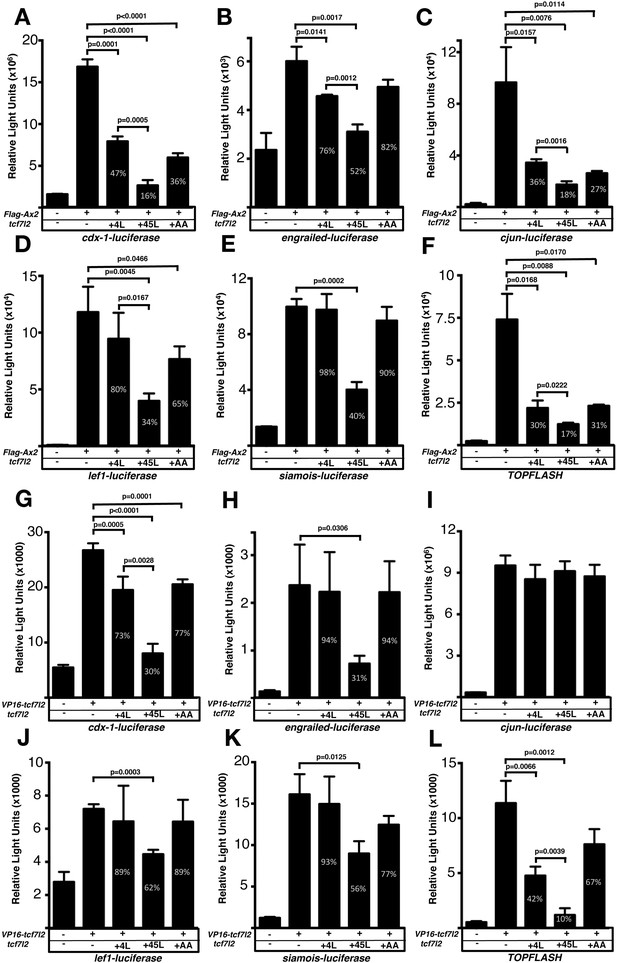
Exon five coding tcf7l2 variant represses Wnt activity induced by FLAG-Ax2 or VP16-TCF7L2 in luciferase assays.
Bar plots showing luciferase reporter assay results expressed in relative light units. HEK293 cells were transiently co-transfected with luciferase reporter constructs indicated beneath the X-axis, (A–F) FLAG-Ax2 (except for first bars), (G–L) VP16-TCF7L2 DNA (except for first bars) and 4L-tcf7l2 DNA (+4L; 3rd bars), or 45L-tcf7l2 DNA (+45L; 4th bars) or tcf7l2-AA DNA (+AA; 5th bars). Control experiments show only background luciferase activity with no transfected plasmids (1st bars). Figures in the bars indicate the percentage size of that bar relative to transfection with either FLAG-Ax2 or VP16-TCF7L2 alone (2nd bar). Error bars are mean ± SD n = 3 (experiments were performed twice), P values from unpaired t tests comparing FLAG-Ax2 or VP16- TCF7L2 control condition with tcf7l2 variant co-transfections. Comparisons with no statistical significance are not marked.
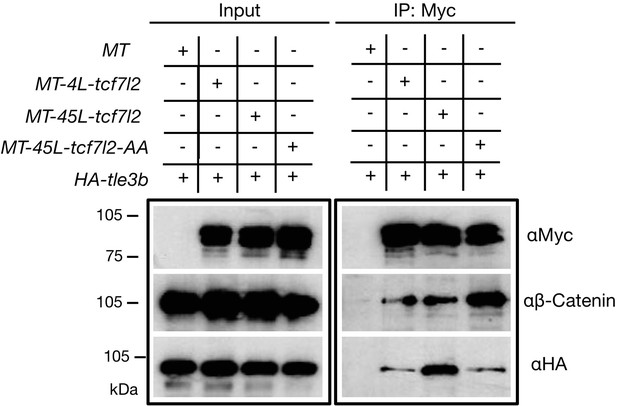
Alternative exon 5 of tcf7l2 enhances affinity with Tle3b.
Protein input (left panel) and anti-Myc immunoprecipitation (IP) eluate western blot (right panel) showing co-inmunoprecipitation of β-Catenin or HA-tagged Tle3b. HEK293 cells were transiently transfected with HA tagged tle3b together with empty myc tag vector (1st lane), MT-4L-tcf7l2 (2nd lane), MT-45-tcf7l2 (3rd lane) and MT-AA-tcf7l2 (4th lane). Left panels show protein input before anti-Myc IP. Right panels show protein eluate from anti-Myc antibody coupled beads. Westernblots were probed with anti-Myc (tagged Tcf7l2 proteins, top panel), anti-βcatenin (middle panel) and anti-HA (tagged Tle3b protein, bottom panel) antibodies. Asterisk shows that the Tcf7l2 form containing exon five shows more intense binding with Tle3b than other Tcf7l2 forms.
-
Figure 5—source data 1
Zebrafish MT-45L-Tcf7l2-BioID2 peptides recovered by LC-MS/MS analyses are shown in bold.
- https://cdn.elifesciences.org/articles/51447/elife-51447-fig5-data1-v1.docx
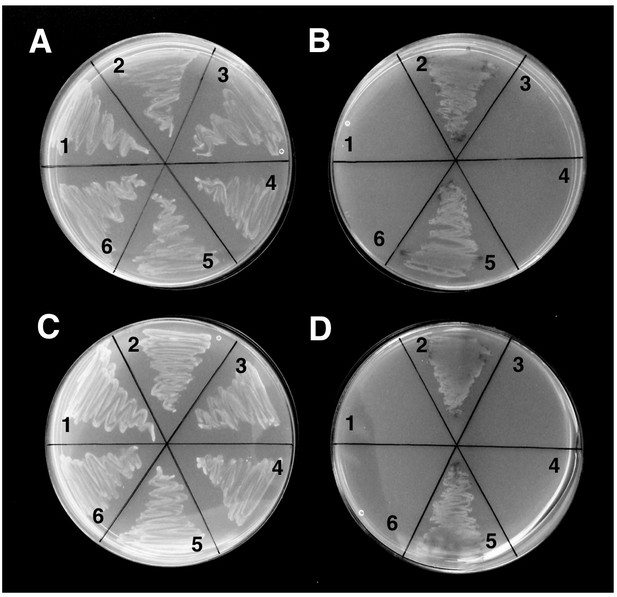
4L-Tcf7l2 and 45L-Tcf7l2 variants interact with β-Catenin and Tle3b in yeast two-hybrid protein interaction assays.
Y2Gold yeast strain was co-transformed with: (1) β-catenin/pGAD and empty pGBK vector, (2) β-catenin/pGAD and 4L-tcf7l2/pGBK (A, B) or 45L-tcf7l2/pGBK (C, D) (3) empty pGAD vector and 4L-tcf7l2/pGBK (A, B) or 45L-tcf7l2/pGBK (C, D) with, (4) dCtle3b/pGAD and Empty pGBK vector, (5) dCtle3b/pGAD and 4L-tcf7l2/pGBK (A, B) or 45L-tcf7l2/pGBK (C, D) (6) empty pGBK and pGAD vectors. and plated in -Leu-Trp dropout selective media agar plates supplemented with Xgal (A, C) or Ade-His-Leu-Trp dropout selective media agar plates supplemented with Aureoblastidin A and X-gal (B, D).
Additional files
-
Supplementary file 1
Supplementary file 1A Analysis of RT-PCR data in Figure 1E and Figure 2B, C showing the expression of alternative tcf7l2 exons 4, 5 and 15 (a) and Tcf7l2 variants (b) through development.
(a) Results in rows 1 to 4 (grey) taken from Figure 1E, rows 5 to 7 (pink) from Figure 2B and rows 8 to 10 (blue) from Figure 2C. (+) and (++) depict relative band intensity in the gels; (-) indicates no signal (b) Tcf7l2 variants expressed during development based on the information in (a). (+) and (-) indicate presence or absence of the variant respectively. (?) indicates that it is not possible to derive a conclusion based on the data available. Analysis of variants that lack both exons 4 and 5 is not included. Supplementary file 1B Analysis of RT-PCR data from Figure 2D-E showing the expression of alternative tcf7l2 exons 4, 5 and 15 (a) and Tcf7l2 variants (b) in adult organs. (a) Results in rows 1 to 4 (grey) taken from Figure 2D-E rows 5 to 7 (pink) from Figure 2F and rows 8 to 10 (blue) from Figure 2G. (+) and (++) depict relative band intensity in the gels; (-) indicates no signal (b) Tcf7l2 variants expressed in adult organs based on the information in (A). (+) and (-) indicate presence or absence of the variant respectively. (?) indicates that it is not possible to derive a conclusion based on the data. Analysis of variants that lack both exons 4 and 5 is not included. Supplementary file 1C Size of the tcf7l2-/-eye is similar to wildtype embryos at 30hpf. Estimated volume in µm3 of the eye of 30hpf fixed embryos coming from a double heterozygous tcf7l1a/tcf7l2 mutant incross. Avg, average; SD, Standard Deviation. Supplementary file 1D Knockdown of tcf7l1b and excision of tcf7l2 exon5 in tcf7l1a-/- mutant embryo compromises eye formation Embryos from female tcf7l1a+/- to male tcf7l1a-/- spawnings were injected with the morpholinos stated in the left column. This pairing scheme leads to 50% of homozygous mutant embryos. Each row represents an individual experiment. Embryos were scored as eyeless when little or no pigmented retinal tissue could be distinguished. Total represents the number of embryos scored in each experiment. Supplementary file 1E Restoration of eye formation by expression of exogenous Tcf7l2 variants in tcf7l1a-/-/tcf7l1b morphant embryos. Tcf7l1a-/- embryos injected with tcf7l1b morpholino and tcf7l1a or the tcf7l2 mRNA variant stated in the first column. Each row represents an individual experiment. Total represents the number of tcf7l1a-/- embryos scored in each experiment. Eye formation was scored as rescued when pigmented retinal tissue was evident. Supplementary file 1F Size of the eye profile area is smaller in moSPtcf7l2 injected embryos at 30hpf. Volume in µm3 of the eye profile of 32hpf fixed embryos from wildtype embryos injected with moC or moSPtcf7l2. Avg, Average; SD, Standard Deviation. Supplementary file 1G Results from luciferase reporter assay experiments expressed in relative light units using FLAG-Ax2 to induce Wnt activity. Avg, Average; SD, Standard Deviation; %, percentage relative to FLAG-Ax2 condition. Supplementary file 1H Results from luciferase reporter assay experiments expressed in relative light units using VP16-TCF7L2 to induce Wnt activity. Avg, Average; SD, Standard Deviation; %, percentage relative to VP16-TCF7L2 condition. Supplementary file 1I Peptides recovered by mass spectrometry and their respective modifications.
- https://cdn.elifesciences.org/articles/51447/elife-51447-supp1-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/51447/elife-51447-transrepform-v1.pdf





