Prickle isoforms determine handedness of helical morphogenesis
Figures
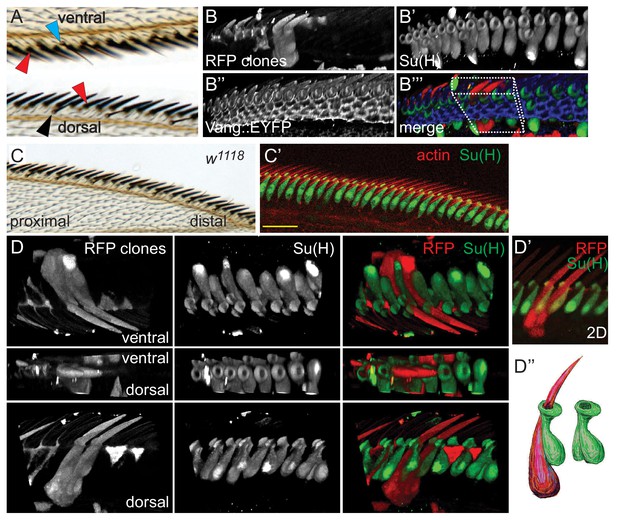
Morphology of wildtype dorsal mechanosensory bristles.
(A) Dorsal and ventral views showing adult dorsal mechanosensory bristles (red arrowheads), ventral chemo- and mechanosensory bristles (blue arrowhead) and dorsal chemosensory bristles (black arrowhead). Bristles are separated by the exoskeleton secreted by cells displaying trichomes (hairs), similar to those in the majority of the wing blade. See also Figure 1—figure supplement 1A for a schematic view. (B) 3D reconstruction of a section of a 36 hr control (w1118) AWM containing two clones expressing cytoplasmic RFP, each labeling two adjacent dorsal mechanosensory bristles. Several clones labeling hair cells are also present in this sample. Costaining with Su(H) marks all socket cells, and Vang::EYFP is present at apical cell junctions. The boxed region is displayed from several angles in panel D). (C–C’) Wildtype adult wing and equivalent region of a 36 hr pupal wing stained for Su(H) and actin. (D–D’) 3D views from different angles of the RFP clone(s) shown in panel B). (D’’) Cartoon interpretation of the sibling shaft-socket pairs from D’). All images throughout are of right wings and are displayed proximal to the left and distal to the right. Scale bars: 20 μm.
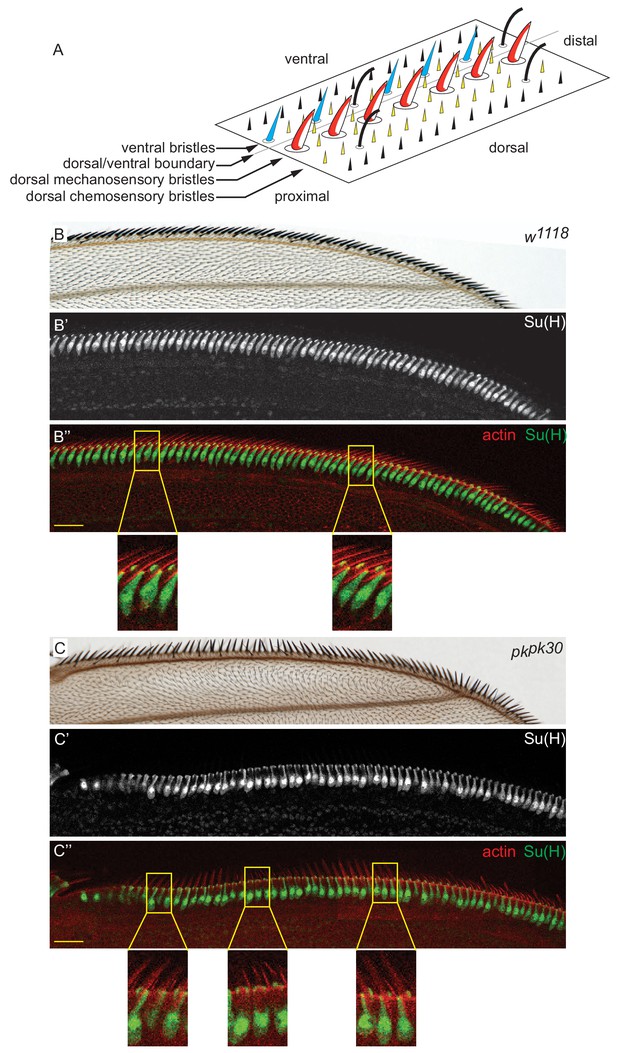
Schematic of the AWM bristles, and expansive views of adult and pupal control (w1118) and pkpk30 bristles.
(A) Schematic drawing showing the organization of the AWM bristles on a conceptually unfolded wing. Dorsal mechanosensory bristles (red and white), ventral mechanosensory bristles (blue) and chemosensory bristles (black). ‘Margin cells’ (yellow) are not a genetically defined subset of wing cells, but we refer to the hair cells near the margin and that express Pksple earlier than other hair cells in this way. In adult wings, hair cells have died, leaving only the cuticle they previously secreted. (B–B’’) Adult control AWM and equivalent region of a 36 hr control AWM stained for Su(H) and actin. Note the uniform relationship of shaft-socket pairs across the length of the AWM. (C–C’’) Similar images of pkpk30 wings. Note the opposite orientations of shaft-socket pairs where shaft polarity is opposite. Scale bars: 20 μm.
Morphology of 36 hr wildtype dorsal mechanosensory bristles displayed in 3D A reconstruction of confocal stacks displaying AWMs stained with Su(H) (green) to mark socket cells and RFP marking two shaft-socket cell pairs.
Note that the software displays the edges of shapes cut by the bounds of the displayed section as bright areas.
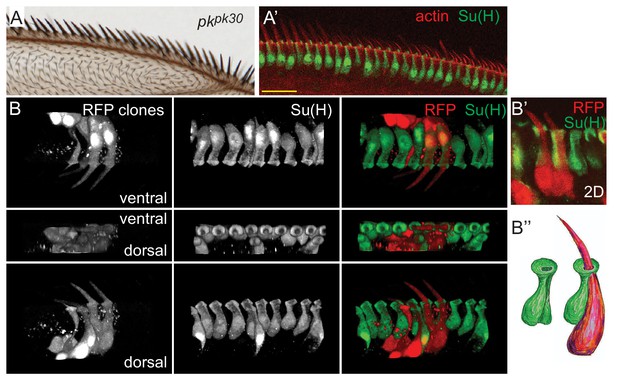
Morphology of pkpk mutant dorsal mechanosensory bristles.
(A–A’) pkpk30 adult wing and equivalent region of a pkpk30 36 hr pupal wing stained for Su(H) and actin. (B–B’) 3D views of RFP clones in a pkpk30 36 hr pupal wing revealing reversed orientation of sibling shaft-socket pairs. Su(H) stains socket cells. (B’’) Cartoon interpretation of shaft-socket pair from B’). Scale bars: 20 μm.
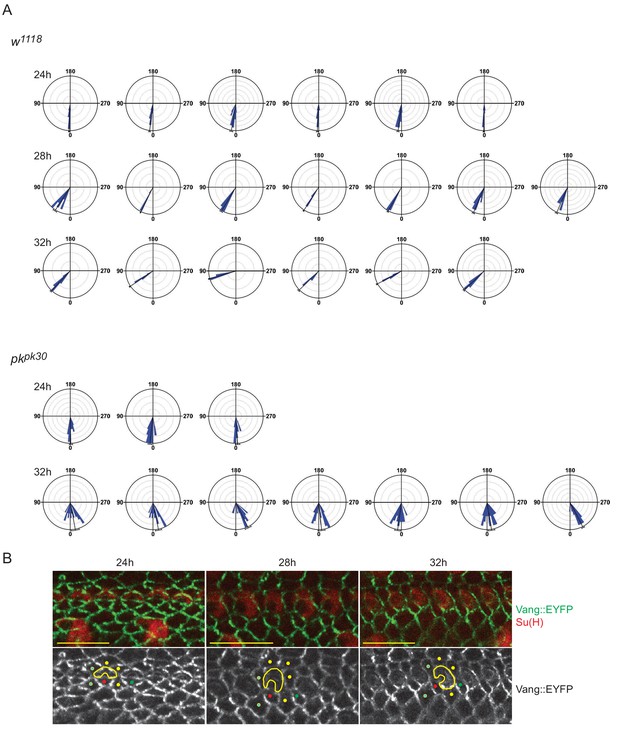
Rose plots for control and pkpk30 socket cell orientations and apical rearrangement of anterior wing margin cells.
(A) Complete set of rose plots for control and pkpk30 socket cell orientations at times sampled (control - 115 sockets from six wings (24 hr), 105 sockets from seven wings (28 hr), 103 sockets from six wings (32 hr); and pkpk30 - 52 sockets from three wings (24 hr), 157 sockets from seven wings (32 hr)). Statistics are given in Table 1. (B) Surface images showing socket cells (Su(H); red) and cells outlined with Vang::EYFP (green) from 24 hr, 28 hr and 32 hr AWMs. Because these cell patterns were highly stereotypical, we could infer stereotyped cell rearrangements. Colored dots mark neighbor cells inferred to rearrange. Note that the cell marked with a red dot appears to rotate with the socket and to insert in between the two cells marked with blue dots, and that it establishes a junction with the next proximal socket. Similarly, the equivalent cell posterior to the next distal socket establishes a junction with the outlined socket cell. Scale bars: 10 μm. Statistical analyses for all genotypes are in Table 2.
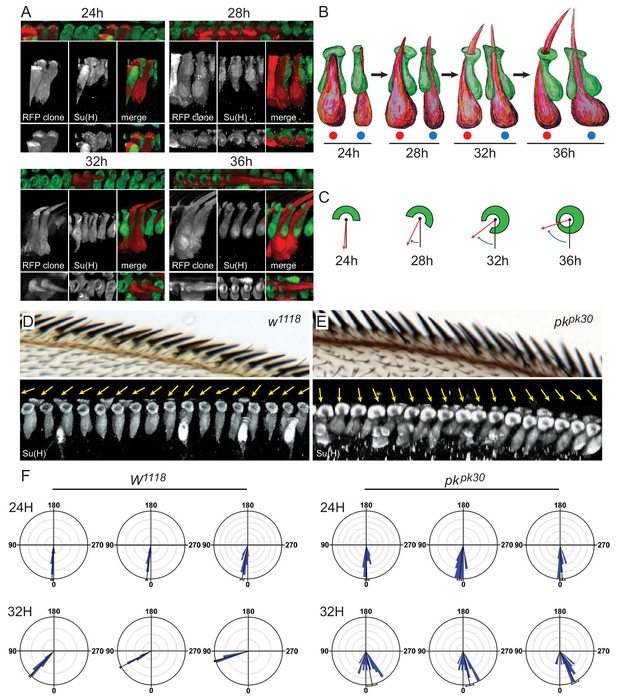
3D images of shaft-socket pairs at different times reveal a clockwise helical growth in control and counterclockwise growth in pkpk30 mutant bristles.
(A) Reconstructed 3D images from varying angles of 24 hr, 28 hr, 32 hr and 36 hr shaft-socket clones marked with RFP and stained with Su(H). (B) Cartoon interpretation of images from panel A), showing the clockwise rotation of the apical aspects of the shaft and socket cells. Views from dorsal (red dots) and proximal (blue dots). (C) Diagrams illustrating scoring of rotation angles. (D–E) Control (w1118) and pkpk30 adult AWMs and corresponding regions from 32 hr pupal wings showing 3D reconstructed images of socket cells stained with Su(H). Orientation angles for these socket cells are indicated by yellow arrows. (F) Quantification of rotation angles for 24 hr and 32 hr control and pkpk30 socket cells. Each rose plot represents an individual wing, with scoring limited to the distal AWM anterior to vein L2 unless otherwise indicated (for complete set of rose plots and description of sample sizes, see the legend for Figure 2—figure supplement 1A). Most variation between individual wings is likely attributable to variation in developmental timing. For detailed description of quantification, see Materials and methods. Statistical analyses for all genotypes are in Table 2.
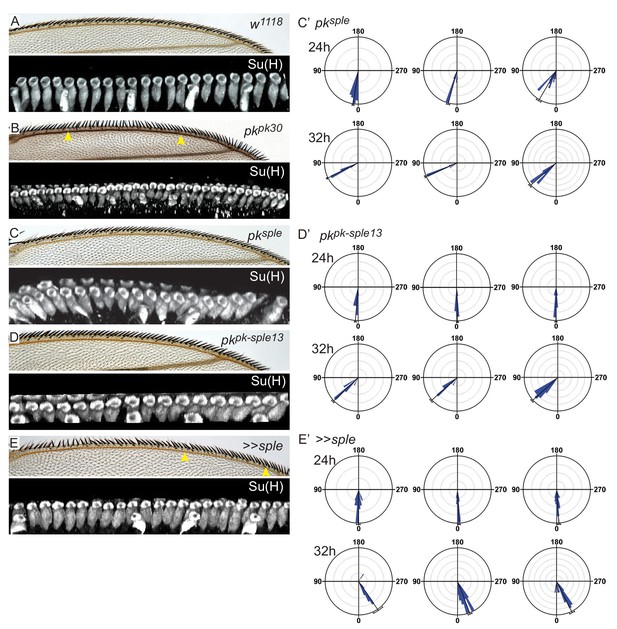
Bristle polarity in adult and socket cell rotation in pupal control (w1118), pkpk30, pksple1, pkpk-sple13, and, MS1096-Gal4 UAS-pksple (>>sple) wings at 24 hr and 32 hr apf.
(A–D) For pkpk30 (panel B) and >>sple (panel E), socket cell images (Su(H)) are from regions corresponding to those demarcated by the arrowheads. (C’, D’, E’) Quantification of socket cell rotation for additional genotypes relevant to Figure 3 (pksple1 - 73 sockets from three wings (24 hr), 72 sockets from four wings (32 hr); pkpk-sple13 - 65 sockets from three wings (24 hr), 63 sockets from three wings (32 hr); and >>sple - 65 sockets from three wings (24 hr), 65 sockets from three wings (32 hr)). Statistical analyses for all genotypes are provided in Table 2.
Morphology of 24 hr wildtype dorsal mechanosensory bristles displayed in 3D A reconstruction of confocal stacks displaying AWMs stained with Su(H) (green) to mark socket cells and RFP marking shaft-socket cell pairs.
Morphology of 28 hr wildtype dorsal mechanosensory bristles displayed in 3D A reconstruction of confocal stacks displaying AWMs stained with Su(H) (green) to mark socket cells and RFP marking shaft-socket cell pairs.
Morphology of 32 hr wildtype dorsal mechanosensory bristles displayed in 3D A reconstruction of confocal stacks displaying AWMs stained with Su(H) (green) to mark socket cells and RFP marking shaft-socket cell pairs.
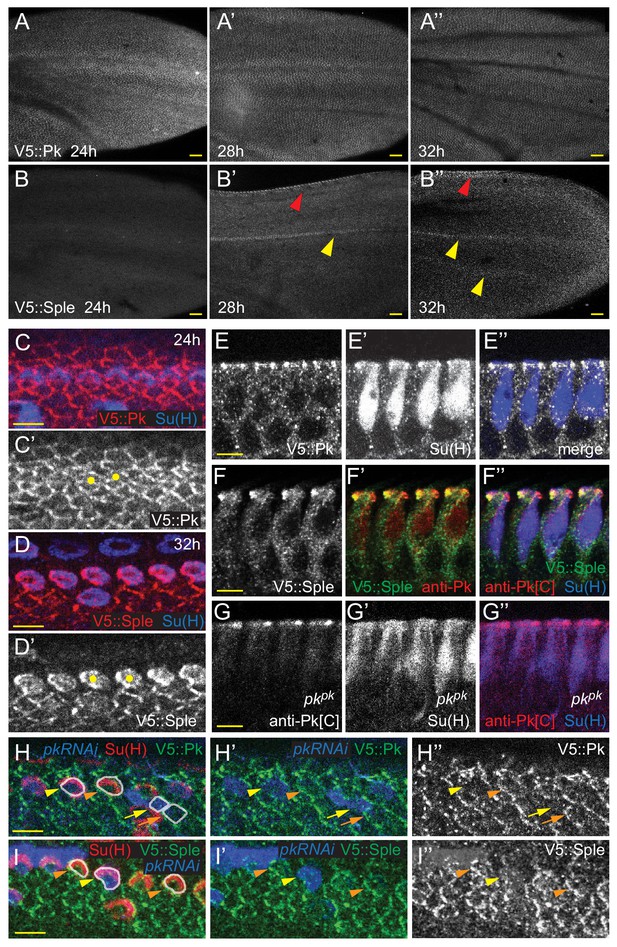
Expression of Pkpk and Pksple isoforms in pupal wings.
(A–B’’) V5::Pk (A–A’’) and V5::Sple (B–B’’) in pupal wings of ages indicated. Red arrowheads mark AWM and yellow arrowheads mark veins L3 and L4. (C–D’) Surface views of V5::Pk at 24 hr (C–C’) or V5::Sple at 32 hr (D–D’) counterstained for Su(H) to locate socket cells. Some socket cell locations are indicated by yellow dots. (E–G’’) Planar sections of socket cells (Su(H)) of 28 hr pupal wings with apical at the top. V5::Pk is at all junctions between socket and margin cells (E–E’’). Pksple (detected with V5::Sple) appears to be localized to the proximal side of control (w1118) socket cells (F–F’’) but to the distal side of pkpk mutant socket cells (detected with anti-Pk[C]; G–G’’). (H–I’) Mosaic expression of V5::Pk (H–H’’) or V5::Sple (I–I’’) in otherwise wildtype wings (28 hr). Cells lacking expression are marked with RFP (blue). Both V5::Pk and V5::Sple localize to the proximal side of expressing (orange arrowheads) but not non-expressing (yellow arrowheads) socket cells, demonstrating their proximal localization. V5::Pk localizes to the proximal side of expressing (orange arrow) but not non-expressing (yellow arrow) margin wing cells. V5::Sple is also proximal in margin wing cells, though no informative clones were captured in this image. Several relevant cells are outlined in (H and I) for clarity. Scale bars: 20 μm (A,B) and 5 μm (C–I).
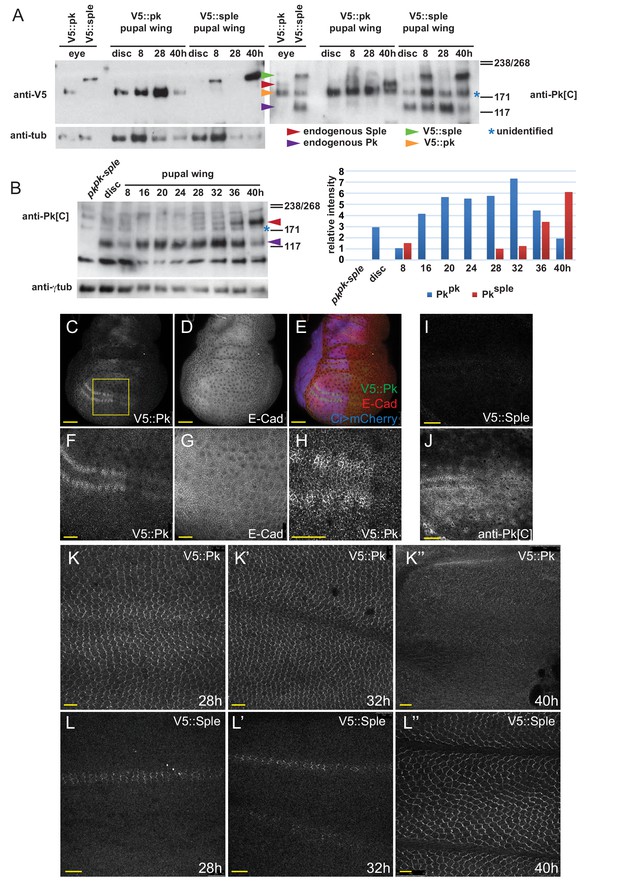
V5::Pk and V5::Sple expression throughout wing development.
(A) Western blot from wing extracts from homozygous V5::Pk and V5::Sple eyes, and wings at various developmental stages, stained with anti-V5 and then anti-Pk[C] antibodies, validating the specific detection and expression of the CRISPR modified isoforms. Anti-Pk common antibody detects both the V5-tagged and endogenous forms of both isoforms. Note that anti-Pk[C] detects an unidentified non-specific band that co-migrates with V5::Pk (asterisk). (B) Time course of wing extracts from control imaginal discs and wings showing early Pkpk expression declining late in development and PkSple expression initially undetectable, with a small amount at 8 hr apf, and then increasing later in pupal wing development. Intensities of Pkpk and PkSple relative to the γtubulin loading control for two independent experiments were averaged and plotted in the accompanying chart. (C–J) V5::Pk and V5::Sple expression in third instar wing discs. V5::Pk is detected throughout, with higher levels in the proneural cells. V5::Sple is not detected. (K–L’’) V5::Pk expression in wing cells declines from 32 hr to undetectable at 40 hr, with vein expression dropping more rapidly, while V5::Sple appears first at the AWM and in vein L3, then in other veins, and is expressed strongly in wing cells exept for veins by 40 hr. Scale bars: 50 μm (C–E), 20 μm (F–J), 10 μm (K,L).
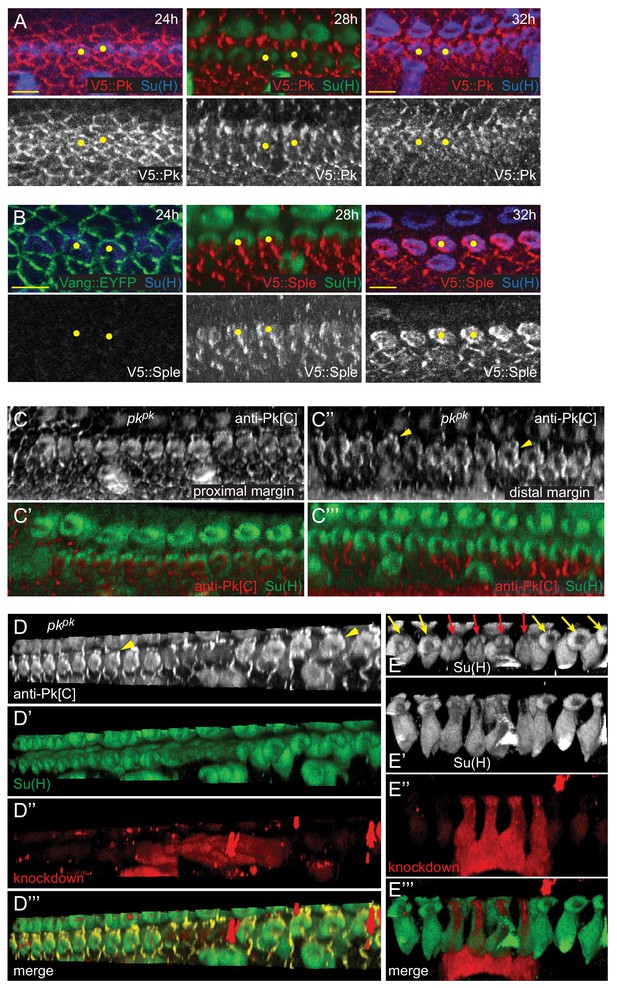
Timing of expression and localization of Pkpk and/or Pksple correlates with handedness of helical rotation.
(A, B) V5::Pk and V5::Sple surface views at the margin of 24 hr, 28 hr and 32 hr pupal wings showing declining expression of Pk, first in socket cells and then in margin cells, and increasing V5::Sple expression, with higher levels in socket cells compared to margin cells. (C–C’’’) Pksple expression in a pkpk mutant wing, detected with anti-Pk[C], showing distal localization in socket cells at the distal end of the AWM where bristle polarity is reversed (C’’,C’’’) and minimal asymmetry in socket cells at the proximal AWM where bristle polarity is not reversed (C,C’). (D–E’’’) A clone of cells (RFP; red) knocked down for pksple in a pkpk mutant wing. Pksple is expressed and distally localized in socket cells with reversed polarity, but in knockdown cells, Pksple is not expressed and those socket cells fail to undergo backward (counterclockwise) rotation. Scale bars: 5 μm (A,B).
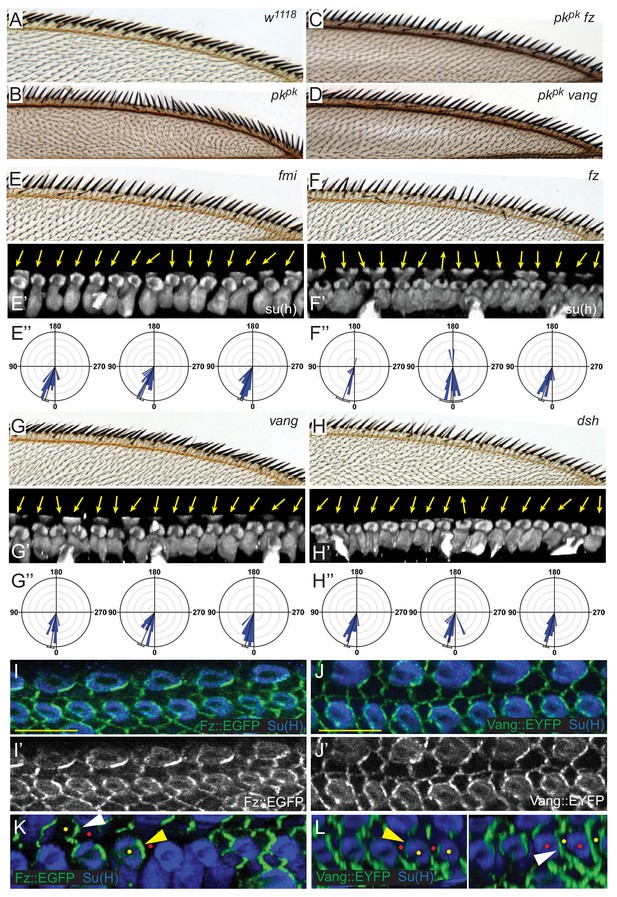
Core PCP components control shaft-socket rotation.
(A–D) Reversed bristle polarity in pkpk mutant compared to control is abrogated in pk fz and pk vang double mutants, demonstrating requirement for core PCP activity for polarity reversal in pkpk mutants (reversed bristle polarity is suppressed in all double mutant wings analyzed (n = 20 of each genotype)).(E–H) While bristle tilt is only mildly disturbed, socket cell rotation is strongly impaired in fmi knockdown, fz, vang and dsh mutant wings. Adult wings, representative socked cell images (32 hr) and quantification of three individual wings (32 hr) for each genotype are shown (fmi knockdown - 97 sockets from four wings; fz mutant - 120 sockets from six wings; vang muant - 114 sockets from six wings; and dsh mutant - 110 sockets from six wings). Statistical analyses for all genotypes are in Table 2. (I–J’) Expression of Fz::EGFP (I–I’) and Vang::EYFP (J–J’) in margin cells. (K–L) Mosaic expression demonstrates distal localization of Fz::EGFP (K) and proximal localization of Vang::EYFP (L) in both socket (yellow) and margin (white arrowheads) cells. Some informative expressing cells (yellow dots) next to non-expressing cells (red dots) are marked. Scale bars: 10 μm. Statistical analyses for all genotypes are in Table 2.
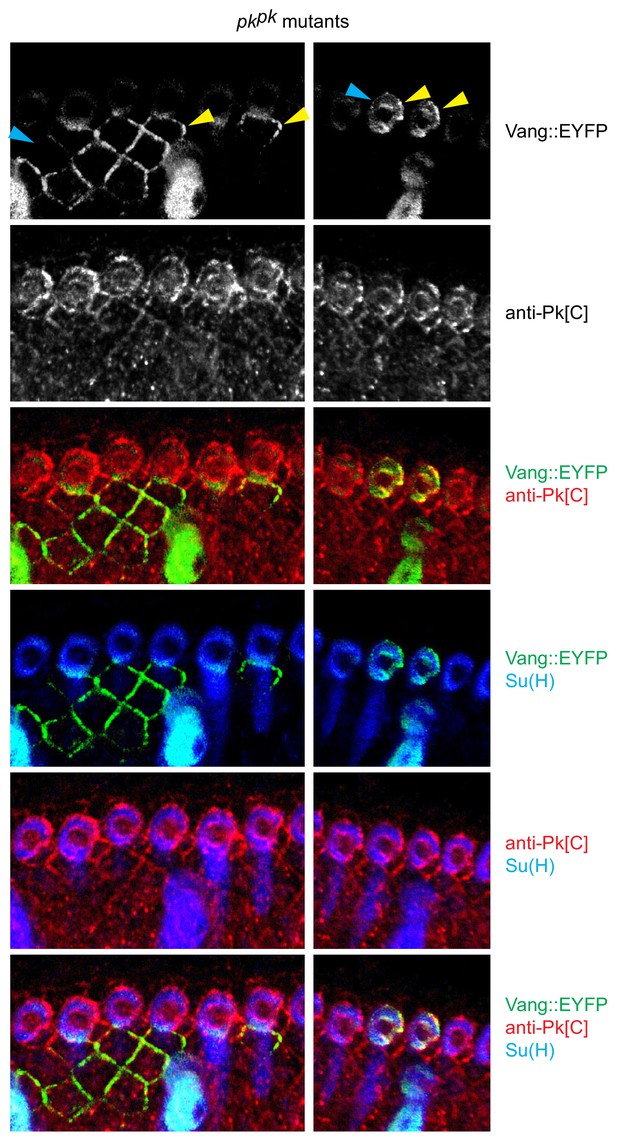
Reversed Vang localization inpkpkmutant socket and margin cells.
Vang::EYFP expression clones were induced in pkpk mutant wings (32h). In the distal region of the AWM, where bristle polarity is reversed, Vang::EFYP is enriched on the distal side of both socket cells and margin cells (yellow arrowheads) and depleted on the proximal sides (blue arrowheads), and colocalizes with Pksple (visualized with anti-Pk[C]), opposite to its proximal localization in control wings (compare to Figure 5L).
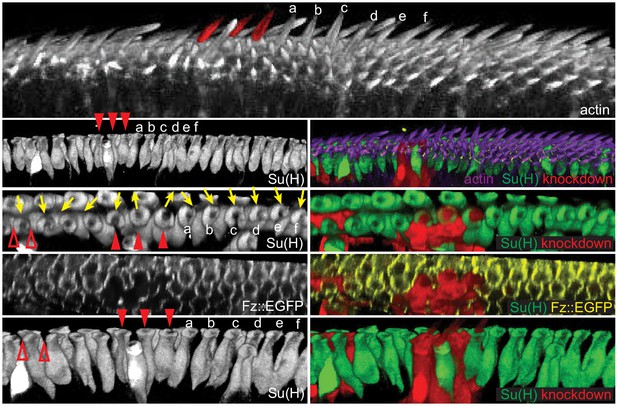
Polarity propagates between bristle and margin cells.
3D reconstructed views of a Fz::EGFP wing (32 hr) with fz knockdown clones, stained for actin to mark wing hairs and bristle shafts, Su(H) to mark socket cells and RFP to indicate knockdown clones. A clone involving three bristles (red arrowheads, false-colored in top image) shows domineering non-autonomy, reversing the polarity of nearby bristles and hairs on the distal side of the clone (bristles marked a-f). The effect on both hair cells and bristles diminishes with distance. A small clone affecting margin cells (red open arrowheads) disrupts the polarity of bristle cells on the distal side of the mutant cells. 20 fz RNAi clones from eight wings were analyzed for cell autonomous and non-autonomous effects. All clones showed cell autonomous polarity disruption and non-autonomous reversal of varying extent depending on the clone size.
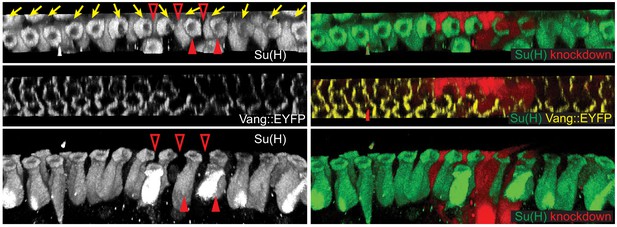
vangRNAi clone showing polarity propagation among bristles at the margin.
3D reconstructed views of a Vang::EYFP wing (32h) with vang knockdown clone, stained for Su(H) to mark socket cells and RFP to indicate knockdown clone. A clone involving 2 bristles (red arrowheads) and several margin cells (open arrowheads) shows domineering non-autonomy, reversing the polarity of bristles on the proximal side of the clone. The effect on bristle polarity diminishes with distance. 20 vang RNAi clones from 7 wings were analyzed for cell autonomous and non-autonomous effects of vang RNAi. All clones showed cell autonomous polarity disruption and non-autonomous reversal of varying extent depending on the clone size.
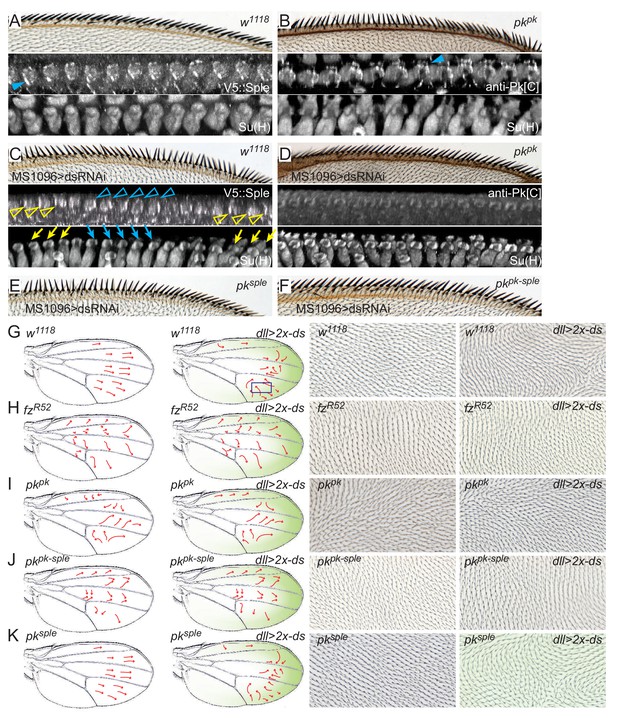
Pkpk and Pksple activity responds to Ds.
(A) A control (w1118) wing showing normally oriented bristles and rotated socket cells, and proximal V5::Sple. (B) A pkpk mutant wing with reversal of distal bristles, counter-rotated socket cells and distal Pksple. (C) Wings knocked down for ds (MS1096-GAL4, UAS-dsRNAi) show variable regions of locally correlated but either reversed or normal polarity. V5::Sple localization varies, corresponding to the local reversed (blue) or normal (yellow) polarity. (D) In pkpk mutant wings in which ds is knocked down, socket cells are minimally rotated, and Pksple, probed with anti-Pk[C] antibody, shows minimal apical localization. (E) pksple wing with ds knocked down. (F) pkpk-sple wing with ds knocked down. Quantification of socket cell rotation for the genotypes shown in C–F are given in Figure 7—figure supplement 1. (G–K) The normal proximal-high and distal-low Ds gradient was reversed in the distal wing by driving two copies of UAS-ds by dll-Gal4 in control (w1118), fzR52/R52, pkpk, pkpk-sple and pksple wings. The approximate gradient of ectopic ds expression is shown in green. Polarity reversal in the distal part of a control wing (G) was reversed (arrows, and images representing boxed regions) and depended on the presence of Fz (H) and Pkpk but not on Pksple (I–K). Wing hair images (G–K) are of the dorsal side. Subjectively similar hair polarity patterns were obtained from ≥13 of 15 wings for each genotype in G-K).
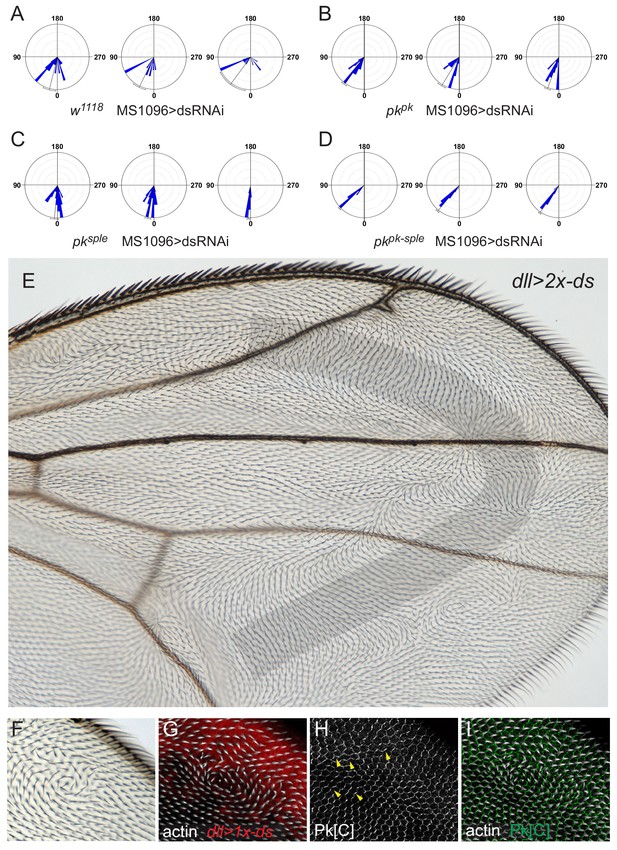
A reversed ectopic Ds gradient re-orients core PCP domains.
(A–D) Quantification of socket cell rotation for the genotypes (32 hr) from Figure 7C–F: w118, MS1096 >dsRNAi ((A), 74 sockets from four wings), pkpk, MS1096 >dsRNAi ((B), 72 sockets from three wings), pksple, MS1096 >dsRNAi ((C), 80 sockets from three wings), and pkpk-sple, MS1096 >dsRNAi ((D), 61 sockets from three wings). (E) Most of the distal portion of a dll >2 x-ds wing (n = 12), with shading indicating the expected steepest portion of the induced gradient. Note that the majority of this region exhibits distal-proximal polarity, consistent with a reversal of polarization rather than a random scrambling of polarity. (F) Re-orientation of core signaling is difficult to appreciate in regions of complete reversal because proteins remain at proximal-distal boundaries in a pattern indistinguishable from that in wildtype. However, in regions where polarity is redirected to an anterior or posterior direction, core proteins localize to the anterior-posterior boundaries. One copy of UAS-ds driven by dll-GAL4 (dll >1 x-ds) causes swirls with regions of anterior-posterior polarity near the distal end of the anterior margin. (G) Swirls form in the region of graded ectopic ds expression, marked with UAS-mCherry (red) in 33 hr pupal wings. (H) Domains of core protein localization, in this case, Pk (Pk[C]; white) are seen to relocalize in response to the ectopic ds expression (arrowheads). (I) Overlay of Pk[C] (green) and hairs (white) shows that Pk expression domains and hair polarity are tightly correlated and coordinately re-oriented. Pk and hair patterns are correlated from all pupal wings analyzed (n = 12) (F–I). Statistical analyses for all genotypes are in Table 2.
Tables
p values for comparison of rotation angles for control W1118 socket cells at different times apf.
| W1118 | 24 hr 5.0° | 28 hr 28.4° |
|---|---|---|
| 24 hr 5.0° | ||
| 28 hr 28.4° | <0.0001 | |
| 32 hr 54.588° | <0.0001 | <0.0001 |
Summary statistics for rotational angles.
CSD = Circular Standard Deviation.
| Genotype | Time | Grand Mean Vector (GM) | Length of Grand Mean Vector (r) | Number of means (wings) | Mean CSD |
|---|---|---|---|---|---|
| W1118 | 24 hr | 5.04° | 0.996 | 6 | 4.14 |
| 28 | 28.368° | 0.995 | 7 | 4.075 | |
| 32 | 54.588° | 0.981 | 6 | 2.940333 | |
| pkpk30 | 24 hr | 0.63° | 0.988 | 3 | 7.781333 |
| 32 hr | 348.482° | 0.946 | 7 | 15.09 | |
| pkpk-sple13 | 24 hr | 1.061° | 0.997 | 3 | 3.687 |
| 32 hr | 47.59° | 0.993 | 3 | 6.582 | |
| pksple1 | 24 hr | 17.8° | 0.979 | 3 | 7.244667 |
| 32 hr | 59.288° | 0.99 | 4 | 4.24425 | |
| MS1096 >> sple | 24 hr | 358.057° | 0.994 | 3 | 5.796 |
| 32 hr | 333.296° | 0.964 | 3 | 13.31267 | |
| fzR52 | 24 hr | 3.351° | 0.998 | 3 | 3.923333 |
| 32 hr | 20.471° | 0.905 | 6 | 23.5668 | |
| dsh1 | 24 hr | 5.684° | 0.996 | 2 | 5.023 |
| 32 hr | 20.144° | 0.944 | 6 | 14.316 | |
| fmi RNAi | 24 hr | 1.15° | 0.996 | 3 | 4.78 |
| 32 hr | 19.291° | 0.978 | 4 | 11.741 | |
| vangstbm6 | 24 hr | 1.51° | 0.967 | 3 | 11.52167 |
| 32 hr | 25.447° | 0.94 | 6 | 14.21117 | |
| w1118 MS1096 > dsRNAi | 32 hr | 16.593° | 0.79 | 4 | 33.10867 |
| pkpk MS1096 > dsRNAi | 32 hr | 29.323° | 0.961 | 3 | 14.37067 |
| pksple MS1096 > dsRNAi | 32 hr | 6.596° | 0.97 | 3 | 12.63867 |
| pkpk-sple MS1096 > dsRNAi | 32 hr | 40.297° | 0.99 | 3 | 6.219667 |
-
Table 2—source data 1
Source data for w1118.
- https://cdn.elifesciences.org/articles/51456/elife-51456-table2-data1-v2.xlsx
-
Table 2—source data 2
Source data for pk30.
- https://cdn.elifesciences.org/articles/51456/elife-51456-table2-data2-v2.xlsx
-
Table 2—source data 3
Source data for pkpk-sple13.
- https://cdn.elifesciences.org/articles/51456/elife-51456-table2-data3-v2.xlsx
-
Table 2—source data 4
Source data for pksple1.
- https://cdn.elifesciences.org/articles/51456/elife-51456-table2-data4-v2.xlsx
-
Table 2—source data 5
Source data for MS1096-GAL4; UAS-pksple.
- https://cdn.elifesciences.org/articles/51456/elife-51456-table2-data5-v2.xlsx
-
Table 2—source data 6
Source data for fzR52.
- https://cdn.elifesciences.org/articles/51456/elife-51456-table2-data6-v2.xlsx
-
Table 2—source data 7
Source data for dsh1.
- https://cdn.elifesciences.org/articles/51456/elife-51456-table2-data7-v2.xlsx
-
Table 2—source data 8
Source data for fmiRNAi.
- https://cdn.elifesciences.org/articles/51456/elife-51456-table2-data8-v2.xlsx
-
Table 2—source data 9
Source data for vangstbm6.
- https://cdn.elifesciences.org/articles/51456/elife-51456-table2-data9-v2.xlsx
-
Table 2—source data 10
Source data for w1118; MS1096-GAL4; UAS-dsRNAi.
- https://cdn.elifesciences.org/articles/51456/elife-51456-table2-data10-v2.xlsx
-
Table 2—source data 11
Source data for pkpk; MS1096-GAL4; UAS-dsRNAi.
- https://cdn.elifesciences.org/articles/51456/elife-51456-table2-data11-v2.xlsx
-
Table 2—source data 12
Source data for pksple; MS1096-GAL4; UAS-dsRNAi.
- https://cdn.elifesciences.org/articles/51456/elife-51456-table2-data12-v2.xlsx
-
Table 2—source data 13
Source data for pkpk-sple; MS1096-GAL4; UAS-dsRNAi.
- https://cdn.elifesciences.org/articles/51456/elife-51456-table2-data13-v2.xlsx
p values for comparison of rotation angles for control and pk-related genotypes at 32 hr apf. ns = not significant.
| 32 hr angles | W1118 54.6° | pksple1 59.3° | pkpk-sple13 47.6° | pkpk30 348.5° |
|---|---|---|---|---|
| W1118 54.6° | ||||
| pksple1 59.3° | (ns) 0.0702 | |||
| pkpk-sple13 47.6° | 0.0143 | 0.0343 | ||
| pkpk30 348.5° | <0.0001 | <0.0001 | 0.0004 | |
| >>Pksple 333.2° | 0.0012 | <0.0001 | 0.001 | (ns) 0.1826 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Drosophila melanogaster) | pkpk-sple13 | Gubb et al., 1999, PMID: 10485852 | BDSC:41790; FLYB:FBal0060943; RRID:BDSC_41790 | FlyBase symbol: pkpk-sple-13 |
| Genetic reagent (Drosophila melanogaster) | pkpk-sple14 | Gubb et al., 1999, PMID: 10485852 | FLYB:FBal0035401 | FlyBase symbol: pkpk-sple-14 |
| Genetic reagent (Drosophila melanogaster) | pkpk30 | Gubb et al., 1999, PMID: 10485852 | BDSC:44229; FLYB:FBal0101223; RRID:BDSC_44229 | FlyBase symbol: pk30 |
| Genetic reagent (Drosophila melanogaster) | pksple1 | Gubb et al., 1999, PMID: 10485852 | BDSC:422; FLYB:FBal0016024; RRID:BDSC_422 | FlyBase symbol: pksple-1 |
| Genetic reagent (Drosophila melanogaster) | vangA3 | Taylor et al., 1998, PMID: 9725839 | FLYB:FBal0093183 | FlyBase symbol: VangA3 |
| Genetic reagent (Drosophila melanogaster) | vangstbm6 | Wolff and Rubin, 1998, PMID: 9463361 | BDSC:6918; FLYB:FBal0062424; RRID:BDSC_6918 | FlyBase symbol: Vangstbm-6 |
| Genetic reagent (Drosophila melanogaster) | fzR52 | Krasnow and Adler, 1994, PMID: 7924994 | FLYB:FBal0004939 | FlyBase symbol: fz23 |
| Genetic reagent (Drosophila melanogaster) | dsh1 | Bloomington Drosophila Stock Center | BDSC:5298; FLYB:FBal0003138; RRID:BDSC_5298 | FlyBase symbol: dsh1 |
| Genetic reagent (Drosophila melanogaster) | UAS-pksple | Bloomington Drosophila Stock Center | BDSC:41780; FLYB:FBti0148928; RRID:BDSC_41780 | FlyBase symbol: P{UAS-sple+}3 |
| Genetic reagent (Drosophila melanogaster) | UAS-pkRNAi | Vienna Drosophila Resource Center | VDRC:v101480; FLYB:FBst0473353; RRID:FlyBase_FBst0473353 | FlyBase symbol: P{KK109294}VIE-260B |
| Genetic reagent (Drosophila melanogaster) | UAS-fmiRNAi | Bloomington Drosophila Stock Center | BDSC:26022; FLYB:FBti0114752; RRID:BDSC_26022 | Flybase symbol: P{TRiP.JF02047}attP2 |
| Genetic reagent (Drosophila melanogaster) | UAS-fzRNAi | Bloomington Drosophila Stock Center | BDSC:34321; FLYB:FBti0140932; RRID:BDSC_34321 | Flybase symbol: P{TRiP.HMS01308}attP2 |
| Genetic reagent (Drosophila melanogaster) | UAS-vangRNAi | Bloomington Drosophila Stock Center | BDSC:34354; FLYB:FBti0140967; RRID:BDSC_34354 | Flybase symbol: P{TRiP.HMS01343}attP2 |
| Genetic reagent (Drosophila melanogaster) | UAS-dsRNAi | Bloomington Drosophila Stock Center | BDSC:32964; FLYB:FBti0140473; RRID:BDSC_32964 | Flybase symbol: P{TRiP.HMS00759}attP2 |
| Genetic reagent (Drosophila melanogaster) | UAS-ds | Matakatsu and Blair, 2004, PMID: 15240556 | FLYB:FBtp0019964 | Flybase symbol: P{UAS-ds.T} |
| Genetic reagent (Drosophila melanogaster) | dll-GAL4 | Bloomington Drosophila Stock Center | BDSC:3038; FLYB:FBti0002783; RRID:BDSC_3038 | Flybase symbol: P{GawB}Dllmd23 |
| Genetic reagent (Drosophila melanogaster) | MS1096-GAL4 | Bloomington Drosophila Stock Center | BDSC:8860; FLYB:FBti0002374; RRID:BDSC_8860 | Flybase symbol: P{GawB}BxMS1096 |
| Genetic reagent (Drosophila melanogaster) | armP-fz::EGFP | Strutt, 2001, PMID:11239465 | FLYB:FBtp0014592 | Flybase symbol: P{arm-fz.GFP} |
| Genetic reagent (Drosophila melanogaster) | actP-vang::EYFP | Strutt, 2002, PMID: 12137731 | FLYB:FBtp0015854 | Flybase symbol: P{Act5C(-FRT)stbm-EYFP} |
| Genetic reagent (Drosophila melanogaster) | actP > CD2>vang::EYFP | Strutt, 2002, PMID: 12137731 | FLYB:FBtp0084387 | Flybase symbol: P{Act5C(FRT.polyA)stbm-EYFP} |
| Genetic reagent (Drosophila melanogaster) | ci-GAL4 | Croker et al., 2006, PMID: 16413529 | FLYB:FBtp0057188 | Flybase symbol: P{ci-GAL4.U} |
| Genetic reagent (Drosophila melanogaster) | UAS-mCherry | Bloomington Drosophila Stock Center | BDSC:38424; FLYB:FBti0147460; RRID:BDSC_38424 | Flybase symbol: P{UAS-mCherry.NLS}3 |
| Genetic reagent (Drosophila melanogaster) | actP > CD2>Gal4 | Bloomington Drosophila Stock Center | BDSC:30558; FLYB:FBti0012408; RRID:BDSC_30558 | Flybase symbol: P{GAL4-Act5C(FRT.CD2).P}S |
| Genetic reagent (Drosophila melanogaster) | UAS-RFP | Bloomington Drosophila Stock Center | BDSC:30558; FLYB:FBti0129814; RRID:BDSC_30558 | Flybase symbol: P{UAS-RFP.W}3 |
| Antibody | goat polyclonal anti-Su(H) | Santa Cruz | Santa Cruz:sc-15183 RRID:AB_672840 | 1/200 (immunolabelling) |
| Antibody | Mouse monoclonal anti-V5 | Thermo-Fisher | Thermo_Fisher:R960-25, RRID:AB_2556564 | 1/200 (immunolabelling) 1/1000 (Western blotting) |
| Antibody | Guinea pig polyclonal anti-Pk[C] | Olofsson et al., 2014, PMID: 25005476 | N/A | 1/800 (immunolabelling) 1/1000 (Western blotting) |
| Antibody | Rat monoclonal anti-dEcad | DSHB | RRID:AB_528120 | 1/200 (immunolabelling) |
| Antibody | Mouse monoclonal anti-γ-Tubulin | Sigma-Aldrich | Sigma-Aldrich: T6557 RRID:AB_477584 | 1/1000 (Western blotting) |
| Recombinant DNA reagent | pCFD4 | Addgene | RRID:Addgene_49411 | CRISPR gRNA backbone |
| Recombinant DNA reagent | pDsRedattp | Addgene | RRID:Addgene_51019 | Donor recombinant DNA backbone |
| Recombinant DNA reagent | pCR-Blunt-II-TOPO | Thermo-Fisher | RRID:Addgene_29705 | Backbone for sub-cloning |
| Sequence-based reagent | pkpk gRNA 1 | This paper | gRNA sequence in PCR primers | ATGGCTCAGGCCCGATCTAG |
| Sequence-based reagent | pkpk gRNA 2 | This paper | gRNA sequence in PCR primers | GTGGATCAACCCCTGGAAAC |
| Sequence-based reagent | pksple gRNA 1 | This paper | gRNA sequence in PCR primers | CTCGTAAATTTAGCTTCGAG |
| Sequence-based reagent | pksple gRNA 2 | This paper | gRNA sequence in PCR primers | AGATGCAATTTGGCCGCCCT |






