An efficient CRISPR-based strategy to insert small and large fragments of DNA using short homology arms
Figures
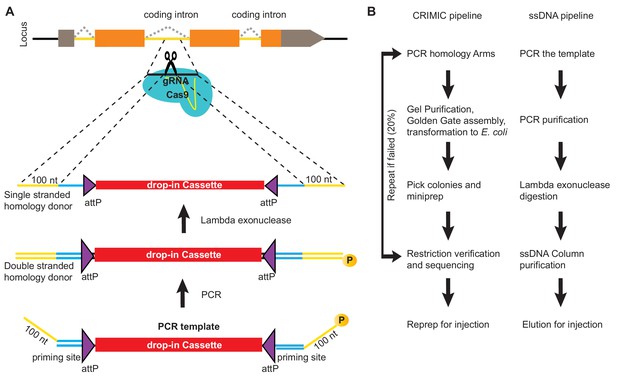
ssDNA pipeline is faster than cloning CRIMIC constructs.
(A) Schematic of PCR-based generation of drop-in ssDNA constructs. Gray boxes, UTRs; orange boxes, coding exons; yellow line, coding introns; black line, outside coding introns and exons. (B) Comparison of donor generation pipelines for CRIMIC and PCR-based drop-in ssDNA homology donors. Making ssDNA donors is about 5X faster than making CRIMIC donors.
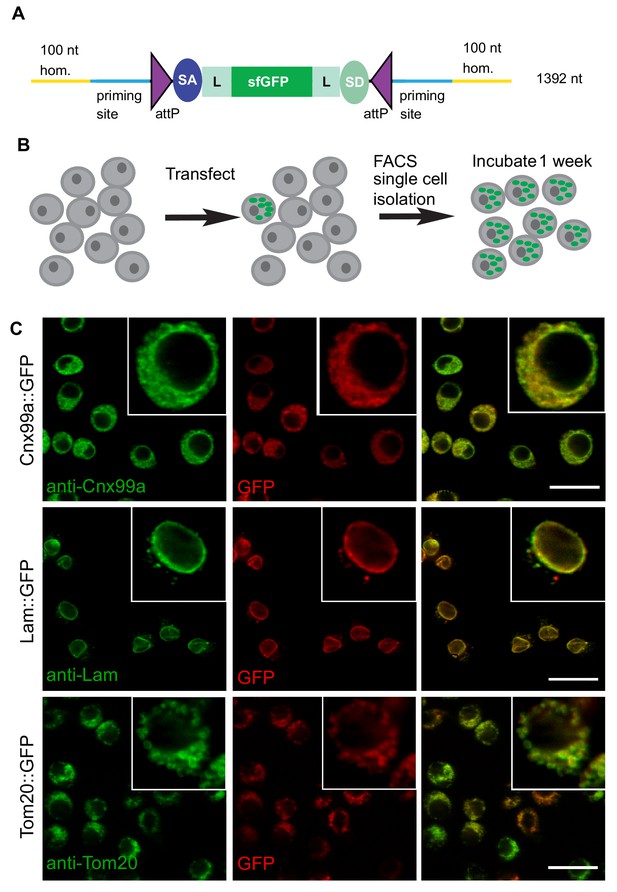
ssDNA homology donors are effective in S2 cells to tag organelles.
(A) Schematic of drop-in cassette encoding for sfGFP artificial exon. Size of the construct including the homology arms is indicated on the right. sfGFP: superfolderGFP; SA: Splice Acceptor of mhc; SD: Splice Donor of mhc; L: flexible linker that consists of four copies of Gly-Gly-Ser. (B) Diagram of steps to isolate cell clones resulting from successful homologous recombination events. (C) Examples of S2R+ cells with organelles marked with GFP. Left panel, antibody staining; middle panel, GFP signal; right panel, the merge.
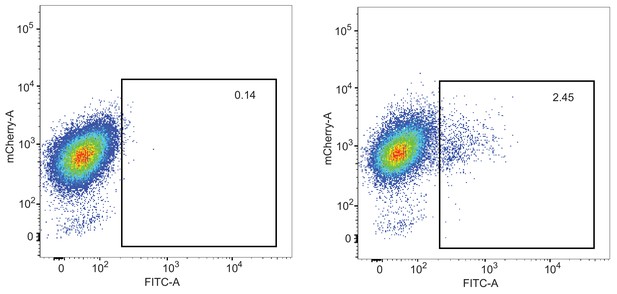
FACS data of control cells (left) and ssDNA knock-in cells (right).
All cell lines express mCherry::Clic, which is present in the parental cell line and thus in these derivatives. Single cell clones with GFP expression levels greater than 2 × 102 were retained.

Detection of subcellular localization of GFP tagged proteins in S2 cells.
Left panels, confocal fluorescence detection of GFP fusion proteins in single-cell isolated clones. The specific proteins tagged by GFP knock-in are indicated. Center panels, confocal fluorescence detection of mCherry::Clic, which is present in the parental cell line and thus in these derivatives. Right panels, merged image.
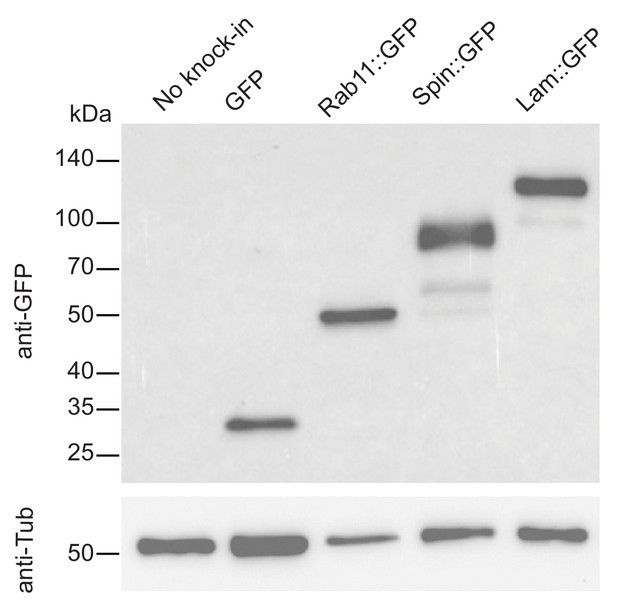
Western blot analysis of tagged proteins observed in S2R+ cells.
Western blot analysis of selected clones using anti-GFP antibody shows expected protein sizes.
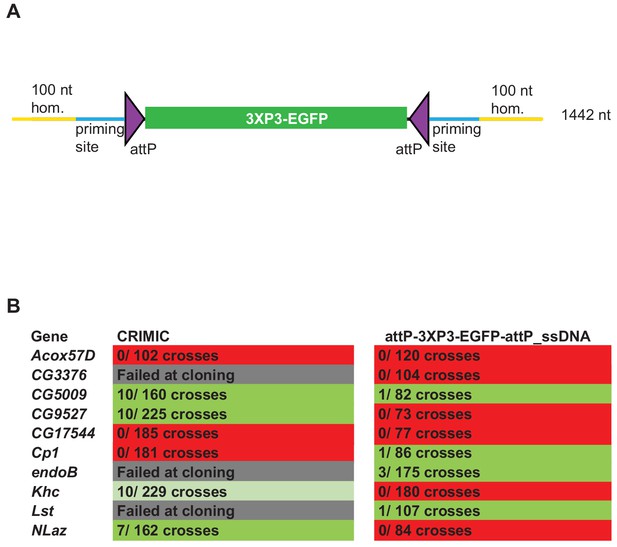
ssDNA constructs are not as efficient as double stranded CRIMIC constructs for fly transformation.
(A) Schematic of drop-in cassette used for fly transformation. Size of the construct including homology arms is indicated on the right. (B) Injection results for the 10 genes selected for comparison of transformation efficacy. Numbers indicate positive events/fertile G0 single fly crosses. Red, no positive events; light green, positive non-confirmed events; dark green, genes with PCR-confirmed events.
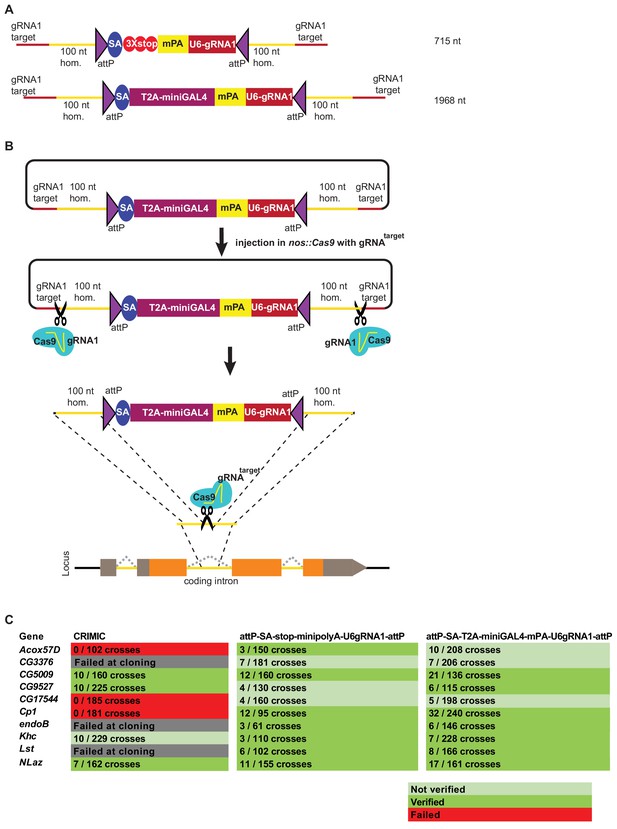
Double stranded DNA synthetic constructs are efficient for fly transformation.
(A) Schematic of synthesized plasmid drop-in donors. mPA indicates minipolyA. (B) Schematic of the donor plasmid followed by linearization by Cas9 in germ cells and integration of donors in vivo. gRNAtarget is gene specific gRNA. (C) Injection results for the 10 genes selected for comparison of transformation efficacy. Numbers indicate positive events/fertile G0 single fly crosses. Red, no positive events; light green, positive non-confirmed events; and dark green, genes with PCR-confirmed events.
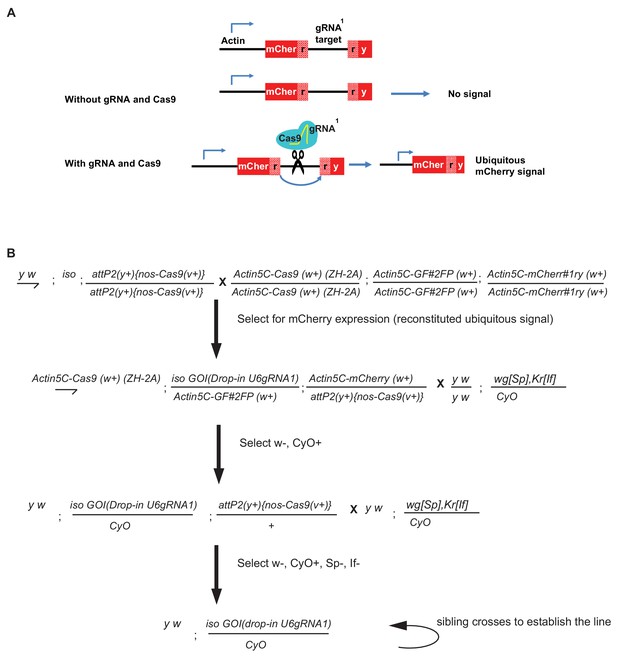
Schematic and crossing scheme for CRISPR gRNA-based dominant marker strategy.
(A) Schematics of the action of gRNA1-based detection of dominant marker. (B) Crossing scheme for screening the transgenics using U6gRNA1 as dominant marker. The crosses for a second chromosome insertion are shown; for insertions on other chromosomes, an appropriate balancer stock would be used in the second and third crosses.
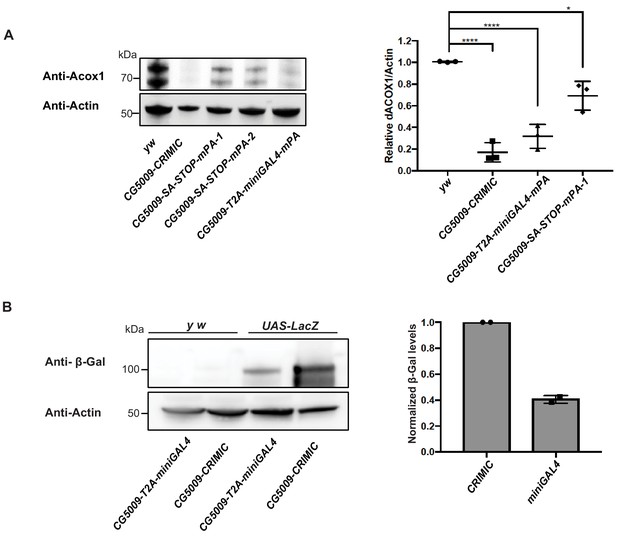
T2A-miniGAL4 gene trap cassette is mutagenic and expresses an active GAL4.
(A) Western blot and quantification of the level of ACOX1 protein in flies homozygous for SA-T2A-miniGAL4-minipolyA, SA-3XSTOP-minipolyA or CRIMIC construct for CG5009. ****p<0.0001, *p<0.01. mPA1 and mPA2 indicate two independent lines of CG5009-SA-3XSTOP-minipolyA. (B) Western blot of β-galactosidase and quantification of heterozygous flies carrying a copy of the miniGAL4 construct compared to heterozygous flies carrying a CRIMIC.
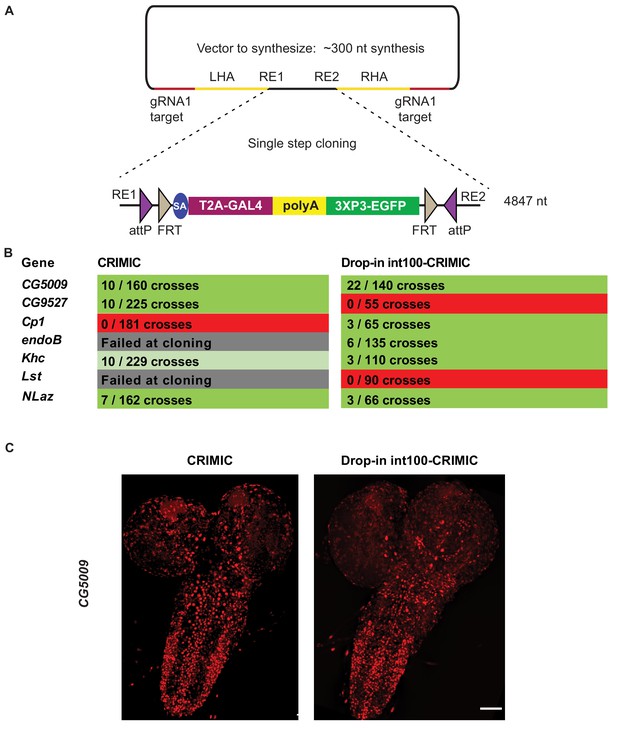
Single step cloning method allows efficient insertion of the CRIMIC cassette in coding introns.
(A) Schematic of a single step cloning vector pUC57. LHA Left Homology Arm, RE1 Restriction Enzyme 1, RE2 restriction Enzyme 2, RHA Right Homology Arm. (B) Injection results for the seven genes selected to estimate transformation efficacy. Numbers indicate positive events/G0 single fly crosses. (C) Third instar larval brain expression domain of CG5009 as determined by crossing conventional CRIMIC or drop-in int100-CRIMIC flies to UAS-NLS-mCherry reporter lines. Scale bar is 100 µm.
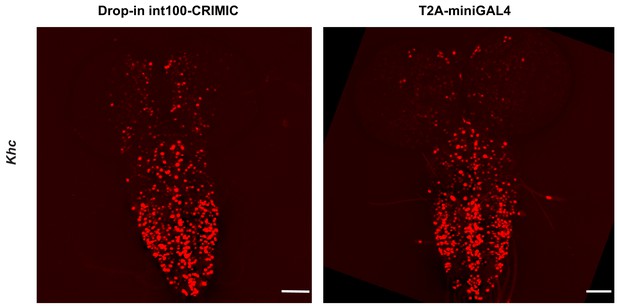
Comparison of expression domain obtained by T2A-miniGAL4 and drop-in int100-CRIMIC.
Third instar larval brain expression domain of Khc as determined by crossing conventional T2A-miniGAL4 or drop-in int100-CRIMIC flies to UAS-NLS-mCherry reporter lines. Scale bars are 100 µm.
Videos
Polo-sfGFP knock-in in S2R+ cells through ssDNA drop-in faithfully reports dynamic localization of Polo throughout the cell cycle.
https://doi.org/10.7554/eLife.51539.008Tables
Summary of ssDNA drop-in mediated homologous recombination in S2R+ cells
https://doi.org/10.7554/eLife.51539.007| Organelle | Fly protein | clones obtained | population imaged | clones imaged | insertion sequence verified | Immunostained | Correct GFP localization | DGRC stock# | S2R+ expression (modENCODE RPKM) |
|---|---|---|---|---|---|---|---|---|---|
| Autophagosomes | Atg8a | 14 | ✔ | ✔ | N | N | N | N | 153 |
| Autophagosomes/aggregates | Ref(2)P | 9 | ✔ | ✔ | Y | a-Ref2P, a-FK2 | N | N | 138 |
| Endoplasmic reticulum (ER) | Calnexin99A | 16 | ✔ | ✔ | Y | a-Cnx99a | Y | 273 | 235 |
| Endoplasmic reticulum (ER), transitional | Sec23 | 30 | ✔ | ✔ | Y | N | * | 294 | 101 |
| Endosomes, early | Rab5 | 9 | ✔ | ✔ | N | N | N | N | 98 |
| Endosomes, recycling | Rab11 | 23 | ✔ | ✔ | Y | N | * | 274 | 302 |
| G-Bodies (cytoplasmic puncta) | Pfk | 14 | ✔ | ✔ | N | N | N | N | 23 |
| Golgi (cis-Golgi) | Gmap | 10 | ✔ | ✔ | Y | a-GMAP | Y | 276, 277 | 15 |
| Golgi (trans-Golgi) | Sec71 | 10 | ✔ | ✔ | N | N | N | N | 8 |
| Golgi (trans-Golgi) | Golgin245 | 1 | ✔ | ✔ | Y | a-Golgin245 | Y | 280 | 27 |
| Kinetochore | Polo | 2 | ✔ | ✔ | Y | N | Y | 275 | 50 |
| Lipid droplets | Seipin | 12 | ✔ | ✔ | N | N | N | N | 9 |
| Lysosomes | spin | 2 | ✔ | ✔ | Y | a-Arl8 | Y | 293 | 112 |
| Lysosomes | Arl8 | 9 | ✔ | ✔ | Y | a-Arl8 | Y | 291 | 78 |
| Mitochondria | Tim17b | 3 | ✔ | ✔ | N | a-ATP5A | N | N | 266 |
| Mitochondria | Tom20 | 17 | ✔ | ✔ | Y | a-ATP5A | Y | 302 | 117 |
| Nuclear membrane, inner | dLBR | 0 | ✔ | N | N | N | N | N | 42 |
| Nuclear membrane, inner | Lamin | 53 | ✔ | ✔ | Y | a-Lamin | Y | 292 | 249 |
| Nucleolus | Fibrillarin | 14 | ✔ | ✔ | Y | a-Fib | Y | 278, 279 | 53 |
| Peroxisomes | Pmp70 | 6 | ✔ | ✔ | N | N | N | N | 26 |
-
* = distribution was as expected, but no antibody available to test by co-stain
Additional files
-
Supplementary file 1
Sequences of drop-in constructs.
- https://doi.org/10.7554/eLife.51539.015
-
Supplementary file 2
Protocol for designing drop-in int100-CRIMIC constructs.
- https://doi.org/10.7554/eLife.51539.016
-
Supplementary file 3
List of oligos and ultramers used in this study.
- https://doi.org/10.7554/eLife.51539.017
-
Transparent reporting form
- https://doi.org/10.7554/eLife.51539.018





