Perturbation of base excision repair sensitizes breast cancer cells to APOBEC3 deaminase-mediated mutations
Figures
A3B activity is not the only determinant of repair-induced mutations.
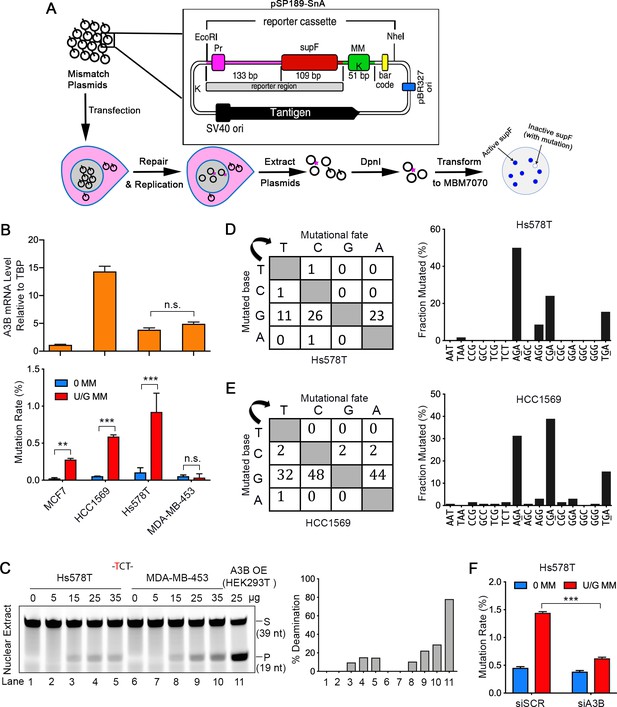
(A) Schematic depicting the shuttle vector assay to detect U/G MM repair-induced mutations. MM, no mismatch or U/G mismatch. K depicts location of KpnI site. (B) Upper panel: qRT-PCR of A3B relative to the housekeeping gene TBP. Lower panel: mutation rate (scored as % of white/total colonies) induced by U/G mismatch repair in MCF7, HCC1569, Hs578T, and MDA-MB-453 breast cancer cell lines. 0 MM, no mismatch; U/G MM, U/G mismatch. Error bars represent s.d., n = 2 for MCF7, HCC1569 and MDA-MB-453 cells; n = 5 for Hs578T cells. **P < 0.01; ***P < 0.001; n.s., no significant difference by two-tailed unpaired Student’s t test. (C) Concentration gradient of in vitro deaminase assay using nuclear extracts from Hs578T and MDA-MB-453 cells against a -TCT-containing fluorescein-labeled single strand oligonucleotide (39 nt). The amounts of total protein used are listed on top of the gel. The right panel shows quantification of the deamination percentage. The deamination activity is specific for -TCT- (Figure 1—figure supplement 1B). The time course deamination is shown in Figure 1—figure supplement 1C. S, substrate; P, product. (D and E) Mutation matrices and 5’-Trinucleotide context of mutations induced by U/G MM repair in Hs578T (D) and HCC1569 (E) cells. C is the most frequently mutated base and 70% of the mutated bases are in a 5’-GA (reverse complement of 5’-TC) motif. (F) A3B deficiency decreases U/G mismatch repair-induced mutagenesis. 0 MM, no mismatch; U/G MM, U/G mismatch. Error bars represent s.d., n = 3. ***P < 0.001 by two-tailed unpaired Student’s t test.

Shuttle vector-based assay of repair-induced mutations and A3 deaminase activity in breast cancer cell lines.
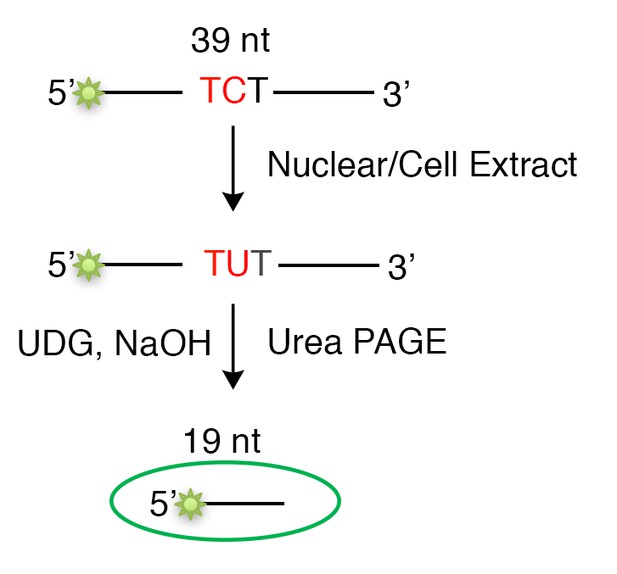
(A) Nicking and ligation controls. The shuttle vector pSP189-SnA contains 2 KpnI restriction sites (marked as ‘K’ on the mismatch plasmid in Figure 1A), one of which is in the mismatch region (MM). Removal of the top strand after nicking by Nt.BbvCI generates a gapped plasmid that migrates as a single band after KpnI treatment. Insertion of either the original (control, 0 MM) or a U-containing oligonucleotide (U/G MM) restores the KpnI site and results in two fragments upon KpnI digestion. Klenow treatment (see Materials and methods part) eliminates residual gapped plasmids, which otherwise are highly mutagenic. (B) In vitro deamination assay by nuclear extracts from four breast cancer cell lines shows specificity on a 39 nt -TCT-containing single strand substrate. An -ACT-containing substrate was used as a negative control. Whole cell extract from HEK293T expressing A3B-3HA (A3B OE) was used as a positive control. S, substrate; P, product. (C) Time course of deamination by nuclear extracts from Hs578T and MDA-MB-453 cells using the -TCT-containing substrate. Whole cell extract from HEK293T expressing A3B-3HA (A3B OE) was used as a positive control. The right panel shows the deamination percentage. S, substrate; P, product. (D) Generation of APOBEC3-mediated mutations. Downstream processing of BER hijacked by MMR exposes a C in a 5’-TC-3’ context on the bottom strand, which is a substrate for A3 deaminase converting C to U. U is copied to A by replication, or if it is removed by a glycosylase to generate an AP site, then copied to an A (the A rule) but also to T or C. Thus, the original G on the top strand will be mutated to T, C, or A. In this scenario the originally introduced U/G is restored to C/G.
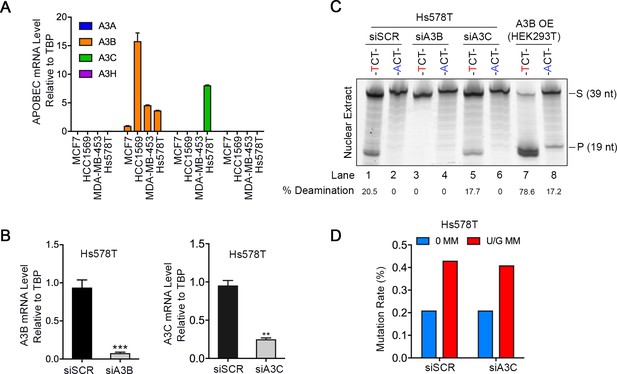
A3B, but not A3C, is correlated with the repair-induced mutagenesis.
(A) qRT-PCR of A3A, A3B, A3C, and A3H relative to the housekeeping gene TBP in MCF7, HCC1569, MDA-MB-453, and Hs578T breast cancer cell lines. Hs578T and MDA-MB-453 have similar A3B levels. A3A, A3C, and A3H are undetectable in all cell lines except for A3C in Hs578T. (B) qRT-PCR shows knockdown efficiency of A3B (left) and A3C (right) by siRNA (10 nM) in Hs578T cells relative to TBP. siSCR, scramble siRNA. Error bars represent s.d., n = 3. **P < 0.01; ***P < 0.001 by two-tailed unpaired Student’s t test. (C) In vitro deamination assay of nuclear extracts from Hs578T cells with A3B or A3C knocked down by siRNA. Fluorescein-labeled single strand oligos (39 nt) with 5’-TCT or 5’-ACT (negative control) were used. Knockdown of A3B but not A3C reduces the deaminase activity of nuclear extracts. The nuclear extract from HEK293T expressing A3B (A3B OE) serves as a positive control. The percentage of deamination (cleaved products relative to total) are listed at the bottom. S, substrate; P, product. (D) A3C knockdown does not affect U/G MM repair-induced mutation rate in Hs578T cells. 0 MM, no mismatch; U/G MM, U/G mismatch. siSCR, scramble siRNA.
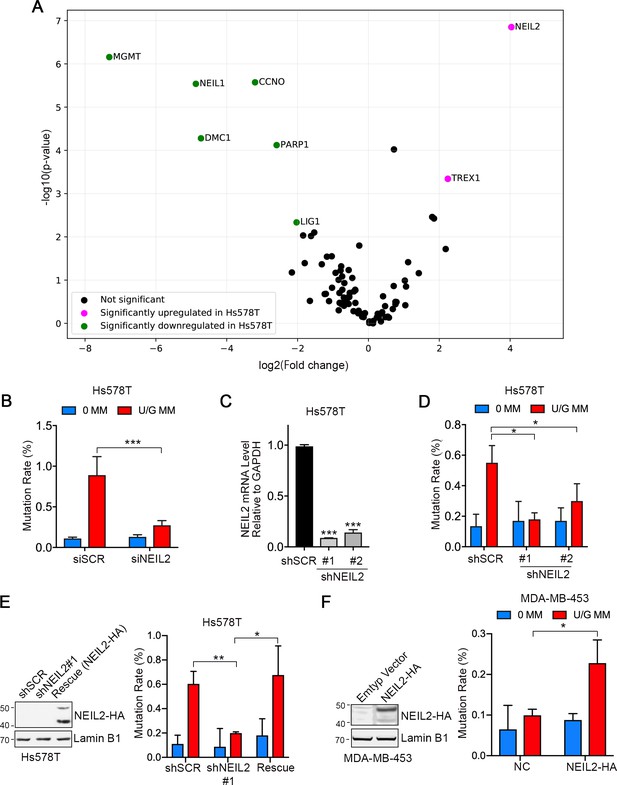
NEIL2 facilitates repair-induced mutagenesis.
(A) Volcano plot of transcription levels of 84 DNA repair enzymes determined by RT2 Profiler PCR Array for Hs578T cells (high mutation rate cell line) relative to MDA-MB-453 cells (low mutation rate cell line). Genes significantly downregulated and upregulated in Hs578T cells are highlighted in green and magenta, respectively. Data were generated from four independent determinations (Figure 2—source data 1). (B) U/G repair-induced mutation using the shuttle vector assay upon NEIL2 knockdown by siRNA in Hs578T cells. NEIL2 depletion decreased U/G MM repair-induced mutagenesis. siSCR, scramble siRNA; 0 MM, no mismatch; U/G MM, U/G mismatch. Error bar represents s.d., n = 3. ***P < 0.001 by two-tailed unpaired Student’s t test. (C) qRT-PCR shows knockdown efficiency of NEIL2 relative to GAPDH in NEIL2-stable-knockdown Hs578T cell lines (shNEIL2#1 and shNEIL2#2). shSCR, scramble shRNA; shNEL2#1 targets NEIL2 3’UTR; shNEIL2#2 targets NEIL2 ORF. Error bars represent s.d., n = 3. ***P < 0.001 by two-tailed unpaired Student’s t test. (D) U/G repair-induced mutation using the shuttle vector assay in NEIL2-stable-knockdown Hs578T cell line. NEIL2 depletion decreased U/G MM repair-induced mutagenesis. 0 MM, no mismatch; U/G MM, U/G mismatch. Error bars represent s.d., n = 3. *P < 0.05 by two-tailed unpaired Student’s t test. (E) Rescue of NEIL2 by NEIL2-HA overexpression vector restores U/G mismatch repair-induced mutation rate in NEIL2-stable-knockdown Hs578T cell line shNEIL2#1 (targets NEIL2 3’UTR). Western blot (left panel) shows NEIL2-HA overexpression for rescue of NEIL2 in shNEIL2-#1 Hs578T cell line. Lamin B1 serves as a loading control. Error bars represent s.d., n = 3. *P < 0.05; **P < 0.01 by two-tailed unpaired Student’s t test. (F) Overexpression of NEIL2-HA (pPM-NEIL2-3’HA) in MDA-MB-453 cells increased U/G MM repair-induced mutation. Western blot (left panel) shows NEIL2-HA overexpression level in MDA-MB-453 cells. Lamin B1 serves as a loading control. Error bar represents s.d., n = 3. *P < 0.05 by two-tailed unpaired Student’s t test.
-
Figure 2—source code 1
Python script for generating the volcano plot in Figure 2A.
qRT-PCR data from RT2 Profiler PCR array of Hs578T and MDA-MB-453 is used as the source data (Figure 2—source data 1).
- https://cdn.elifesciences.org/articles/51605/elife-51605-fig2-code1-v2.py
-
Figure 2—source data 1
Original data for DNA repair enzymes screening.
RT2Profiler PCR screening for 84 human DNA repair genes.
- https://cdn.elifesciences.org/articles/51605/elife-51605-fig2-data1-v2.docx
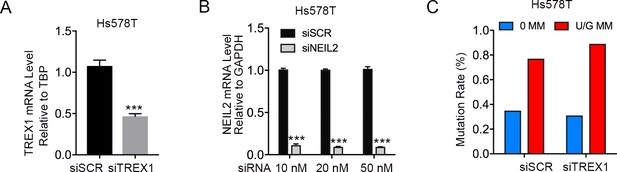
TREX1 knockdown does not affect repair-induced mutagenesis.
(A) qRT-PCR of TREX1 (10 nM siRNA) relative to the housekeeping gene TBP in Hs578T cells. siSCR, scramble siRNA. Error bars represent s.d., n = 3. ***P < 0.001 by two-tailed unpaired Student’s t test. (B) qRT-PCR shows knockdown efficiency of NEIL2 relative to GAPDH by siRNA (final concentration 10 nM, 20 nM, and 50 nM) in Hs578T cells. Error bars represent s.d., n = 3. ***P < 0.001 by two-tailed unpaired Student’s t test. (C) siRNA knockdown of TREX1 does not affect U/G MM repair-induced mutagenesis in Hs578T cells. 0 MM, no mismatch; U/G MM, U/G mismatch.
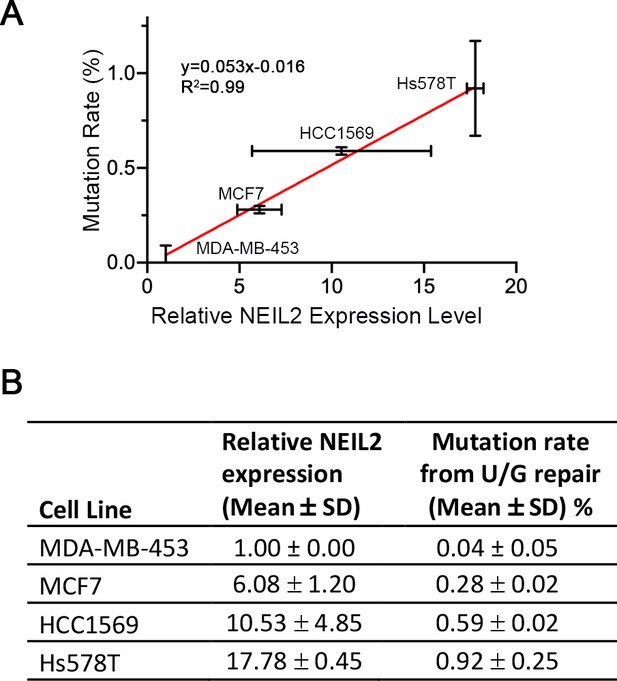
Repair-induced mutation rate is positively correlated with NEIL2 expression in breast cancer cells.
(A) U/G mismatch repair-induced mutation rate as a function of the NEIL2 expression (qRT-PCR) in MCF7, HCC1569, and Hs578T relative to that in MDA-MB-453 cells. Error bars represent s.d., n = 2. (B) Original data for (A). The mutation rate data was taken from Figure 1B. The NEIL2 expression data for Hs578T and MDA-MB453 was taken from Figure 2A and Figure 2—source data 1. NEIL2 expression data for MCF7 and HCC1569 were also obtained using the RT2Profiler PCR array. The relative NEIL2 expression level was normalized to that in MDA-MB-453 cells.
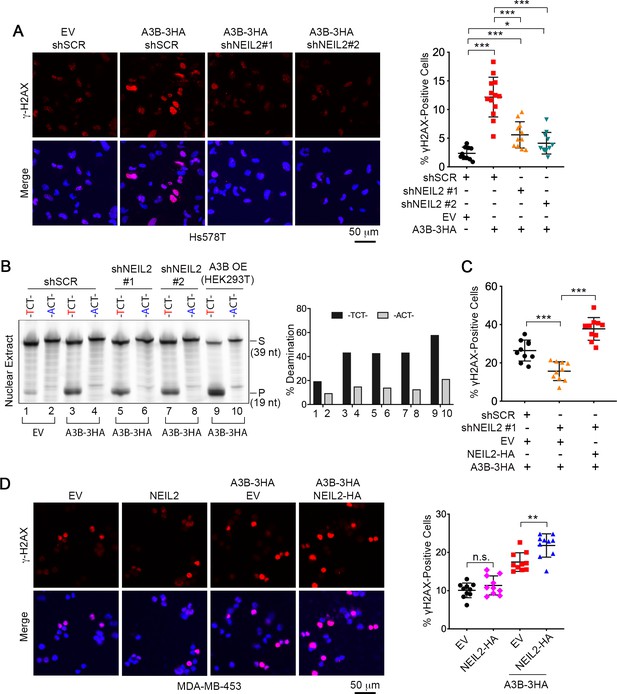
NEIL2 participates in A3B-mediated genomic DNA damage.
(A) Immunostaining of γH2AX foci in NEIL2-stable-knockdown Hs578T cell lines (shNEIL2#1 and shNEIL2#2) transfected with A3B-3HA. NEIL2 knockdown decreases the A3B-mediated γH2AX foci. EV, empty vector; shSCR, scramble shRNA. Scale bar, 50 μm. Right panel: Percentage of γH2AX foci, showing mean ± s.d., in at least 10 randomly selected microscopic fields in two replicate experiments for each condition. ***P < 0.001 by two-tailed unpaired Student’s t test. (B) In vitro deamination assay of nuclear extracts from NEIL2-stable-knockdown Hs578T cell lines with or without A3B-3HA expression. The substrate was a fluorescein-labeled single-stranded oligonucleotide (39 nt) containing -TCT- or -ACT- (negative control). Nuclear extract from HEK293T expressing A3B-3HA (A3B OE) was used as a positive control. NEIL2 knockdown does not affect A3B deaminase activity. Right panel: Quantifications of the cleaved products relative to total DNA loaded onto gel. S, substrate; P, product. (C) Quantification of the percentage of cells with γH2AX foci in NEIL2-stable-knockdown Hs578T cell line (shNEIL2#1) in the absence or presence of a NEIL2 expression vector pcDNA3.1(+)-NEIL2-3’HA. NEIL2 restoration increases A3B-triggered γH2AX foci. Data are represented as mean ± s.d. (n = 10 randomly selected microscopic fields in two replicate experiments). ***P < 0.001 by two-tailed unpaired Student’s t test. The corresponding images of γH2AX foci are shown in Figure 3—figure supplement 1C. (D) Immunostaining of γH2AX foci in MDA-MB-453 cells overexpressing A3B-3HA and NEIL2-HA. Percentage of cells with γH2AX foci is shown in the right panel. EV, empty vector. Scale bar, 50 μm. Data are represented as mean ± s.d. (n = 10 randomly selected microscopic fields in two replicate experiments). **P < 0.01; n.s., no significant difference by two-tailed unpaired Student’s t test.
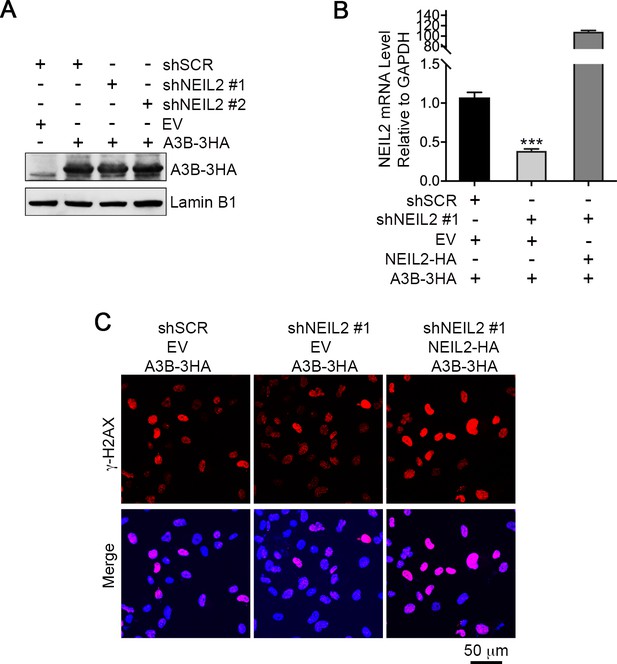
NEIL2 is required for A3B-mediated genomic DNA damage.
(A) Western blot of A3B-3HA expression in NEIL2-stable-knockdown Hs578T cell lines shNEIL2#1 and shNEIL2#2. LaminB1 serves as a loading control. shSCR, scramble shRNA. EV, empty vector. (B) qRT-PCR shows NEIL2 rescue efficiency in NEIL2-stable-knockdown Hs578T cell line shNEIL2#1 (targets NEIL2 3’UTR) with NEIL2-HA overexpression. Error bars represent s.d., n = 3. ***P < 0.001 by two-tailed unpaired Student’s t test. (C) Immunostaining of γH2AX foci in NEIL2-stable-knockdown Hs578T cell line shNEIL2#1 with rescue by exogenous expression of NEIL2-HA. shSCR, scramble shRNA; shNEL2#1 targets NEIL2 3’UTR; EV, empty vector. Scale bar, 50 μm. The quantification data is shown in Figure 3C.
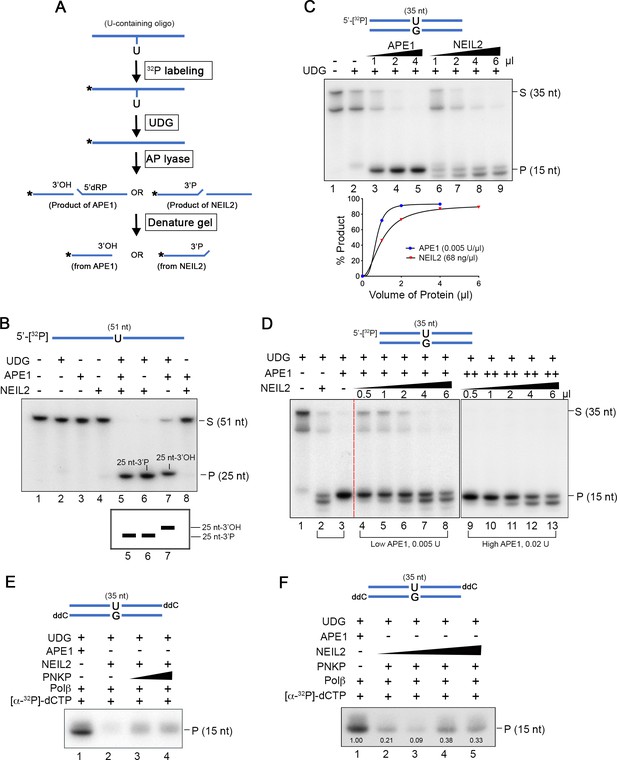
NEIL2 outcompetes APE1 at AP sites and the NEIL2 product is poor Polβ substrate.
(A) Diagram of AP lyase assay. (B) NEIL2 outcompetes APE1 on AP sites generated from 5’-[32P]-U-containing ssDNA (51 nt) by UDG. Amounts of NEIL2-His6 or APE1 are just sufficient to completely cleave the AP site (lane 7 of Figure 4—figure supplement 3A,B). The NEIL2 product retains a 3’P and migrates faster than 3’OH-terminated APE1 product. When both NEIL2 and APE1 are present, only the NEIL2 product is generated (lane 5). S, substrate; P, product. (C) Concentration gradient and product accumulation curves of APE1 and NEIL2 on 5’-[32P]-U-containing dsDNA (35 nt) in the presence of UDG. The volumes of NEIL2 (68 ng/µl) and APE1 (0.005 U/µl) used are listed in the figure. (D) NEIL2 competes with APE1 on 5’-[32P]-U-containing dsDNA (35 nt) in the presence of UDG. The reactions contained either 0.005 U (Low APE1) or 0.02 U APE1 (High APE1) and increasing amounts of NEIL2-His6. Lanes 2 and 3 contain respectively 4 μl NEIL2 (68 ng/μl) and 0.02 U APE1. The APE1 cleavage pattern was converted to the NEIL2 pattern with increasing amounts of NEIL2. S, substrate; P, product. (E) Incorporation of [α-32P]-dCTP by Polβ for products generated by APE1, NEIL2, and NEIL2 and PNKP on di-deoxynucleotide (ddC)-modified oligonucleotide in the presence of UDG. P, product. (F) Incorporation of [α-32P]-dCTP by Polβ in the presence of UDG and APE1 as a function of NEIL2 (68 ng/μl, 0, 1, 2, 5, 10 μl) and constant PNKP (127 ng/μl, 2 μl). As the NEIL2 product increasingly dominates the reaction, Polβ incorporation decreases. The number under each band gives the intensity relative to that in the first lane. P, product.
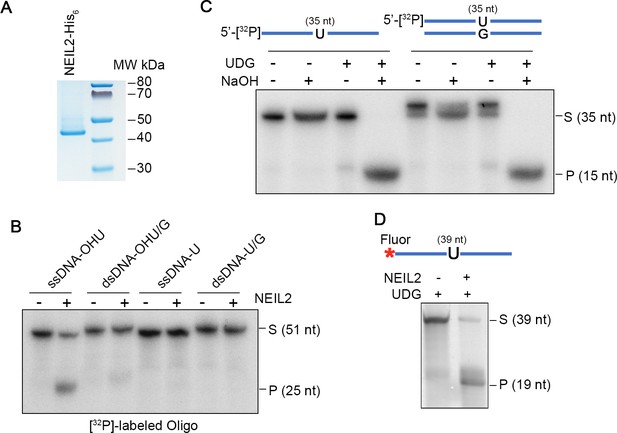
Purification and activity of NEIL2.
(A) Purified NEIL2-His6 (50 ng) from E. coli was subjected to NuPAGE and stained with Coomassie blue. (B) Activity of purified NEIL2-His6 on 5’-[32P]-labeled oligonucleotides (51 nt) containing: hydroxyuracil (OHU), OHU/G, U, or U/G. ssDNA, single-stranded DNA; dsDNA, double-stranded DNA. S, substrate; P, product. (C) Validation of UDG-generated AP sites from 5’-[32P]-labeled single-stranded and double-stranded oligonucleotides (35 nt) by treatment with NaOH. AP sites are lysed by alkali treatment. (D) NEIL2 cleaves Fluorescein (Fluor)-labeled U-containing single strand oligonucleotide (39 nt) in the presence of UDG. S, substrate; P, product.

Reaction mechanisms for the APE1 and NEIL2-PNKP pathways.
Glycosylase UDG removes U and generates an AP site on U/G mismatch (only top strand is shown). For the APE1 pathway, APE1 hydrolyzes the deoxyribose-phosphate backbone and generates a 5’-deoxyribophosphate (5’dRP) and a 3’-hydroxyl (3’OH). DNA Polymerase β (Polβ) adds a C to the 3’OH and removes the 5’dRP. For the NEIL2-PNKP pathway, NEIL2 catalyzes β,δ-elimination step by step at the AP site and generates a 5’-phosphate (5’P) and a 3’P. PNKP phosphatase hydrolyzes 3’P to generate a 3’OH primer for Polβ.

Concentration gradients of APE1 and NEIL2 on AP sites.
(A and B) Concentration gradient of APE1 (0, 0.25, 0.5, 1, 2.5, 5, and 10 U) and NEIL2-His6 (0, 0.68, 1.36, 3.4, 6.8, 13.6, and 34 ng) on 5’-[32P]-labeled U-containing ssDNA (51 nt) in a 10 µl-reaction in the presence of UDG. A 25-nt product is generated. The percentage of product relative to the starting substrate is shown on the right. S, substrate; P, product. (C) Concentration gradient of APE1 and NEIL2 on [32P]-U-containing dsDNA (51 nt) in the presence of UDG. The volumes of NEIL2 (68 ng/µl) and APE1 (0.005 U/µl) used are listed in the figure. S, substrate; P, product.

Purification and activity assays of PNKP and Polβ.
(A) Purified Polβ-His6 (17 ng) and PNKP-His6 (127 ng) from E. coli were subjected to PAGE and stained with Coomassie blue. (B) Incorporation of [α-32P]-dCTP by Polβ using APE1-generated product. ddC, di-deoxynucleotide; P, product. (C) Schematic of the preparation of S (substrate) and subsequent enzymatic reactions for testing PNKP activity. (D) Efficiency of oligonucleotide labeling, annealing, and ligation leading to S indicated in (C). (E) Fpg (NEB, 1 U) completely digested S and the 3’ phosphate was completely removed by PNKP (12.7 ng and 127 ng, lanes 2 and 3), or by T4 PNK (NEB, 0.1 U and 1 U, lanes 7 and 8). NEIL2 (272 ng) only partially digested S and its 3’P was resistant to the PNKP phosphatase (lanes 4 and 5).
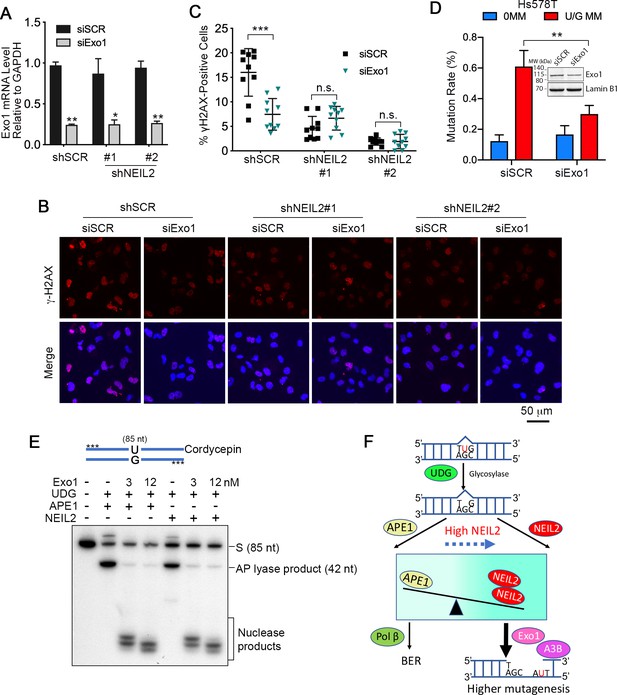
Exo1 generates single-stranded substrates vulnerable to A3B and DNA damage from NEIL2 products.
(A) qRT-PCR analysis of siRNA knockdown of Exo1 in NEIL2-stable-knockdown Hs578T cell lines (shNEIL2#1 and shNEIL2#2). shNEL2#1 targets NEIL2 3’UTR; shNEIL2#2 targets NEIL2 ORF; shSCR, scramble shRNA; siSCR, scramble siRNA. Error bars represent s.d., n = 3. *P < 0.05; **P < 0.01 by two-tailed unpaired Student’s t test. (B) Immunostaining of γH2AX foci in Hs578T cells depleted of Exo1 by siRNA in NEIL2-stable-knockdown Hs578T cell lines. A3B-3HA was expressed in these cell lines for 48 h before immunostaining. Scale bar, 50 μm. (C) Quantification of the percentage of cells with γH2AX foci from (B). Data are represented as mean ± s.d. (n=10 randomly selected microscopic fields). ***P < 0.001; n.s., no significant difference by two-tailed unpaired Student’s t test. (D) Effect of Exo1 depletion on U/G MM repair-induced mutation rate in Hs578T cells. Insert, western blot analysis of Exo1 knockdown efficiency by siExo1 (20 nM siRNA). Lamin B1 serves as a loading control. 0 MM, no mismatch; U/G MM, U/G mismatch. **P < 0.01 by two-tailed unpaired Student’s t test. (E) APE1 and NEIL2 scission products serve equally well as Exo1 substrates. 5’-phosphorothioate-modified (denoted by asterisk) U-containing oligo (85 nt) was 3’-labeled with cordycepin and used as the substrate for UDG, APE1 or NEIL2, and Exo1(human Exo1 protein). Nuclease products are bracketed. (F) Model depicting NEIL2-diversion of BER to Exo1-mediated resection to generate single-stranded A3B substrate.
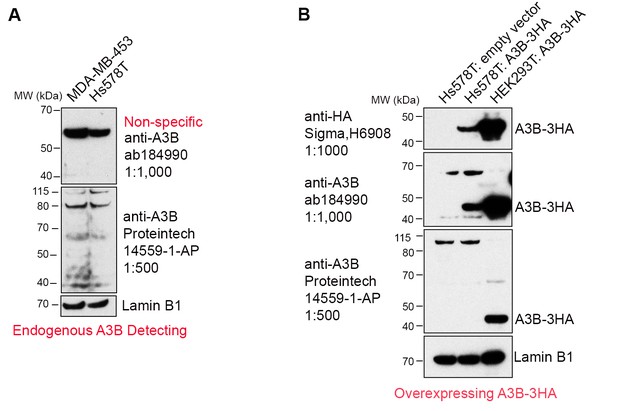
Commercially available anti-A3B antibodies do not recognize endogenous A3B protein.
(A) 100 μg whole cell lysate from MDA-MB-453 and Hs578T cells were subject to western blotting and detected by anti-A3B antibodies shown. Lamin B1 serves as a loading control. (B) 100 μg whole cell lysate from Hs578T cells overexpressing empty vector or A3B-3HA were subject to western blotting and detected by anti-A3B antibodies shown. Both antibodies recognize overexpressed A3B-3HA but not endogenous A3B. HEK293T overexpressing A3B-3HA lysate detected by anti-HA antibody was used as a positive control. LaminB1 serves as a loading control.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo-sapiens) | Hs578T, breast cancer | ATCC | HTB-126, RRID:CVCL_0332 | Authenticated by ATCC |
| Cell line (Homo-sapiens) | MDA-MB-453, breast cancer | ATCC | HTB-131, RRID:CVCL_0418 | Authenticated by ATCC |
| Cell line (Homo-sapiens) | MCF7, breast cancer | ATCC | HTB-22, RRID:CVCL_0031 | Authenticated by ATCC |
| Cell line (Homo-sapiens) | HCC1569, breast cancer | ATCC | CRL-2330, RRID:CVCL_1255 | Authenticated by ATCC |
| Cell line (Homo-sapiens) | LentiX-293T | Clontech | 632180 | Mycoplasma free |
| Strain, strain background (Escherichia coli) | MBM7070 (lacZuag_amber) | Gift from Dr Michael Seidman (NIH) | Electroporation | |
| Strain, strain background (Escherichia coli) | Rosetta (DE3) | Millipore | 70954–4 | Protein expression |
| Antibody | anti-HA (Rabbit polyclonal) | Sigma-Aldrich | H6908, RRID:AB_260070 | WB (1:1,000) |
| Antibody | anti-Exo1 (Rabbit polyclonal) | Proteintech | 16253–1-AP, RRID:AB_2278140 | WB (1:1000) |
| Antibody | anti-Lamin B1(Rabbit polyclonal) | abcam | ab16048, RRID:AB_10107828 | WB (1:2,000) |
| Antibody | anti-Rabbit IgG-Peroxidase | Sigma-Aldrich | A0545, RRID:AB_257896 | WB (1:10,000) |
| Antibody | anti-Mouse IgG-Peroxidase | Sigma-Aldrich | A4416, RRID:AB_258167 | WB (1:5,000) |
| Antibody | anti-γH2AX | Cell Signaling | 2577, RRID:AB_2118010 | IF (1:800) |
| Antibody | Alexa Fluor 568 anti-Rabbit IgG Secondary Antibody | Invitrogen | A-11036, RRID:AB_143011 | IF (1:500) |
| Sequenced-based reagent | siSCR (negative control) | Dharmacon | UAGCGACUAAACACAUCAA | |
| Sequenced-based reagent | siA3B | Dharmacon | J-017322-08-0005 | A3B knockdown |
| Sequenced-based reagent | siA3C | Dharmacon Muckenfuss et al., 2006 | AAGCCAACGAUCGGAACGAAA | |
| Sequenced-based reagent | siNEIL2 | Dharmacon | GCGAGGAUGAUUCUGAGUA | |
| Sequenced-based reagent | siTREX1 | Dharmacon | ACAAUGGUGACCGCUACGA | |
| Sequenced-based reagent | siExo1 | Dharmacon | CAAGCCUAUUCUCGUAUUU | |
| Commercial assay or kit | A3A_TaqMan | ThermoFisher Scientific | Hs02572821_s1 | |
| Commercial assay or kit | A3B_TaqMan | ThermoFisher Scientific | Hs00358981_m1 | |
| Commercial assay or kit | A3C_TaqMan | ThermoFisher Scientific | Hs00819353_m1 | |
| Commercial assay or kit | A3H_TaqMan | ThermoFisher Scientific | Hs00419665_m1 | |
| Commercial assay or kit | NEIL2_TaqMan | ThermoFisher Scientific | Hs00979610_g1 | |
| Commercial assay or kit | Exo1_TaqMan | ThermoFisher Scientific | Hs01116190_m1 | |
| Commercial assay or kit | TREX1_TaqMan | ThermoFisher Scientific | Hs03989617_s1 | |
| Commercial assay or kit | GAPDH_TaqMan | ThermoFisher Scientific | Hs02786624_g1 | |
| Commercial assay or kit | TBP_TaqMan | ThermoFisher Scientific | Hs00427620_m1 | |
| Recombinant DNA reagent | pLKO.1 | addgene | RRID:Addgene_10878 | shRNA construction |
| Recombinant DNA reagent | pMDLg/pRRE | addgene | RRID:Addgene_12251 | lentivirus packaging |
| Recombinant DNA reagent | pRSV-Rev | addgene | RRID:Addgene_12253 | lentivirus packaging |
| Recombinant DNA reagent | pMD2.G | addgene | RRID:Addgene_12259 | lentivirus packaging |
| Transfected construct (Homo-sapiens) | phAPOBEC3B-HA (A3B-3HA) | NIH AIDS Reagent Program | 11090 | A3B overexpression |
| Transfected construct (Homo-sapiens) | pPM-NEIL2-3’HA | abm | PV028075 | NEIL2 rescue in mutation rate assay |
| Transfected construct (Homo-sapiens) | pcDNA3.1(+)-NEIL2-3’HA | this paper | NEIL2 rescue in γH2AX assay | |
| Transfected construct (Homo-sapiens) | pET22b-NEIL2 | Gift from Dr Tapas Hazra (U of Texas) | NEIL2 expression and purification from E. coli | |
| Transfected construct (Homo-sapiens) | pET28a(+)-PNKP | this paper | For PNKP expression and purification | |
| Transfected construct (Homo-sapiens) | pET28a(+)-Polβ | this paper | For Polβ expression and purification |