Relationship between simultaneously recorded spiking activity and fluorescence signal in GCaMP6 transgenic mice
Figures
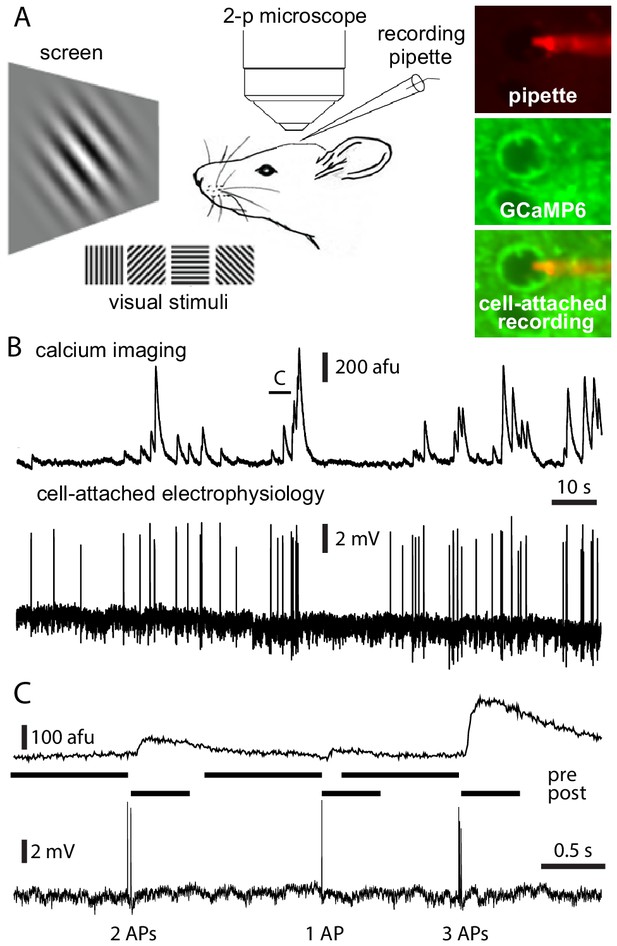
Simultaneous calcium imaging and electrophysiology in vivo.
(A) Experimental design. (B) Fluorescence and Vm traces from an exemplar Emx1-s neuron. (C) 5 s of data from the neuron in panel B, showing a 2 AP, a 1 AP, and a 3 AP event. Pre- and post-AP exclusion windows, used to separate events, are illustrated for each event. AP, action potential.
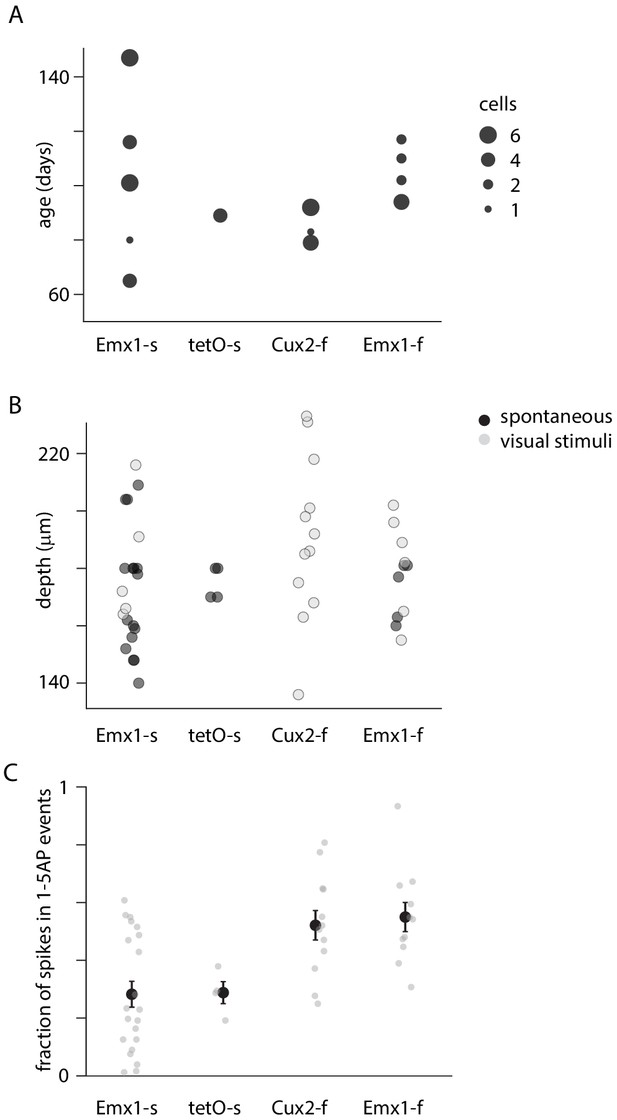
Mouse age, cell depth, and firing rate.
(A) Mouse age. (B) Cell depths. (C) For each neuron, percentage of action potentials (APs) contained within 1–5 AP events.
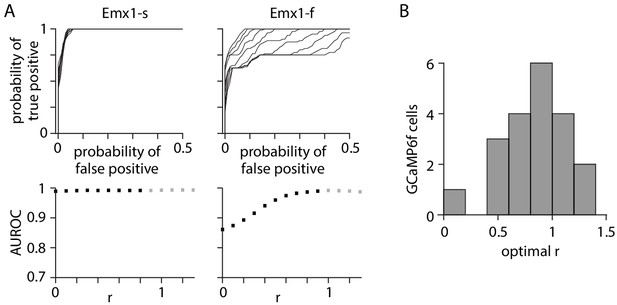
Neuropil subtraction optimized for 1 AP (action potential) detection.
(A) Effect of changing neuropil subtraction on detection for exemplar GCaMP6s and GCaMP6f neurons. Upper plots: family of receiver operating characteristic (ROC) curves. Each curve illustrates detection probability for true APs against probability of false positives as detection threshold is changed, for 1 AP events. False positives were calculated from time windows with no APs. Each ROC curve represents a different value of r. Lower plots: area under the ROC curve as a function of r. Gray symbols represent value of r for which r * Fneuropil(t) was greater than Fcell_measured(t), resulting in a negative F0 and inversion of the ΔF/F trace. (B) Distribution of r values for 20 GCaMP6f neurons.
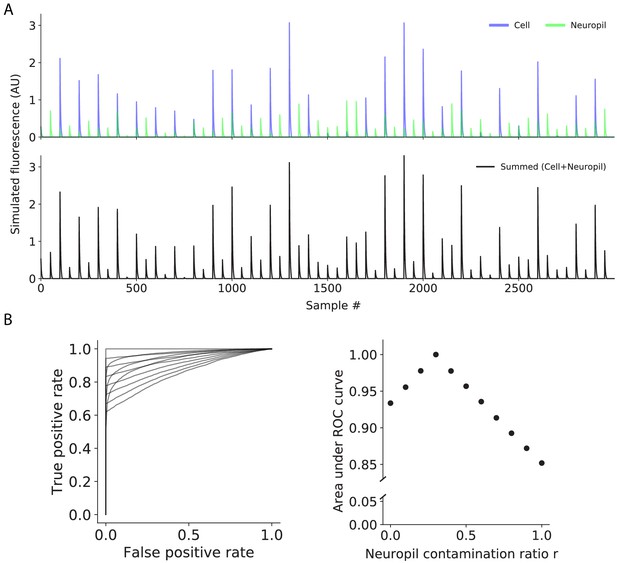
Simulated effect of neuropil subtraction on event detection.
(A) Simulated cell and neuropil traces. The neuropil trace contained transients that were (1) associated with cell transients and (2) between cell transients, and amplitudes were scaled by the neuropil contamination ratio r (r = 0.3) relative to the cell amplitudes. (B) (Left) Receiver operating characteristic (ROC) curves for classifying cell amplitudes, where r was varied from 0 to 1 for neuropil correction. The detection threshold was defined as the xth percentile of noise amplitudes (amplitudes of the neuropil trace that were between cell transients), where 1–x represented the false positive probability, and the detection probability (true positive probability) was the fraction of estimated cell amplitudes (amplitudes of the summed trace that were associated with cell transients) above the detection threshold. (Right) Area under ROC curves as a function of r.
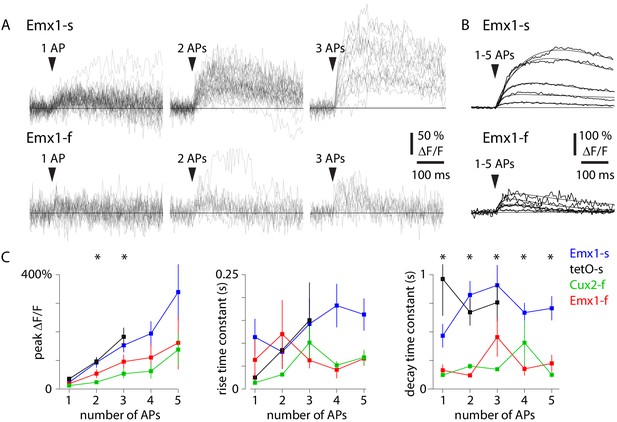
Action potential (AP)-evoked calcium transients in four mouse lines.
(A) Example fluorescence traces (ΔF/F, with r = 0.82 neuropil subtraction) for 1 AP, 2 AP, and 3 AP events for an exemplar Emx1-s neuron (forty-two 1 AP events, thirty-eight 2 AP events, sixteen 3 AP events) and an exemplar Emx1-f neuron (twenty-three 1 AP events, twelve 2 AP events, eleven 3 AP events). (B) Mean fluorescence traces and fits (sum of two exponentials) for 1–5 AP events for the two neurons in A. (C) Mean ± SEM peak DF/F, rise time constant, and decay time constant for four mouse lines. Number of neurons for 1–5 AP events were: 15, 16, 14, 9, 6 for Emx1-s; 4, 4, 4, 0, 0 for tetO-s; 10, 10, 10, 5, 4 for Emx1-f; 9, 9, 7, 4, 2 for Cux2-f. Asterisks indicate differences between mouse lines (p<0.05, one-way ANOVA).
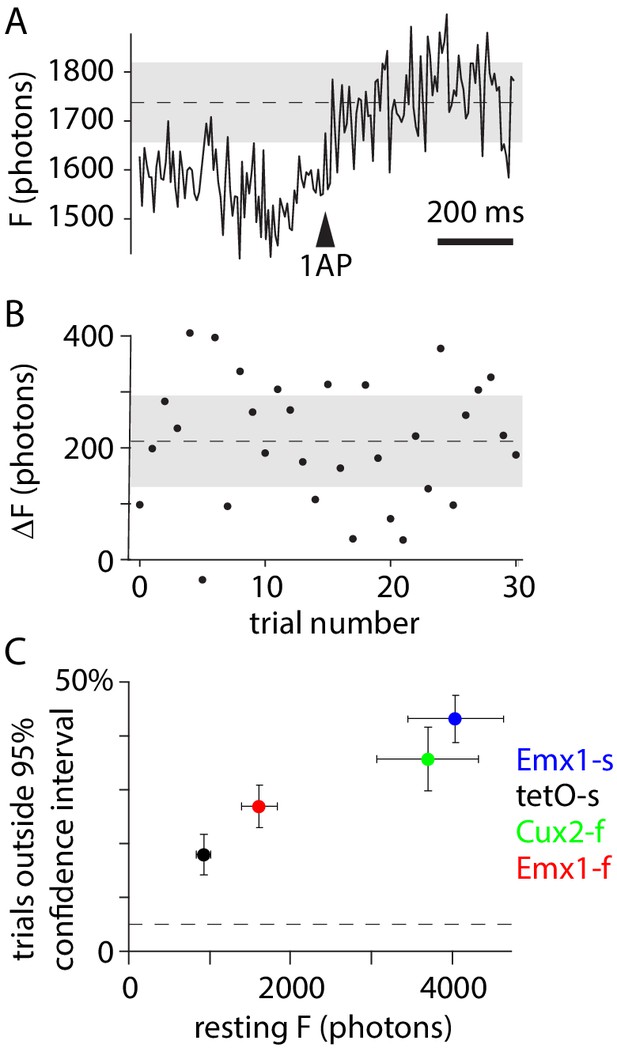
Trial-to-trial variability of 1 AP (action potential) events.
Fraction of trials in which the peak of the fluorescence transient exceeded the range expected from Poisson noise, a measure of trial-to-trial variability. (A) Example 1 AP event for an Emx1-s neuron, with fluorescence in units of photons. Dashed line: mean, across trials, of peak fluorescence. Gray: 95% confidence interval for Poisson-distributed noise. (B) Amplitudes of 1 AP events for 30 trials. Dashed line and gray area: mean and 95% confidence interval. (C) Mean ± SEM percentage of trials with peak ΔF outside the 95% confidence interval, for each mouse line. In all four mouse lines, peak fluorescence exceeded the 95% confidence interval for Poisson-distributed noise in >>5% of 1 AP trials. The fraction of trials outside the confidence interval correlated with resting fluorescence, likely the result of different illumination intensities across experiments.
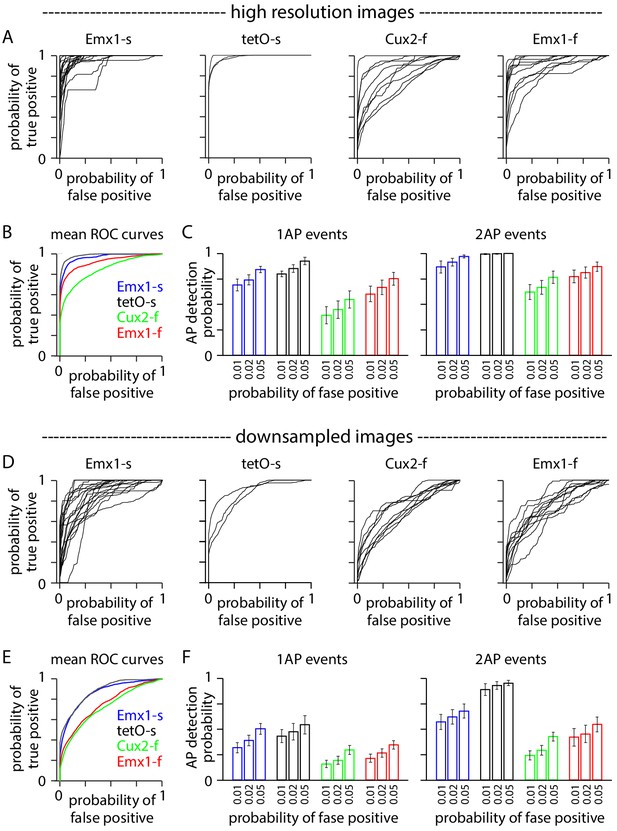
Downsampling affects event detection.
(A–C) Event detection from images at high spatial and temporal resolution. (A) Receiver operating characteristic (ROC) curves for 1 AP events in 42 neurons, organized by mouse line. Neuropil subtraction was performed with r = 0.8 where possible (see 'Materials and methods'). Numbers of neurons were 18 Emx1-s, 3 tetO-s, 11 Cux2-f, 9 Emx1-f. (B) Mean ROC curves for the four mouse lines. (C) Event detection probabilities for 1 AP and 2 AP events. Bars represent mean ± SEM. (D–F) Equivalent plots for the same neurons after downsampling.
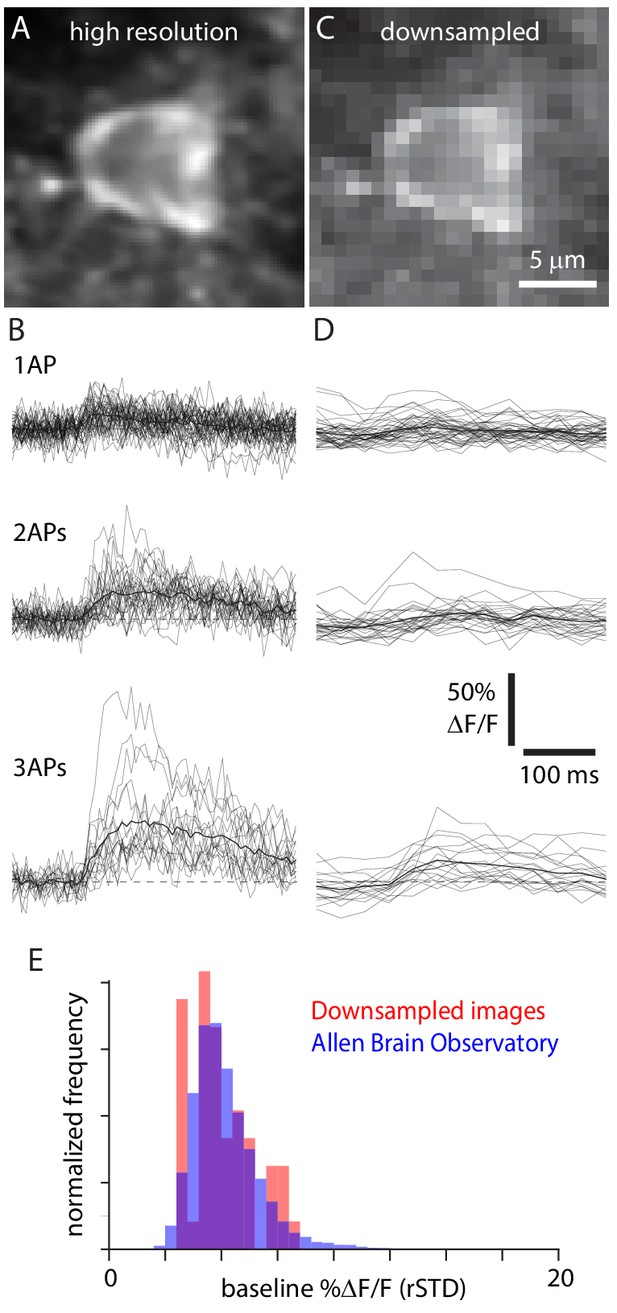
Downsampling mimics large field-of-view images.
(A) Original, high spatial and temporal resolution GCaMP6f image of an exemplar Cux2-f neuron. (B) 1 AP, 2 AP, and 3 AP traces for the same neuron. Thin lines, individual trials; thick line, mean. Dashed line, ΔF/F = 0. (C) Downsampled image of the same neuron. (D) Traces from the downsampled neuron. (E) Normalized distribution of baseline noise (robust standard deviation [rSTD]) for 48 downsampled neurons (red) and 11,816 layer 2/3 neurons from Emx1-f and Cux2-f mice in the Allen Brain Observatory (blue).
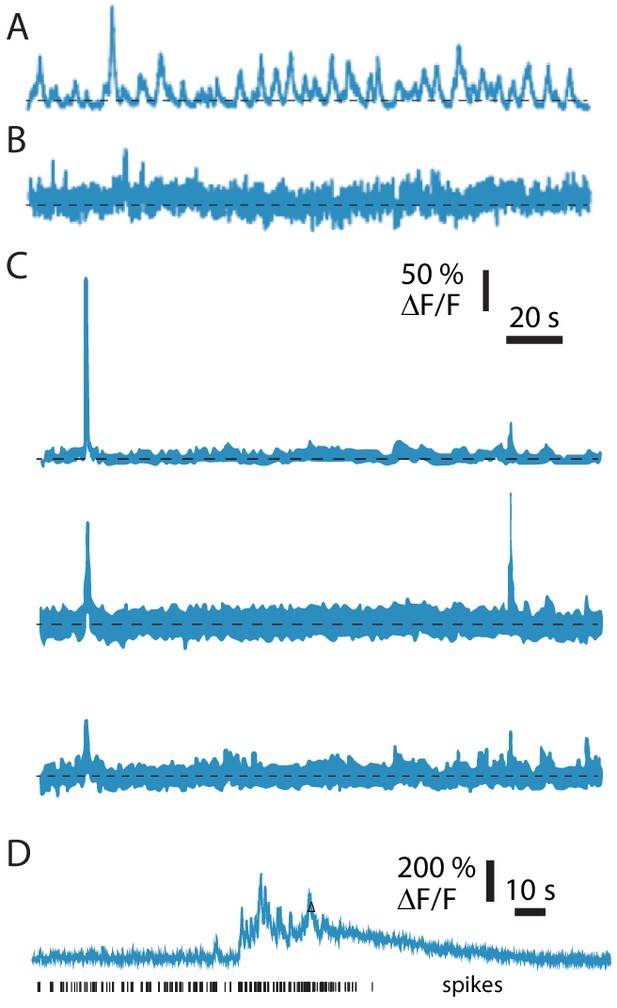
Example fluorescence traces from image quality control procedures, implemented during image downsampling.
(A) Median of traces extracted from an example segmentation in which only the neuron of interest was found in each image during segmentation. The trace is significantly different from noise (KS test, p=1.8×10−40). (B) Median of traces extracted from an example segmentation in which only the neuron of interest was found in each image during segmentation. The trace is not significantly different from noise (KS test, p=0.072). (C) Medians of traces extracted from an example segmentation in which three regions of interest were identified. Each trace is significantly different from noise (KS tests, p=7.7×10−152, 1.6 × 10−19, 2.2 × 10−25). The sum of these three cluster medians had the highest correlation with the firing rate and was therefore used as the ΔF/F trace for this recording in the dataset. Scale bars apply to A–C. (D) Example fluorescence trace with an abrupt and sustained rise in fluorescence and in spike rate and subsequent loss of spiking activity. This neuron was manually removed from the dataset.
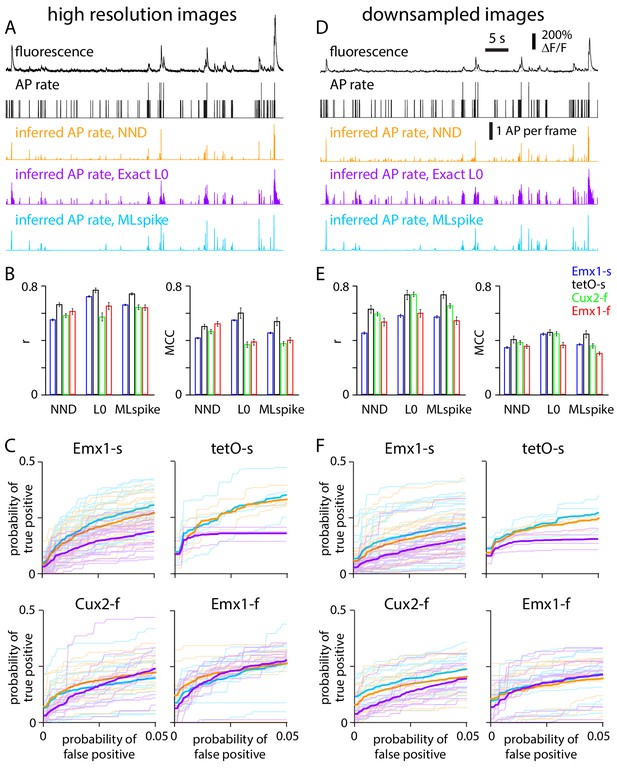
Performance of spike inference algorithms on high-resolution and downsampled images.
(A) Results from an exemplar Cux2-f neuron, at high resolution. Fluorescence and action potential (AP) rate from electrophysiology (black). Below, APs per image frame estimated with three spike inference algorithms: MLspike (blue), Exact L0 (purple), and non-negative deconvolution (NND, orange). (B) Pearson correlation coefficient (r) and Matthews correlation coefficient (MCC) for the three algorithms for each mouse line. 300 ms bins. (C) Receiver operating characteristic (ROC) curves, reporting probabilities of detecting true and false events in each time bin. Thin lines: individual neurons. Thick lines: mean across neurons. 300 ms bins. (D–F) Equivalent plots for downsampled images.
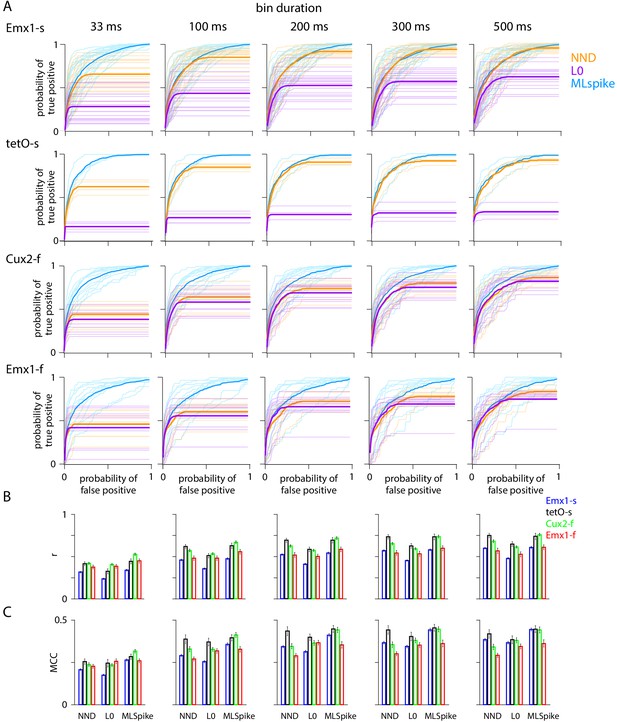
Effect of bin duration on measures of spike inference algorithm performance.
(A) Receiver operating characteristic (ROC) curves for MLspike (blue), Exact L0 (purple), and non-negative deconvolution (NND) (orange) algorithms after binning of spike probabilities into 33, 100, 200, 300, and 500 ms bins, for downsampled images resampled to 150 Hz. (B and C) Pearson correlation coefficients (r) and Matthews correlation coefficients (MCC) for each bin duration.
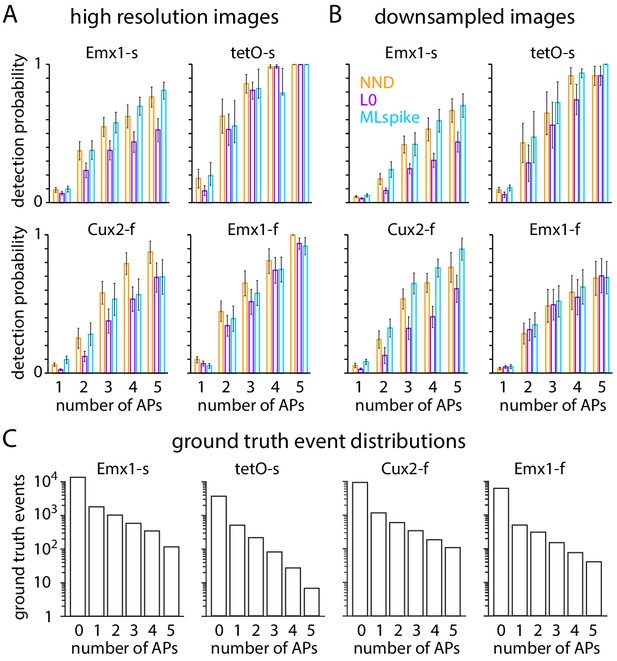
Performance of blind spike inference algorithms for 1–5 AP (action potential) events.
(A) Mean ± SEM detection probabilities at 1% false positive probability for high-resolution images. (B) Mean ± SEM detection probabilities at 1% false positive probability for downsampled images. (C) Frequency of 0–5 AP events for 250 ms bins for each mouse line.
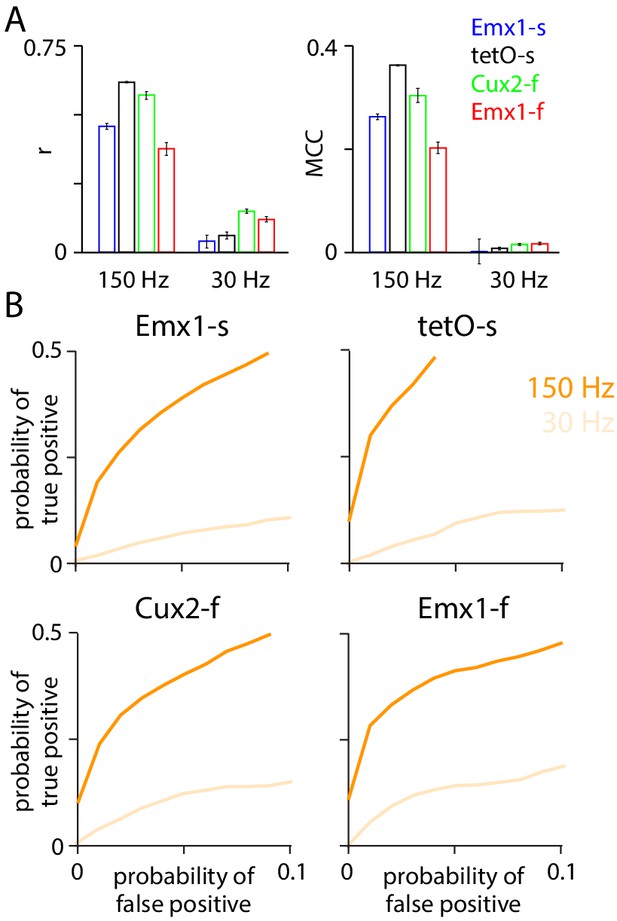
Upsampling enhances performance of unconstrained non-negative deconvolution (NND).
Performance of the NND algorithm when applied to fluorescence traces from downsampled images (frame rate 30 Hz) and to the same fluorescence traces after upsampling to 150 Hz. 100 ms bins. (A) Pearson r and Matthews correlation coefficient (MCC). (B) Receiver operating characteristic (ROC) curves.
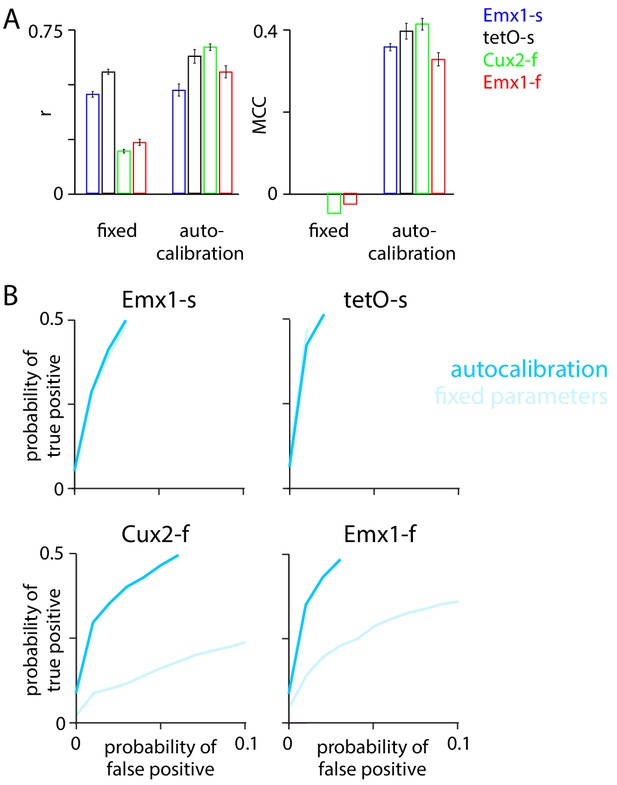
Autocalibration enhanced performance of MLspike.
Performance of MLspike with and without use of the autocalibration procedure, applied to downsampled images, with fluorescence traces upsampled to a frame rate of 158 Hz. 100 ms bins. (A) Pearson r and Matthews correlation coefficient (MCC). (B) Receiver operating characteristic (ROC) curves.
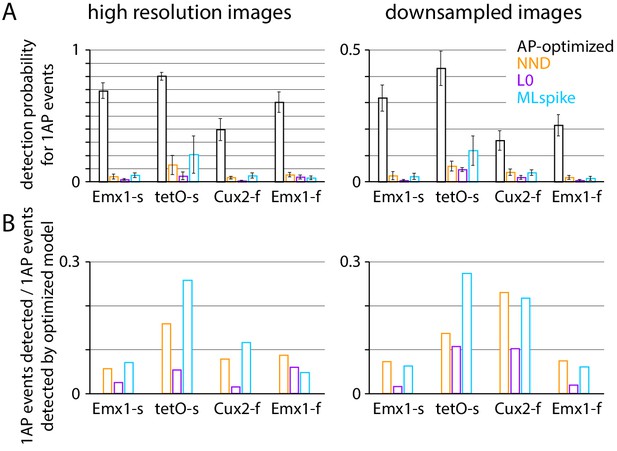
Comparison of spike inference with blind and ground truth action potential (AP)-optimized algorithms.
(A) Mean ± SEM 1 AP event detection probability at 1% false positive probability for ground truth-optimized and blind algorithms (used in Figure 4 and in Figures 6 and 7, respectively). Traces were selected as for Figure 4. (B) 1 AP events detected by each blind algorithm as a fraction of those detected by the ground truth-optimized algorithm, at 1% false positive probability.
Tables
Dataset.
Sample size for each mouse line (see also Figure 1—figure supplement 1).
| Mouse line | Acronym | GECI | Mice | Cells | Recording duration | APs per neuron |
|---|---|---|---|---|---|---|
| Emx1-IRES-Cre; Camk2a-tTA;Ai94 | Emx1-s | GCaMP6s | 5 | 21 | 241 ± 32 s | 478 ± 121 |
| Camk2a-tTA; tetO-GCaMP6s | tetO-s | GCaMP6s | 1 | 4 | 347 ± 108 s | 348 ± 71 |
| Cux2-CreERT2; Camk2a-tTA;Ai93 | Cux2-f | GCaMP6f | 3 | 12 | 300 ± 79 s | 484 ± 112 |
| Emx1-IRES-Cre; Camk2a-tTA;Ai93 | Emx1-f | GCaMP6f | 4 | 11 | 201 ± 23 s | 219 ± 38 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent Mus musculus | B6.129S2-Emx1tm1(cre)Krj/J, Emx1-IRES-Cre | Jackson Laboratory | RRID:IMSR_JAX:005628 RRID:MGI:2684610 | |
| Genetic reagent Mus musculus | B6(Cg)-Cux2tm3.1(cre/ERT2)Mull/Mmmh, Cux2-CreERT2 | MMRRC | RRID:MMRRC_032779-MU RRID:MGI:5014172 | |
| Genetic reagent Mus musculus | B6.Cg-Tg(Camk2a-tTA)1Mmay/DboJ, Camk2a-tTA | Jackson Laboratory | RRID:IMSR_JAX:007004 RRID:MGI:2179066 | |
| Genetic reagent Mus musculus | B6;DBA-Tg(tetO-GCaMP6s)2Niell/J, tetO-GCaMP6s | Jackson Laboratory | RRID:IMSR_JAX:024742 RRID:MGI:5553332 | |
| Genetic reagent Mus musculus | B6;129S6-Igs7tm93.1(tetO-GCaMP6f)Hze/J, Ai93(TITL-GCaMP6f) | Jackson Laboratory | RRID:IMSR_JAX:024103 RRID:MGI:5558086 | |
| Genetic reagent Mus musculus | B6.Cg-Igs7tm94.1(tetO-GCaMP6s)Hze/J, Ai94(TITL-GCaMP6s) | Jackson Laboratory | RRID:IMSR_JAX:024104 RRID:MGI:5607576 | |
| Software, algorithm | MATLAB R2016b | http://www.mathworks.com/products/matlab/ | RRID:SCR_001622 | |
| Software, algorithm | Python 3.7.4 | http://www.python.org/ | RRID:SCR_008394 | |
| Software, algorithm | LabVIEW 2015 | http://www.ni.com/labview/ | RRID:SCR_014325 |





