Muscle-derived Myoglianin regulates Drosophila imaginal disc growth
Figures
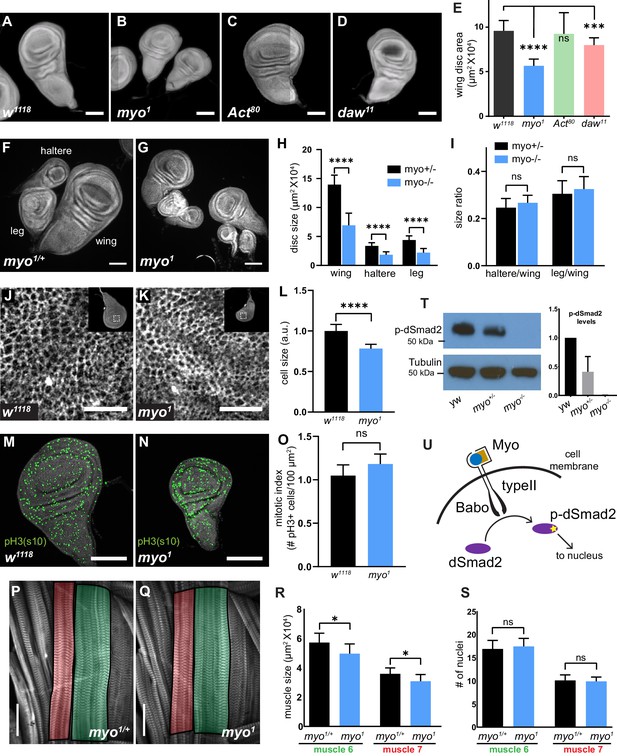
Myoglianin regulates size of imaginal discs and activates dSmad2.
(A–D) DAPI staining of wing imaginal discs from late third instar larvae of various TGFβ ligand mutants. Scale bar, 100 µm. (E) Wing disc size of myo mutants (blue bar) are approximately 40% smaller than control, Activinβ mutant discs (C) are the same size as controls (green bar), and dawdle mutant discs (D) are ~16% smaller (red bar) (n > 20 per group). (F–I) Wing, leg, and haltere imaginal discs of myo1 null mutants are 50% smaller vs heterozygous myo1/+ controls. (H) Size of various imaginal discs, normalized to controls. (I) Size ratio of haltere/wing disc (.25) or leg/wing disc (.3) are the same in myo mutants vs controls. (J–K) Phalloidin staining showing the apical actin-belt of epithelial cells in wing discs; each cell has a distinct actin belt that defines the edges of a cell (scale bar = 10 µm, n > 5 per group). Single confocal section is shown with the whole disc in insert with the magnification of the pouch region outlined. (L) Based on cell density, myo mutant discs cells are 12% smaller vs control. (M–N) Phospho-Histone H3-pSer10 (pH3s10) staining to mark mitotic cells in early third instar wing discs, showing no difference in number of mitotic cells in myo mutants (scale bar = 100 µm, n = 5 per group). Number of mitotic cells per 100 µm2 is plotted (O). (P–Q) Phalloidin staining showing the late 3rd instar larvae body wall muscle #6 (green highlight) and #7 (red highlight) (scale bar = 100 µm, n = 10). Muscle area (R) and number of nuclei per fiber (S) is plotted. myo mutant muscles are 13% smaller (p=0.02) yet have the same number of nuclei per fiber (S). (T) Western blot analysis of phospho-dSmad2 levels in wing discs, with quantification from 3 separate blots. myo mutants lack p-dSmad2 signal indicating loss of canonical signaling. (U) Model for canonical TGFβ signaling in wing imaginal disc via Myo/Babo/dSmad2. C-terminal activation of dSmad2 (yellow star) results in regulation in expression of downstream target genes which is required for proper growth of imaginal discs.
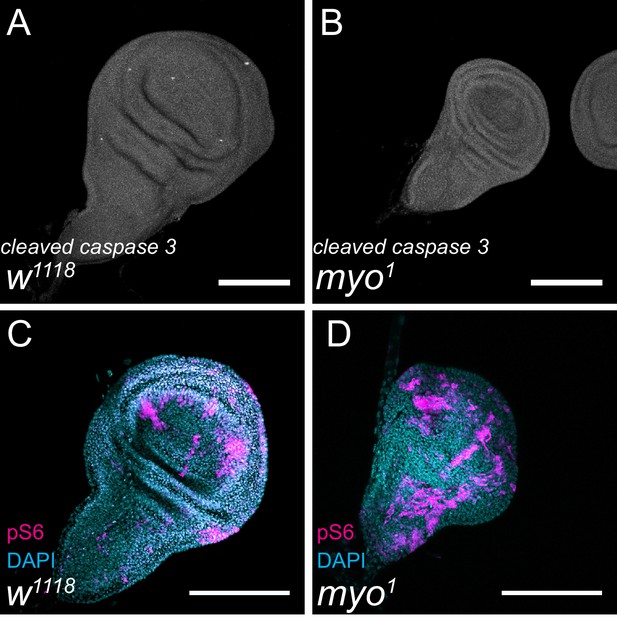
myo mutants show no defects in cell death or cell growth.
(A-B) Third instar larval wing discs stained for cleaved-caspase 3 shows no increase in cell death of myo mutant discs. (C-D) Early third instar (90–96 hr AEL) larval wing discs stained for phospho-S6, downstream of TOR signaling, shows no defect in cell growth patterns in myo mutant discs. Scale bar 100 µm, n = 5–10 per group.
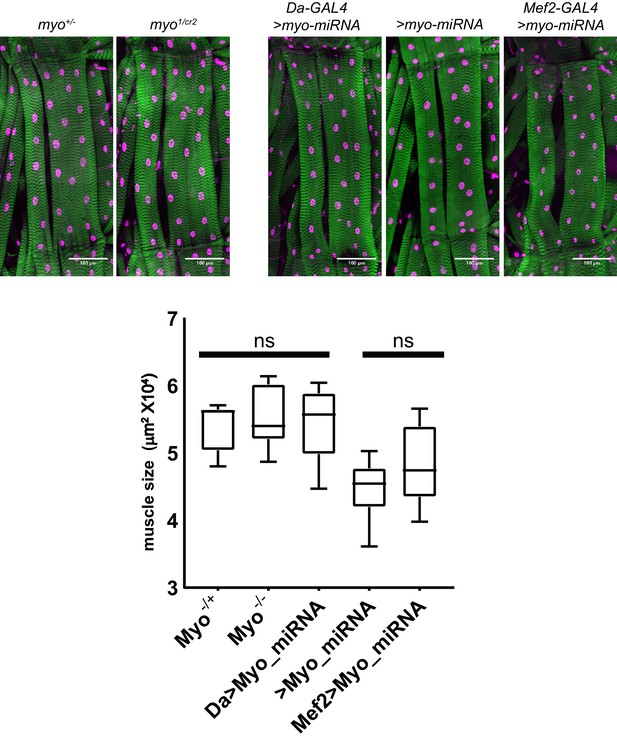
Trans-heterozygote myo mutants do not have larger muscles.
Third instar larval fillets stained with phalloidin (green) and DAPI (magenta) show no difference in muscle size for trans-heterozygotes myo1 and myoCR2 alleles vs heterozygote controls, scale bar = 100 µm, n = 8–14. The size of muscle 6 in segment A2 is quantified. Knocking down myo in muscles using mef2-GAL4 does not significantly increase muscle size vs no GAL4 control. Knocking down myo uibiquitously using da-GAL4 does result in larger muscles, however no larger than the genetic nulls.
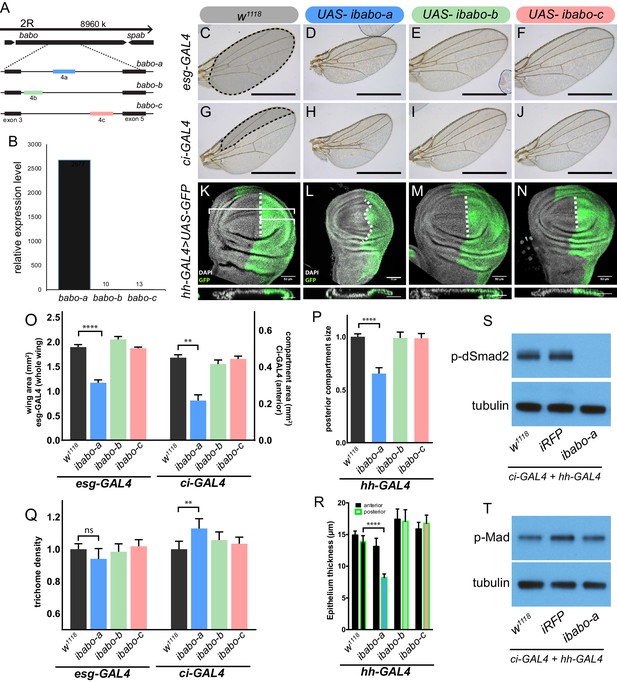
Babo-A regulates wing disc growth.
(A) Cartoon illustrating the gene region and splice isoforms of TGFβ/Activin type-I receptor babo. The three splice isoforms are the result of 3 different mutually exclusive 4th exons; the remaining exons (not shown) are the same for all babo isoforms. (B) qPCR analysis of babo isoform expression from late 3rd instar wing imaginal discs identifies only babo-a expression. Expression level is relative to Rpl32, the expression level of babo-b and babo-c are indicated by the numbers above their respective bars. (C–J) Adult wings of animals where specific babo isoforms were knocked down in the entire wing blade (esg-GAL4) (C–F) or one compartment (ci-GAL4) (G–J). Knockdown of babo-a by esg-GAL4 results in a smaller wing blade (D) and by ci-GAL4 results in a smaller anterior compartment (H), scale bar 1 mm, n = 5–12 per group. Size measurements of tissue size (indicated by shading in C and G) are quantified in O. (K–N) Imaginal discs from late 3rd instar larvae where specific babo isoforms were knocked down in only the posterior compartment by hh-GAL4, marked with GFP, the size of the posterior compartment is measured relative to the entire wing pouch as illustrated by the brackets, the anterior/posterior boundary is highlighted by the dashed line, scale bar = 50 µm, n = 3–6 per group. (O) Quantification of tissue size from C-J. Knockdown of babo-a by esg-GAL4 and ci-GAL4 results in 40% and 50%, respectively, smaller tissues (O, blue bars). (P) Quantification of the relative size of the posterior compartment, data normalized to controls. Knockdown of babo-a by hh-GAL4 results in a 35% reduction of the posterior compartment relative to the overall size of the wing disc. (Q) Quantification of trichome density in the affected region of adult wings from C-J, measurements are normalized to their respective GAL4 x w1118 controls. Knockdown of babo-a using esg-GAL4 does not affect trichome density, whereas knockdown with ci-GAL4 results in higher trichome density, indicating smaller cells (P, blue bar). (R) Epithelium thickness was quantified from orthogonal views of the imaginal discs from K-N. Knockdown of babo-a results in a 29% decrease in epithelium thickness of the posterior compartment. (S–T) Western blot analysis of phospho-dSmad2 and phospho-Mad in L3 imaginal discs. Knockdown of babo-a throughout the imaginal discs using both ci-GAL4 and hh-GAL4 leads to loss of phospho-dSmad2 signal (S), but not phospho-Mad signal (T). Tubulin is shown as a loading control. n = 10 per lane.
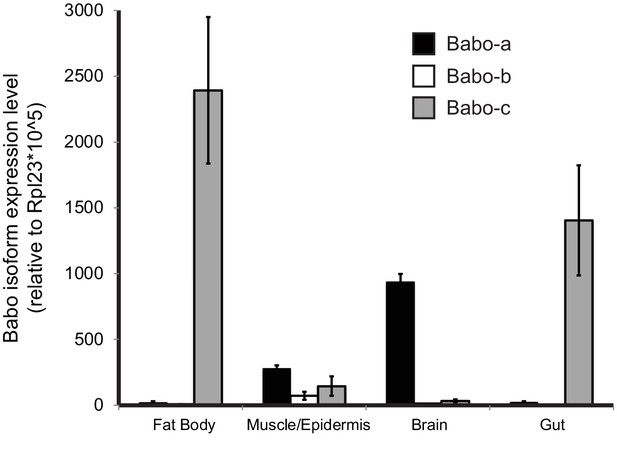
qPCR analysis of Babo isoform expression in various tissues from late third instar larvae.
Fat-body and gut express only babo-c, whereas brain samples predominantly express babo-a. The carcass sample produces detectable amounts of all three isoforms. This sample contains muscle, cuticle, and associated cells, precluding assignment of isoform expression to cell types. Expression levels are relative to Rpl23.
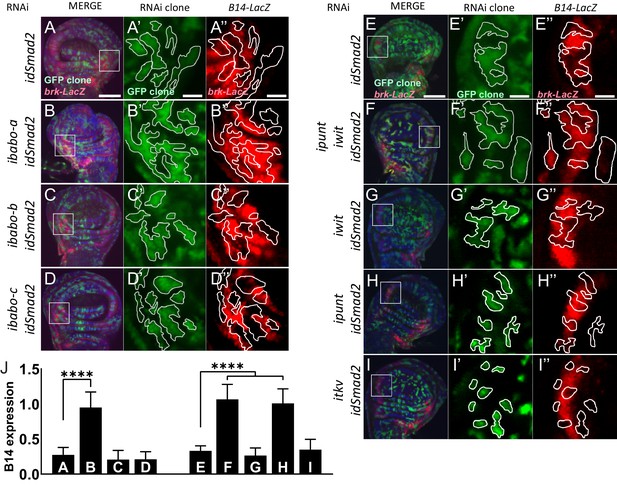
Babo-A and Punt are required for non-canonical silencing of brk.
In the background of brk-LacZ reporter line random GFP+ (green) clones were induced by heat shock 48 hr prior to analysis (see methods for details). GFP+ marked clones also express RNAi for the indicated genes to the left of respective panels. Clones at the lateral edges of the disc were analyzed for patterning defects of the brk reporter. (A-D) Analysis of Babo isoforms. (E-I) Analysis of Type-II receptors. (A-I) Maximum intensity projections of wing imaginal discs stained with DAPI (blue) and anti β-gal (red), and fluorescent clone marker (green), scale bar = 100 µm. (A’-I’) Single confocal section of higher magnification of insert in (A-I) showing GFP+ marked clone expressing RNAi for indicated genes. Clones are outlined with white lines, scale bar = 25 µm. (A’’-I’’) Same confocal section as in (A’-I’) with outlined clones showing brk-LacZ reporter expression pattern, scale bar = 25 µm. Brk reporter is ectopically silenced in dSmad2 RNAi clones (A and E). This ectopic silencing is rescued by the concomitant RNAi of babo-a alone (A’’ vs B’’) but not babo-b or babo-c (A’’ vs C’’ or D’’). dSmad2 RNAi dependent brk reporter silencing (E’’) can be rescued by the concomitant RNAi of both Type-II receptors punt and wit (E’’ vs F’’). RNAi for wit does not rescue brk reporter (E’’ vs G’’). RNAi for punt completely rescues the brk reporter (E’’ vs H’’). brk silencing in dSmad2 RNAi clones at the lateral edge of the disc is not mediated by thickveins (tkv, BMP Type-I receptor), since concomitant RNAi for tkv does not rescue ectopic brk silencing (E’’ vs I’’), indicating that brk silencing in dSmad2 RNAi clones occurs via activation of TGFβ/Activin signaling. (J) Quantification of relative brk-LacZ reporter expression in GFP+ clones (n = 9 clones per condition).
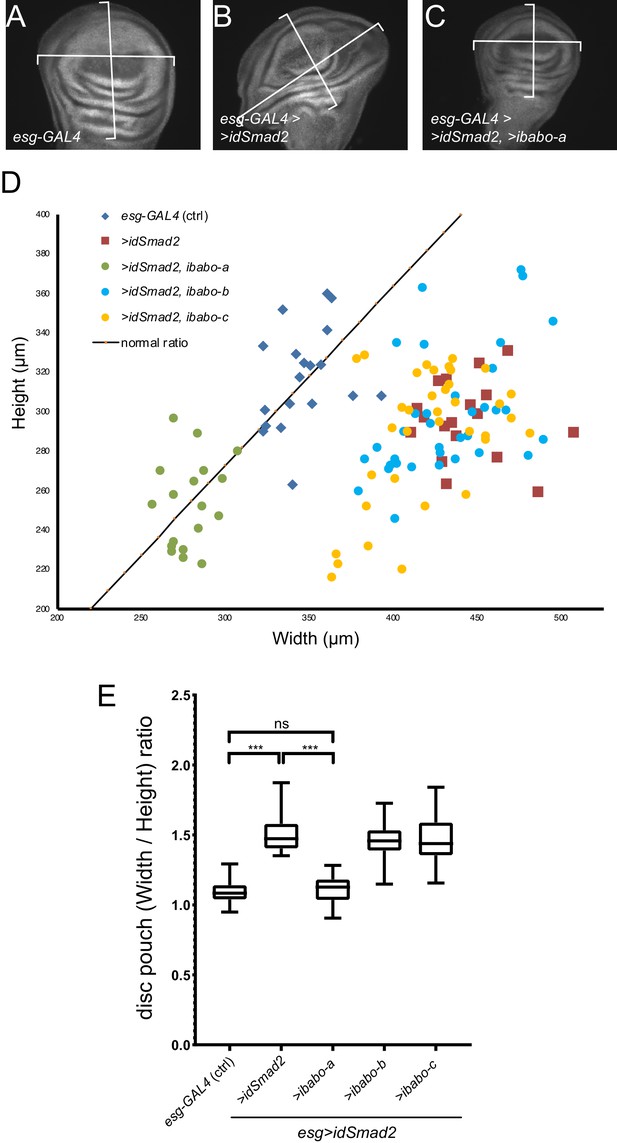
RNAi for babo-a rescues wing disc overgrowth due to dSmad2 RNAi.
(A) Representative image of wild-type (control) imaginal disc. The width/height ratio is generally 1.1 (black line in D) for controls (blue diamonds in D). (B) RNAi for dSmad2 throughout the disc using esg-GAL4 leads to ectopic brk silencing which results in overgrowth of the tissue along the anterior/posterior axis (widening) (red squares in D). (C) Simultaneous RNAi for dSmad2 and babo-a rescues the widening phenotype where the W/H ratio is back to 1.1 (green circles in D). (D) The size of all wing discs measured are depicted on a width by height chart. (E) Quantification of the width/height ratio of each disc as illustrated in A-C. Control samples (blue diamonds) have an average ratio of 1.1; RNAi for dSmad2 (red squares) leads to widening with a ratio of 1.5; RNAi for babo-a (green circles) alone rescues the widening back to wild type ratio of 1.1. These discs are smaller than wild type due to loss of canonical Baboon activity (Figures 1 and 2). There is no effect on widening by knocking down either babo-b or babo-c (blue and yellow circles vs red squares).
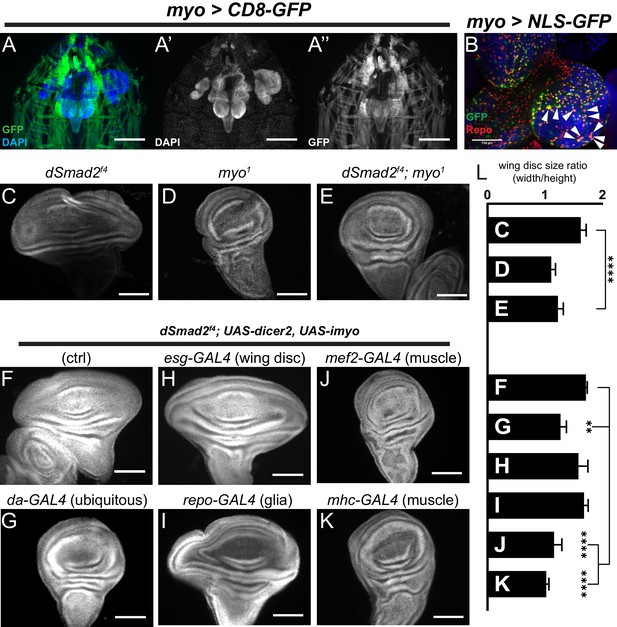
Muscle-derived Myo is required for non-canonical Babo activity in imaginal discs.
(A) 3rd instar larval fillet showing myo-GAL4 driving mCD8-GFP (green), DAPI (blue) counterstain to image all tissues in the field of view. DAPI channel (A’) shows muscle and cuticle-associated cell nuclei, as well as the brain and several imaginal discs. GFP channel (A’’) represents the expression pattern of the myo-GAL4 reporter transgene. Note that the wing imaginal discs does not express detectable GFP, scale bar = 500 µm. (B) Higher magnification of 3rd instar larval brain from myo >NLS GFP, and co-stained with anti-Repo marking glial cells. Arrowheads indicate overlap of myo-GAL4 expression in a subset of Repo+ glial cells, scale bar = 100 µm. (C–E) myo is epistatic to dSmad2, and rescues the disc overgrowth phenotype, scale bar = 100 µm, n > 5. dSmad2f4 mutant discs (C) are overgrown, however myo mutant discs (D) retain the normal shape of the tissue. The dSmad2f4;myo double mutant discs (E) are similar to myo mutant discs, demonstrating rescue of the overgrowth phenotype, and that Myo can function in non-canonical Babo activation. (F–K) Muscle specific RNAi of myo is sufficient to rescue wing disc overgrowth phenotype of dSmad2 mutants, scale bar = 100 µm, n > 5 per group. RNAi of myo ubiquitously using da-GAL4 rescues the wing disc overgrowth (F vs G), phenocopying double dSmad2;myo double mutants (E vs G). RNAi of myo in either wing discs (esg-GAL4) or glia (repo-GAL4) is unable to rescue disc overgrowth (F vs H or I). RNAi of myo in larval muscles using mef2-GAL4 or MHC-GAL4 rescues the disc overgrowth phenotype (F vs J or K), which phenocopies ubiquitous myo RNAi (G vs J or K). (L) Quantification of wing shape widening in C-K. dSmad2 wide wing is rescued by myo (C vs E) and knockdown of myo in muscles (F vs J or K).
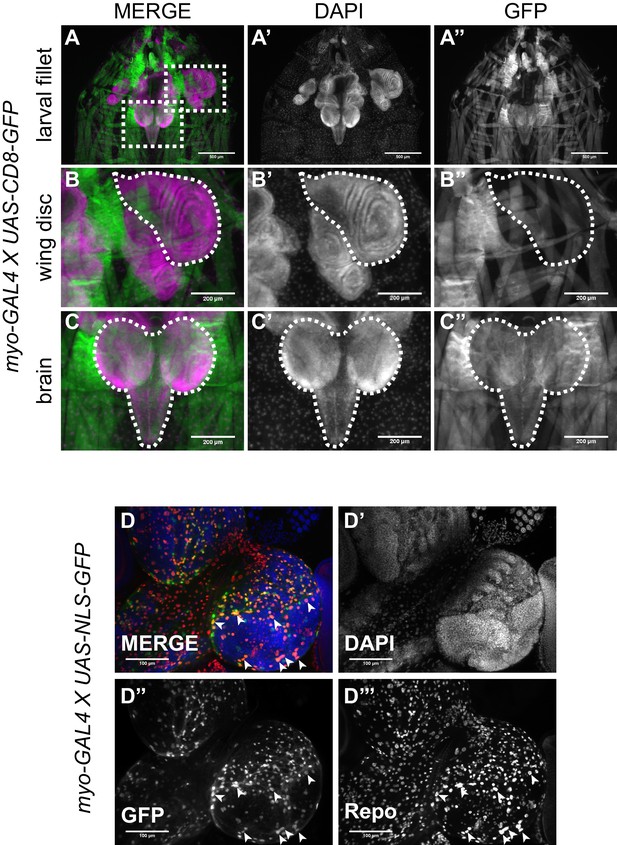
Higher magnification of larval filet showing myo-GAL4 expression pattern.
(A) Fillet of myo >CD8 GFP larvae showing expression pattern of the myo-GAL4 line, scale bar = 500 µm. Image is the same as in Figure 4, dashed boxes are shown in higher magnification in B-C. (B) High magnification of the wing imaginal disc from A, scale bar = 200 µm. No GFP signal is observed in the wing disc in B’’, the dashed outline of the wing disc shows where the wing disc should be in B’’. (C) High magnification of the larval brain from A, scale bar = 200 µm. GFP signal can be detected in the optic lobes as well as the ventral ganglion of the brain in C’’. (D) Split channel image of the image from Figure 4B, scale bar = 100 µm. Arrowhead show overlap for the myo >NLS GFP signal and the Repo+ cells.
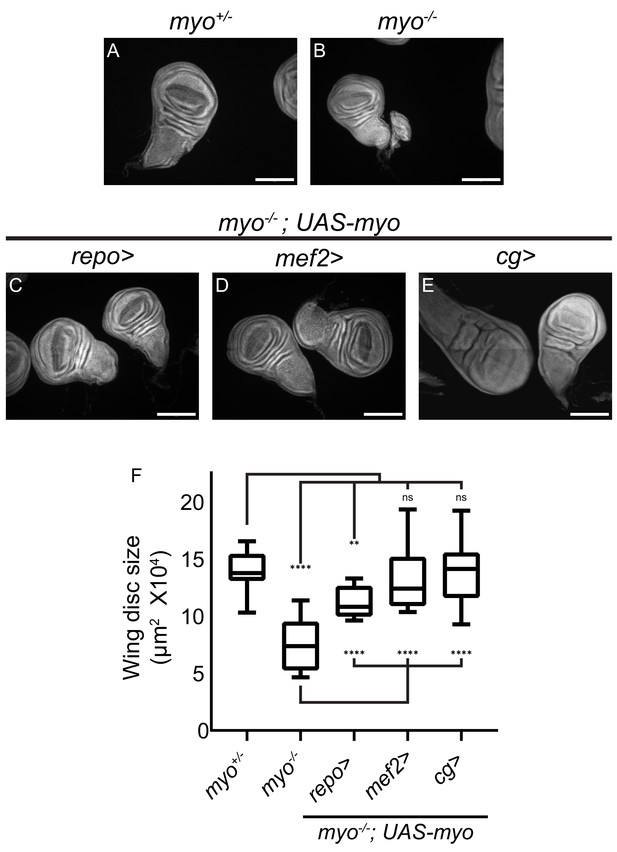
Overexpression of myo in muscles or fat body rescues wing disc size of myo mutants.
(A–B) Myo mutant discs (B) are smaller than heterozygous controls (A), and (C–E) wing discs from myo overexpression rescue experiments, scale bar = 100 µm, n > 19. Myo overexpression in glial cells using Repo-GAL4 partially rescues wing disc size (B vs C). Overexpression in muscles completely rescued myo mutant discs back to wild type size (B vs D vs A). Myo is not normally expressed in the fat body (Figure 4). However, ectopic overexpression in the fat body was sufficient to rescue discs back to wild-type size (B vs E vs A). (F) Quantification of wing disc size from (A–E).
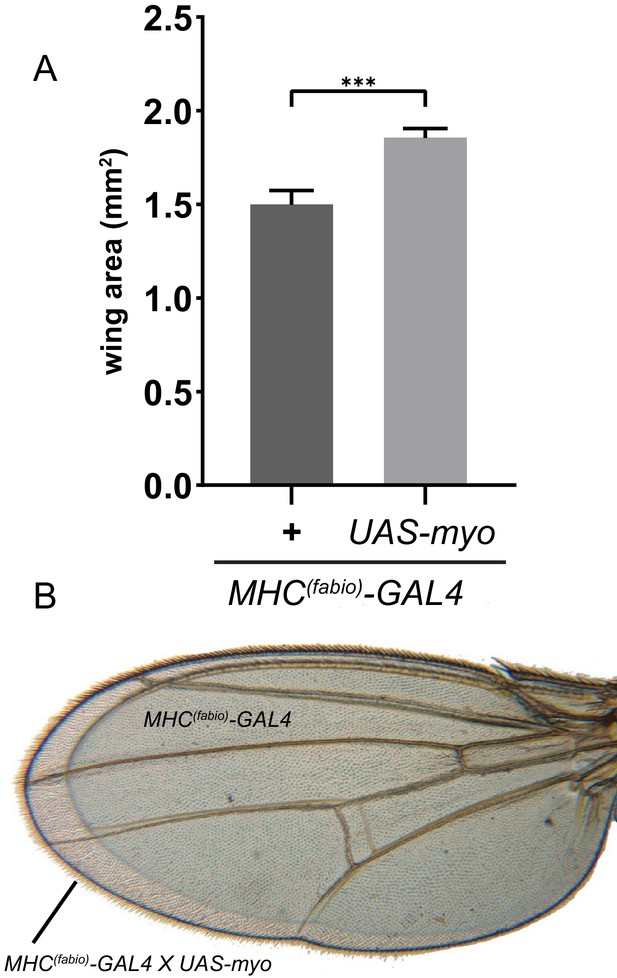
Ectopic Myo expression in muscle produces larger wings.
(A) Muscle specific myo overexpression in addition to endogenous levels increases final wing size of adults. (B) Representative wings from A of control (smaller wing) vs MHC >myo (larger wing) superimposed to show difference in size.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (D. melanogaster) | w1118 | BloomingtonDrosophilaStock Center | RRID:BDSC_5905 | |
| Genetic reagent (D. melanogaster) | myo1 | Awasaki et al., 2011 | FLYB:FBal0267082 | Synonym: myoΔ1 |
| Genetic reagent (D. melanogaster) | Actβ80 | Zhu et al., 2008 | FLYB:FBal0217392 | Synonym: Actβed80 |
| Genetic reagent (D. melanogaster) | daw11 | Serpe and O'Connor, 2006 | FLYB:FBal0212326 | |
| Genetic reagent (D. melanogaster) | myoCR2 | This paper | See Materials and methods | |
| Genetic reagent (D. melanogaster) | dSmad2F4 | Peterson et al., 2012 | FLYB:FBal0268512 | Synonym: SmoxF4 |
| Genetic reagent (D. melanogaster) | babo-a miRNA | Awasaki et al., 2011 | FLYB:FBtp0071178 | Also a new allele at VK37; see Materials and methods |
| Genetic reagent (D. melanogaster) | babo-b miRNA | Awasaki et al., 2011 | FLYB:FBtp0071179 | |
| Genetic reagent (D. melanogaster) | babo-c miRNA | Awasaki et al., 2011 | FLYB:FBtp0071180 | |
| Genetic reagent (D. melanogaster) | Myo-GAL4 | Awasaki et al., 2011 | FLYB:FBtp0071181 | |
| Genetic reagent (D. melanogaster) | esg-GAL4 | BloomingtonDrosophilaStock Center | FLYB:FBal0098823 | Lab stock is esg-GAL4/Cyo-GFP |
| Genetic reagent (D. melanogaster) | ci-GAL4 | Herman Steller | FLYB:FBtp0022318 | Source obtained from G Morata |
| Genetic reagent (D. melanogaster) | hh-GAL4 | Herman Steller | FLYB:FBal0121962 | Source obtained from G Morata |
| Genetic reagent (D. melanogaster) | da-GAL4 | BloomingtonDrosophilaStock Center | FLYB:FBal0042573 | |
| Genetic reagent (D. melanogaster) | myo miRNA | Awasaki et al., 2011 | FLYB:FBtp0071174 | |
| Genetic reagent (D. melanogaster) | Mef2-GAL4 | BloomingtonDrosophilaStock Center | FLYB:FBal0052385 | |
| Genetic reagent (D. melanogaster) | idSmad2 | Peterson et al., 2012 | FLYB:FBal0268514 | |
| Genetic reagent (D. melanogaster) | B14-LacZ | Müller et al., 2003 | FLYB:FBal0148007 | |
| Genetic reagent (D. melanogaster) | iPunt | This paper | See Materials and methods | |
| Genetic reagent (D. melanogaster) | iWit | This paper | See Materials and methods | |
| Genetic reagent (D. melanogaster) | iTkv | Peterson et al., 2012 | FLYB:FBal0268515 | |
| Genetic reagent (D. melanogaster) | iMyo | This paper | See Materials and methods | |
| Genetic reagent (D. melanogaster) | Repo-Gal4 | BloomingtonDrosophilaStock Center | FLYB:FBal0127275 | |
| Genetic reagent (D. melanogaster) | Mhc-Gal4 | BloomingtonDrosophilaStock Center | FLYB:FBti0160475 | P{Mhc-Gal4.K}2 |
| Genetic reagent (D. melanogaster) | Mhc(f)-GAL4 | N. Perrimon | FLYB:FBti0012514 | P{GAL4-Mhc.W}MHC-82 |
| Genetic reagent (D. melanogaster) | Cg-Gal4 | BloomingtonDrosophilaStock Center | FLYB:FBal0104726 | |
| Genetic reagent (D. melanogaster) | UAS-Myo-7D3 | Gesualdi and Haerry, 2007 | FLYB:FBtp0039710 | |
| Genetic reagent (D. melanogaster) | iRFP | BloomingtonDrosophilaStock Center | BDSC:41554 | |
| Genetic reagent (D. melanogaster) | iGFP | BloomingtonDrosophilaStock Center | BDSC:67852 | |
| Antibody | p-dSmad2 (rabbit monoclonal) | Cell Signaling Technology | Cat# 3108; RRID:AB_490941 | WB (1:1000); detects phosphorylated dSmad2 |
| Antibody | Tubulin (mouse monoclonal) | Sigma-Aldrich | Cat# T9026 | WB (1:1000) |
| Antibody | p-Mad (rabbit polyclonal) | Peterson et al., 2012 | WB (1:500); detects phosphorylated Mad | |
| Antibody | Repo (mouse monoclonal) | DSHB | Cat# 8D12; RRID:AB_528448 | IF(1:100) |





