Regulation of mRNA translation by a photoriboswitch
Figures
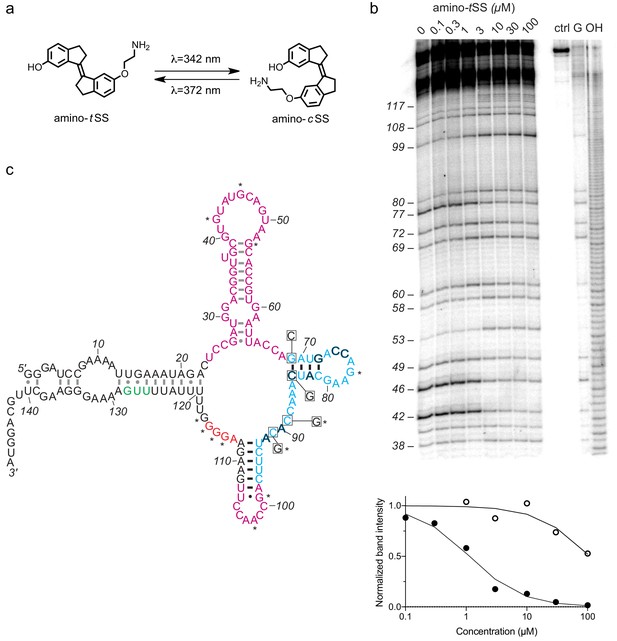
An amino-tSS-responsive aptamer.
(a) Amino-tSS isomerizes from trans to cis conformation when exposed to 342 nm light, and back to the trans isoform at 372 nm. (b) RNase T1 probing of Were-1 structure. Right lanes contain a control with undigested RNA (ctrl), a T1-digested sequencing control (G), and a hydroxide-mediated partial digestion ladder (OH) of the RNA. The left lanes show partial T1 digestion in the presence of increasing amino-tSS at concentrations indicated above the gel image. The probing shows clear ligand-dependent changes—both increases (e.g. G53, G99, G114-117) and decreases (e.g. G 42, (G46, G77)—interspersed throughout the sequence. Below, an apparent KD of 1.1 µM was calculated based on the change in band intensity with increasing amino-tSS (dark, filled circles) for nucleotide G46, normalized to a control band (G72). Additionally, a KD of 108 µM was calculated based on the change in band intensity with increasing amino-cSS (open circles) for the same nucleotide and control (Figure 1—figure supplement 4g), suggesting high specificity for amino-tSS. An average KD value of 1.5 µM amino-tSS was calculated for changes in nucleotides G42, G46, G77, and G80. (c) Secondary structure prediction of Were-1 derived from all structural probing data in absence of the ligand (see also Figure 1—figure supplement 6, Figure 1—figure supplement 7, Figure 1—figure supplement 8, Figure 1—figure supplement 9). Partially randomized regions (light blue), the Shine-Dalgarno sequence (red), the start codon (green), and the 3' terminus sequence are derived from the B. subtilis mswA SAM-I riboswitch. The 5' part of the aptamer and the loop sequence (A98–U107) of the expression platform (pink) were selected from random regions of the starting pool (Figure 1—figure supplement 1). Outlined dark letters are positions where the selected sequence differs from the B. subtilis riboswitch expression platform. Boxed positions were mutated to the indicated nucleotides to identify regions of structural and functional importance. Black base-pairs indicate stems that do not change in the presence of amino-tSS, and asterisks (*) indicate nucleotide positions that do change in the presence of amino-tSS.
-
Figure 1—source data 1
Derivation of dissociation constants from T1 probing data.
- https://cdn.elifesciences.org/articles/51737/elife-51737-fig1-data1-v3.xlsx
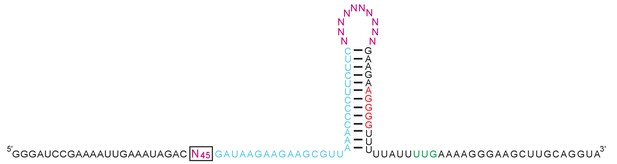
In vitro selection pool design for a photoriboswitch.
An RNA pool was derived from B. subtilis mswA SAM-I riboswitch by replacing its ligand-binding domain with a 45-nucleotide random sequence (pink, boxed), partially randomizing (at a 15% level) its anti-terminator hairpin and the 5' half of the terminator hairpin (light blue), replacing the terminator-helix tetraloop sequence (UUAU) with a random decamer (pink), and retaining the 3' half of the terminator hairpin and its translation initiation sequences (red – Shine-Dalgarno sequence; green – start codon). The pool’s constant regions are shown in black.
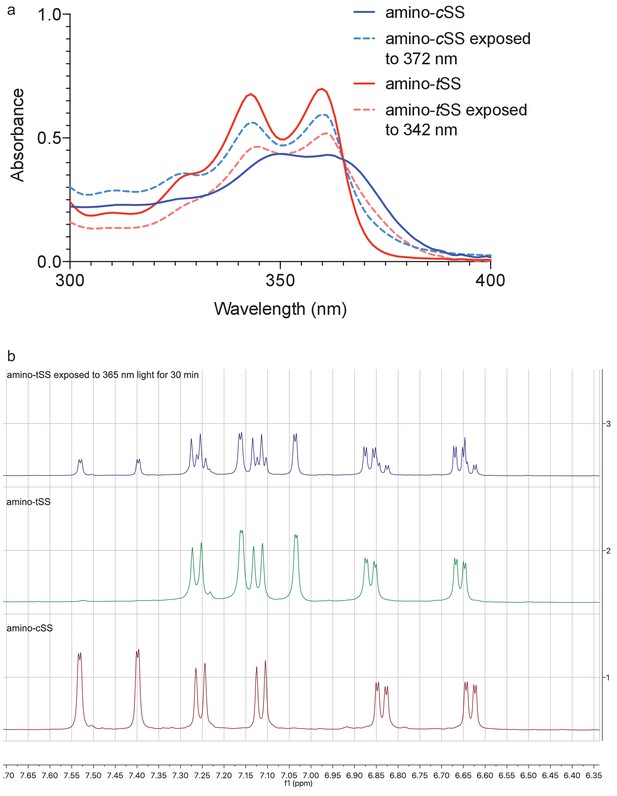
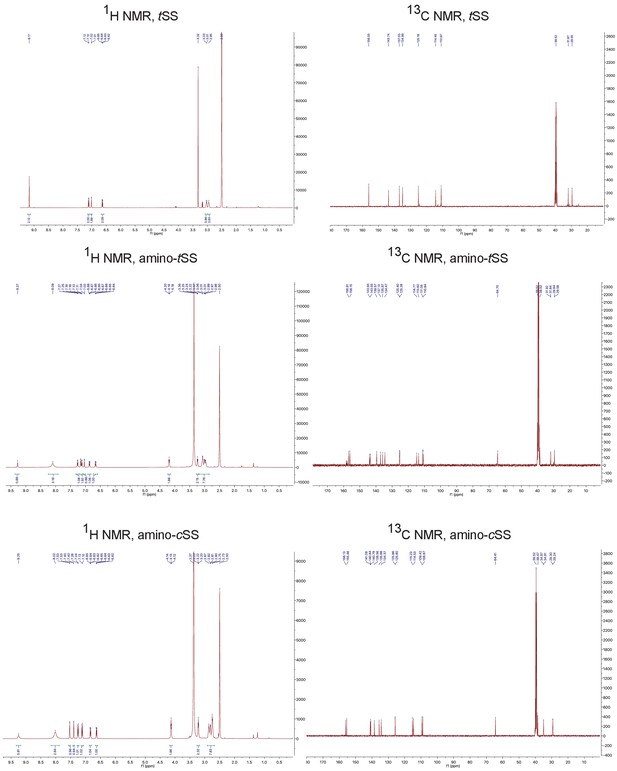
UV absorption and 1H NMR spectroscopy of E/Z isomerization of amino-tSS and amino-cSS.
(a) UV-Vis spectra of amino-tSS (red) and amino-tSS after 3 hr exposure to 342 nm (red, dashed), and amino-cSS (blue) and amino-cSS after 1.5 hr exposure to 372 nm (blue, dashed). Photo isomerization was performed in 30 mM DMSO solutions and subsequently diluted 1000 times for UV-Vis spectra measurement. (b) 1H NMR of amino-tSS, amino-cSS, and E/Z isomerization. At the photostationary state (amino-tSS exposed to 365 nm) the ratio of amino-tSS to amino-cSS is approximately 2:1 as determined by NMR.
-
Figure 1—figure supplement 2—source data 1
UV spectra of amino-tSS and amino-cSS.
- https://cdn.elifesciences.org/articles/51737/elife-51737-fig1-figsupp2-data1-v3.xlsx
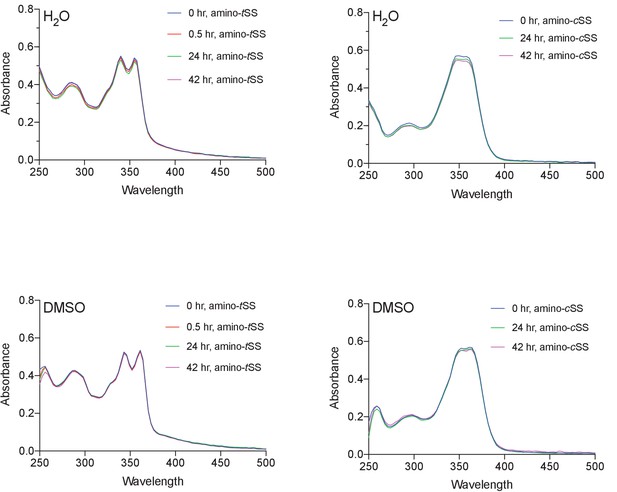
Stability of amino-tSS and amino-cSS in water and DMSO analyzed by UV-vis spectroscopy.
Both isoforms of the ligand are relatively stable in either solvent for the indicated times.
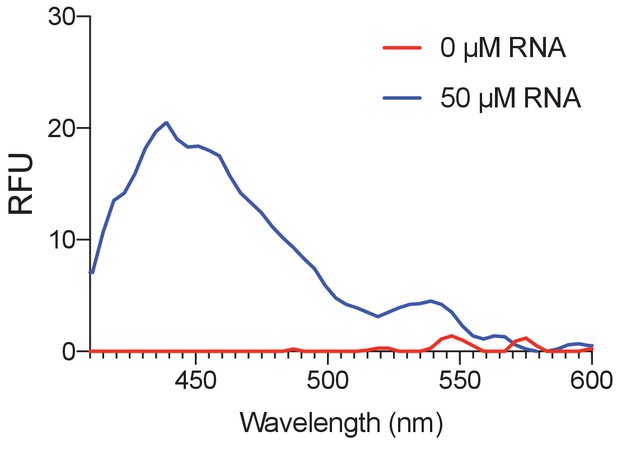
Fluorescence of Were-1 bound to amino-tSS.
Fluorescence emission of 100 nM amino-tSS incubated with purified Were-1 RNA showing an increase in presence of Were-1 and suggesting RNA affinity for the target ligand. Emission spectra were collected using an excitation of 365/10 nm.
-
Figure 1—figure supplement 5—source data 1
Fluorescence spectrum of RNA–bound amino-tSS.
- https://cdn.elifesciences.org/articles/51737/elife-51737-fig1-figsupp5-data1-v3.xlsx
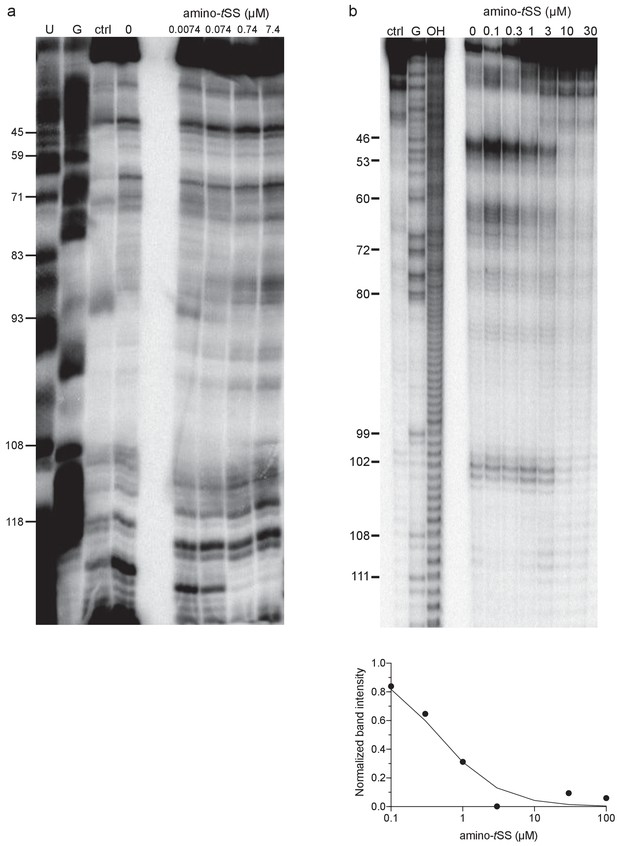
Structural probing of Were-1 in presence of amino-tSS using SHAPE and S1 nuclease digestion.
(a) SHAPE analysis of Were-1. Left lanes show reverse transcriptase (RT) stops due to ddA (‘U’) and ddC (‘G’) incorporation for sequence reference, followed by a no-acylation RT control (ctrl) lane, and SHAPE reactions with amino-tSS at concentrations indicated above the gel image. (b) S1 digestion of Were-1. Left lanes contain a control with no RNA (ctrl), RNase T1 digestion for sequence reference, and a partial hydrolysis lane (OH). The right lanes show S1 digestion in the presence of increasing amino-tSS at concentrations indicated above the gel image. Analysis of the band intensity change at positions A44-G46 revealed an apparent KD value of 0.4 µM (below).
-
Figure 1—figure supplement 6—source data 1
Derivation of apparent Kd values from RNA probing data.
- https://cdn.elifesciences.org/articles/51737/elife-51737-fig1-figsupp6-data1-v3.xls

Structural probing of Were-1 in presence of amino-tSS, amino-cSS, tDHS, tS, and SAM by terbium (III) footprinting.
Left lanes show RNase T1 digestion for sequence reference, and partial hydrolysis (OH). Middle lanes show Tb3+ footprinting in the presence of increasing amino-tSS at concentrations indicated above the gel image. Right lanes show control reactions with Were-1 in the presence of 30 µM controls, as labeled above the gel image. Right, a KD value of 4.8 µM was calculated based on the change in intensity with increasing amino-tSS at nucleotides A113 and U107.
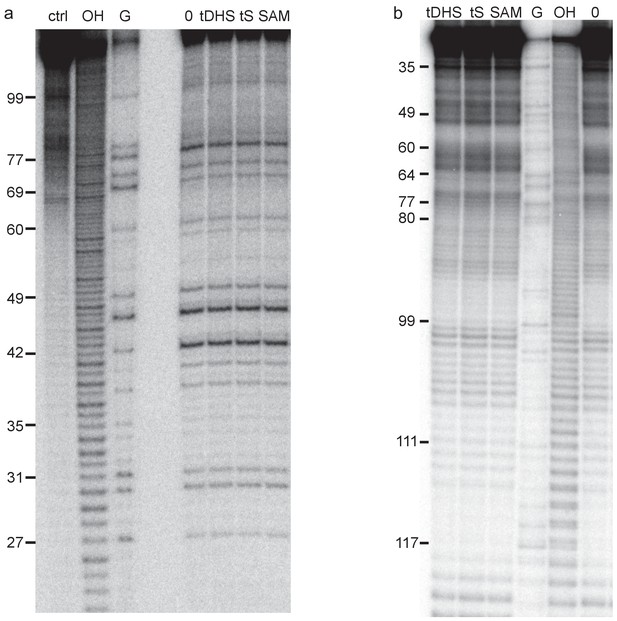
Structural probing of Were-1 in presence of tDHS, tS, and SAM by T1 nuclease digestion and in-line probing.
(a) T1 digestion controls. Left lanes contain a control with no RNA, partial hydrolysis (OH), and RNase T1 digestion for sequence reference. Right lanes indicate no ligand (0), followed by Were-1 in the presence of 30 µM controls, as labeled above the gel image. (b) In-line probing of Were-1 with 30 µM controls, as labeled above the gel image, followed by RNase T1 digestion for sequence reference, partial hydrolysis (OH), and a no ligand control (0).
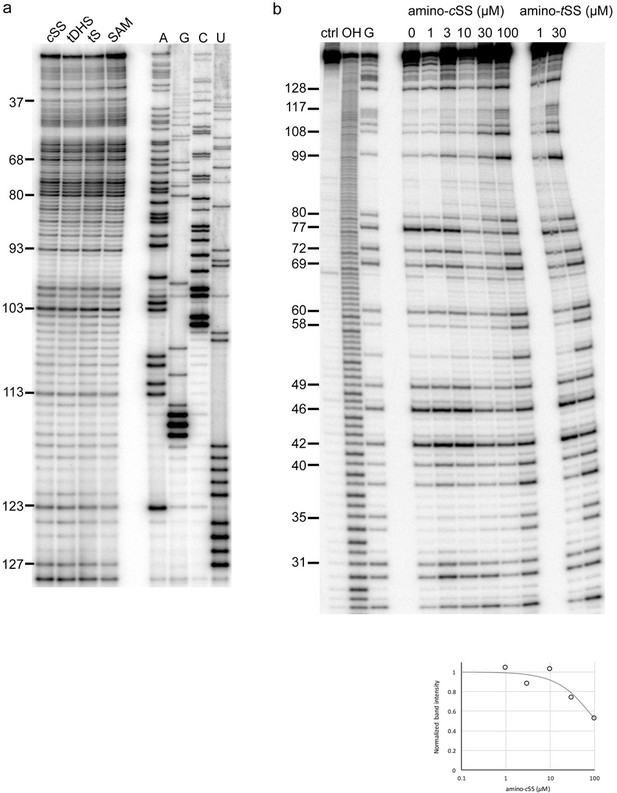
Structural probing of Were-1 in presence of amino-cSS, tDHS, tS, and SAM by SHAPE, and amino-cSS titration by T1 nuclease digestion.
(a) SHAPE of Were-1 in presence of control small molecules. Left lanes show RT stops for Were-1 in the presence of 30 µM amino-cSS, tDHS, tS, and SAM. Right lanes show RT stops due to ddT (‘A’), ddC (‘G’), ddG (‘C’), and ddA (‘U’) incorporation for sequence reference. (b) T1 digestion of Were-1 in the presence of increasing amino-cSS. Left lanes show undigested RNA, partial hydrolysis, and a G-specific sequencing lane. Middle lanes show Were-1 digestion in the presence of increasing amino-cSS and the right lane show amino-tSS experiments at low and high concentrations, for direct comparison of the ligand-induced structural probing. At high concentration (100 µM) amino-cSS mimics the bound profile of amino-tSS. The apparent KD calculation for cSS is shown below the gel image and in Figure 1b, revealing a KD of 108 µM.
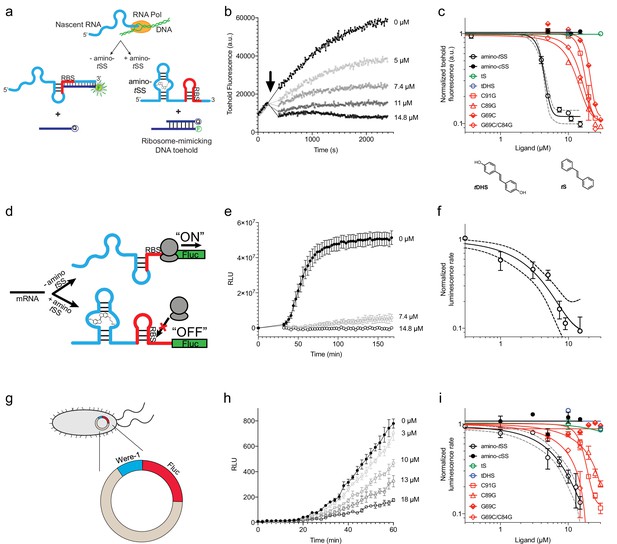
Translation regulation by the Were-1 riboswitch.
(a) Schematic of co-transcriptional binding of Were-1 RNA to amino-tSS in the presence of a toehold-reporter complex. In absence of amino-tSS, the transcribed RNA exposes the ribosomal binding site (RBS), enabling binding of the complementary region of the toehold reporter, displacing the quencher strand, and producing a fluorescence signal. In presence of amino-tSS, the RNA binds the ligand, sequestering the RBS and preventing displacement of the quencher strand. (b) Co-transcriptional response of Were-1 to different concentrations of amino-tSS using the toehold reporter. Initial transcriptions of Were-1 without ligand show identical increase in toehold fluorescence for all samples. When amino-tSS is added (arrow), a dose-dependent decrease in fluorescence is observed. (c) Response (± SEM; n = 81) of Were-1 (black, open circles), and its variants (red) C89G (triangles), C91G (squares), G69C (half-shaded diamonds), and G69C/C84G (open diamonds), in the presence of amino-tSS shows a shift in dose-dependence for single mutations, particularly G69C, and partial recovery of activity for the G69C/C84G double mutant. Were-1 shows no response in the presence of amino-cSS (black circles), trans-stilbene (tS, green, open circles) and trans-4,4-dihydroxystilbene (tDHS, blue, open circles). Structures of tDHS and tS are shown below the graph. (d) Schematic of amino-tSS-dependent inhibition of protein expression in vitro using a Were-1-firefly luciferase (Were-1-Fluc) construct. In absence of the ligand, the RBS is exposed and luciferase is translated, whereas in presence of amino-tSS, the RBS is sequestered, abrogating Fluc expression. (e) In vitro translation of the Were-1-Fluc construct. Robust luminescence is observed when no ligand is present, but the signal is significantly lower in presence of amino-tSS. (f) Response (± SEM; n = 58) of the Were-1–regulated protein expression to amino-tSS. (g) Schematic of the Were-1-Fluc construct incorporated into a bacterial plasmid. (h) Were-1–controlled Fluc gene expression in E. coli. Bioluminescence is observed in absence of amino-tSS, and progressively diminished with increasing amino-tSS. (i) Expression of Were-1-Fluc (± SEM; n = 257) in vivo (black, open circles), and its variants (red) C89G (triangles), C91G (squares), and G69C/C84G (open diamonds), in the presence of amino-tSS, show a dose-dependent response. Were-1 mutant G69C (half-shaded diamond) and Were-1 in the presence of amino-cSS (black circles), trans-stilbene (green, open circles) and trans-4,4-dihydroxystilbene (blue, open circles) showed no change in bioluminescence, whereas the G69C/C84G double mutant shows restoration of activity similar to wild-type levels. Note, dose-response graphs (c, f, i) are on a log-log scale. The apparent amino-tSS IC50s are 3.9 ± 0.2, 2.5 ± 1.0, and 5.3 ± 1.1 µM for the toehold (c), in vitro translation (f), and in vivo expression (i), respectively. Dashed lines correspond to the 95% confidence interval of the binding model.
-
Figure 2—source data 1
Kinetic data for Were-1 activity.
- https://cdn.elifesciences.org/articles/51737/elife-51737-fig2-data1-v3.xlsx
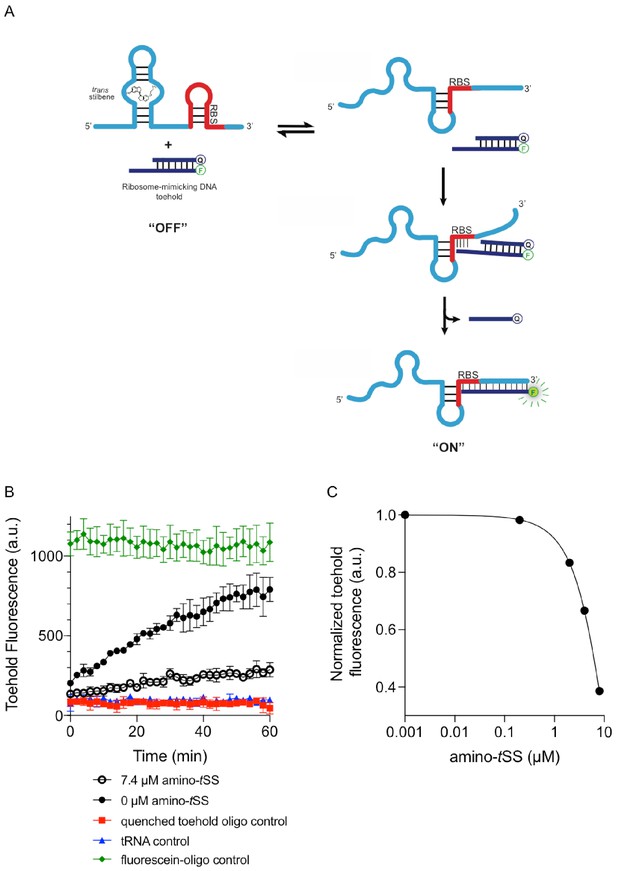
Binding PAGE-purified Were-1 RNA to a ribosome-mimic.
(a) Schematic of strand displacement, where a DNA duplex containing a toehold complementary to the Shine-Dalgarno sequence of Were-1 is displaced when no amino-tSS is present, producing a fluorescence. (b) The presence of amino-tSS prevents the quencher strand release, suppressing toehold fluorescence (± SD) over time. Control reactions were performed using the unquenched fluorophore oligo (green) and the quenched toehold oligo (red) for fully quenched fluorescence to provide a window for maximum fluorescence, and a tRNA (blue) lacking the Shine-Dalgarno sequence as a negative control. (c) Dose-response curve of PAGE-purified Were-1 RNA in the presence of increasing concentrations of amino-tSS and analyzed by the toehold-fluorescence assay. Half maximum fluorescence is observed at 6.3 µM amino-tSS.
-
Figure 2—figure supplement 1—source data 1
Time course of toehold binding by Were-1.
- https://cdn.elifesciences.org/articles/51737/elife-51737-fig2-figsupp1-data1-v3.xlsx
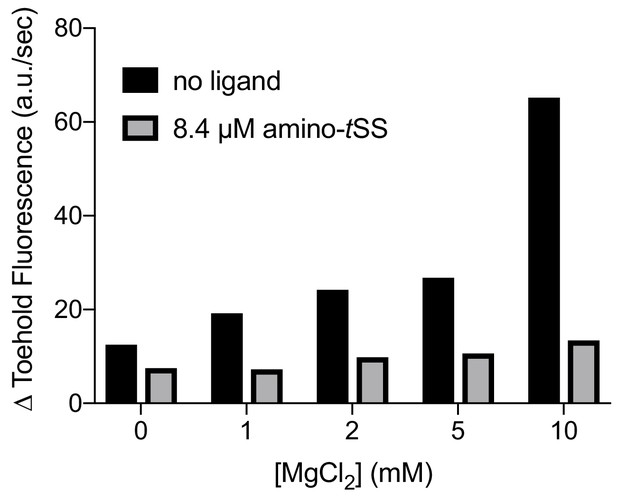
Co-transcriptional binding of a ribosome-mimic in vitro under various Mg2+ conditions.
Whereas the rate of transcription increases with magnesium (black), the bound state (gray) does not change significantly, suggesting that co-transcriptional binding is minimally affected by magnesium concentration.
-
Figure 2—figure supplement 2—source data 1
Effect of Mg (II) on toehold binding by Were-1 in absence and presence of amino-tSS.
- https://cdn.elifesciences.org/articles/51737/elife-51737-fig2-figsupp2-data1-v3.xlsx
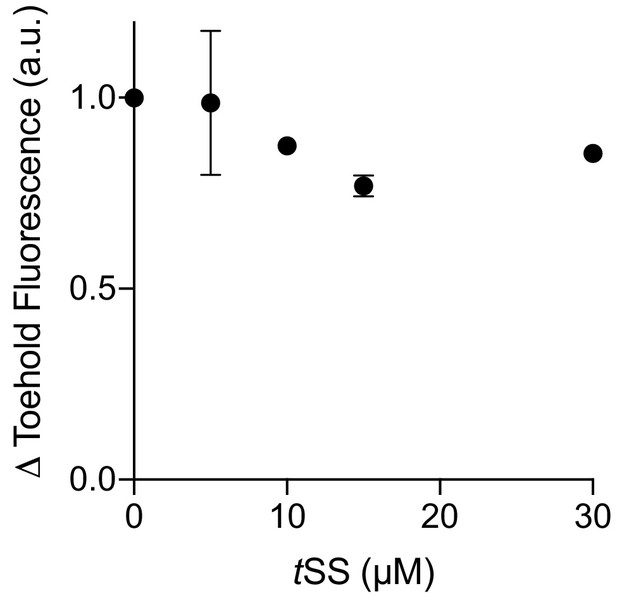
Co-transcriptional Were-1 binding of a ribosome-mimic in vitro in the presence of tSS (in 10% DMSO).
A minor dose-response is observed in the presence of increasing tSS concentrations, but full inhibition, as seen for amino-tSS, is not observed, possibly due to low solubility of tSS.
-
Figure 2—figure supplement 3—source data 1
Effect of tSS in Were-1 binding using the toehold assay.
- https://cdn.elifesciences.org/articles/51737/elife-51737-fig2-figsupp3-data1-v3.xlsx
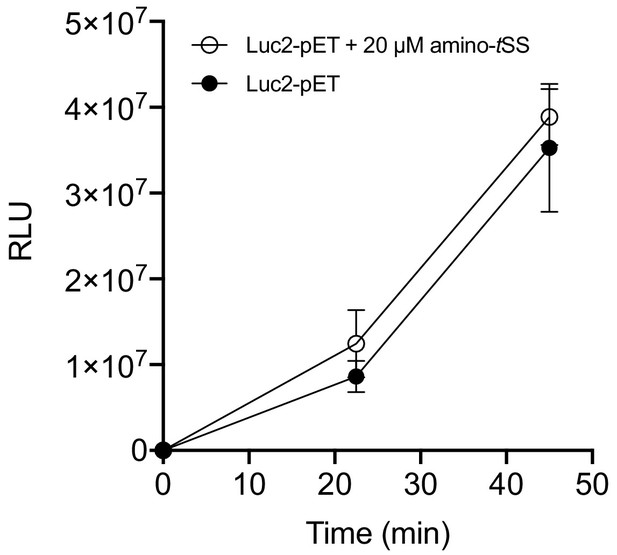
Translation of a control plasmid, Luc2-pET, lacking the Were-1 riboswitch is not inhibited in vitro by amino-tSS.
There is no significant difference in Luc2-pET luminescence (± SD) in the presence and absence of amino-tSS, suggesting that amino-tSS does not inhibit the in vitro translation system.
-
Figure 2—figure supplement 4—source data 1
Lack of inhibition of Were-1-independent expression system by amino-tSS.
- https://cdn.elifesciences.org/articles/51737/elife-51737-fig2-figsupp4-data1-v3.xlsx
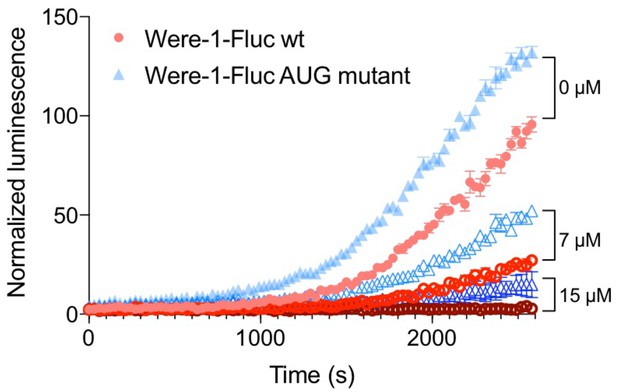
The effect of the canonical start codon on Were-1-Fluc expression in vivo.
Comparison of luciferase expression (± SEM) of Were-1-Fluc wild-type (wt; red) and Were-1-Fluc containing an AUG start codon (Were-1-Fluc AUG mutant; blue), in place of the pool-derived UUG minor start codon, with increasing amino-tSS concentrations. Bioluminescence was overall higher in the Were-1-Fluc AUG mutants for all conditions but retained the wt dose-dependent response.
-
Figure 2—figure supplement 5—source data 1
Comparison of protein expression by amino-tSS in Were-1 variants containing UUG or AUG start codon.
- https://cdn.elifesciences.org/articles/51737/elife-51737-fig2-figsupp5-data1-v3.xls
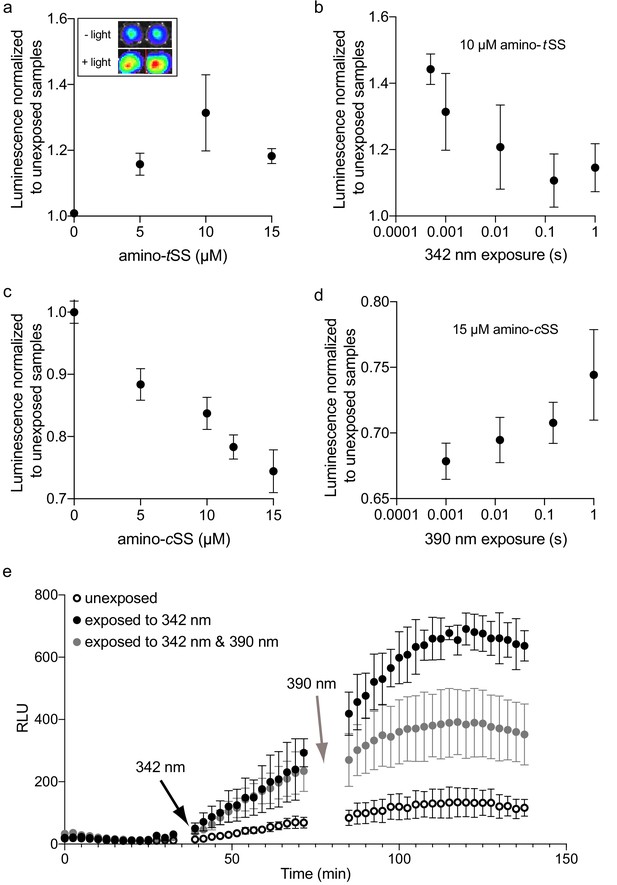
Regulation of luciferase expression by the Were-1 photoriboswitch in vivo.
(a) Normalized amino-tSS–dependent bioluminescence (± SEM) of the Were-1-Fluc construct after 1 ms exposure of 342 ± 5 nm light (Φq = 1.4*10−2 W/cm2). The largest change in expression was observed in the presence of 10 µM amino-tSS. Inset shows the light–dependent bioluminescence of the bacterial cultures at 10 µM ligand. (b) Were-1 regulation of luciferase expression (± SEM) in vivo at various exposure times in presence of 10 µM amino-tSS. (c) Normalized bioluminescence of the Were-1-Fluc E. coli incubated with amino-cSS after 1 s exposure of 390 ± 9 nm light (Φq = 5.5*10−2 W/cm2) showing progressively lower protein expression at higher amino-cSS concentrations, presumably due to higher production of amino-tSS after photoisomerization. (d) Change of Fluc expression after photoisomerization of 15 µM amino-cSS at 390 ± 9 nm for various exposure times, showing largest photoswitching at 1 ms exposure. (e) Regulation of luciferase expression (± SEM) by the Were-1-Fluc construct before exposure, after a 1 ms pulse of 342 ± 5 nm light (black arrow), which resulted in increased bioluminescence (black and gray circles) compared to control (empty circles). Samples exposed to 0.5 ms of 390 ± 9 nm light (gray arrow) showed gradual inhibition of new protein production (gray) compared to samples that were only exposed to 342 nm (black).
-
Figure 3—source data 1
Photoswitching of Were-1.
- https://cdn.elifesciences.org/articles/51737/elife-51737-fig3-data1-v3.xls

Co-transcriptional binding of a ribosome-mimic in vitro under amino-tSS photoisomerization conditions.
(a) Initial transcription of Were-1 without ligand was induced by adding ribonucleoside triphosphates (rNTPs) to a transcription mix, producing robust toehold fluorescence increase (see Figure 2b for examples of co-transcriptional toehold binding). Upon adding 11 µM amino-tSS (arrow), fluorescence growth decreased, suggesting that Were-1 is bound to the ligand and prevents toehold binding. The sample was then exposed to 342 nm (Φq = 6.8*10−5 W/cm2) for 60 s (indicated by '342 nm' under the graph), causing photoisomerization of the ligand, and resulting in increased toehold fluorescence slope. Were-1 transcription reaction was subsequently exposed to 372 nm light (Φq = 1*10−4 W/cm2) for 60 s, to switch the ligand from cis to trans conformation, which resulted in decrease of toehold fluorescence slope. This was repeated two more times, as indicated, until the transcription reaction plateaued, and all toehold was expended. The data suggest that the system can be regulated multiple times in one reaction. Error bars represent fluorescence slope error. (b) Kinetics of co-transcriptional toehold fluorescence after irradiation with short pulses of 342 ± 10 nm light (Φq = 1.4*10−2 W/cm2) with exposure lengths indicated to the right of the graph. Experiments with 10 (red hues) and 15 µM (blue hues) amino-tSS are shown. Error bars indicate average deviation of two experiments.
-
Figure 3—figure supplement 1—source data 1
Photoswitching of Were-1 binding by the DNA toehold.
- https://cdn.elifesciences.org/articles/51737/elife-51737-fig3-figsupp1-data1-v3.xlsx

Were-1 photoregulates translation in vitro.
In vitro transcription-translation reaction of Were-1-Fluc mRNA in presence of amino-tSS (left column) was irradiated at 342 nm for 60 s (Φq = 6.8*10−5 W/cm2), resulting in an increase in luciferase production (± SEM). The reaction was then irradiated at 372 nm for 60 s (Φq = 1*10−4 W/cm2), resulting in a decrease in luciferase expression rate (± SEM; Φq = 1*10−4 W/cm2) over time.
-
Figure 3—figure supplement 2—source data 1
Photoregulation of Were-1-regulated translation in vitro.
- https://cdn.elifesciences.org/articles/51737/elife-51737-fig3-figsupp2-data1-v3.xlsx
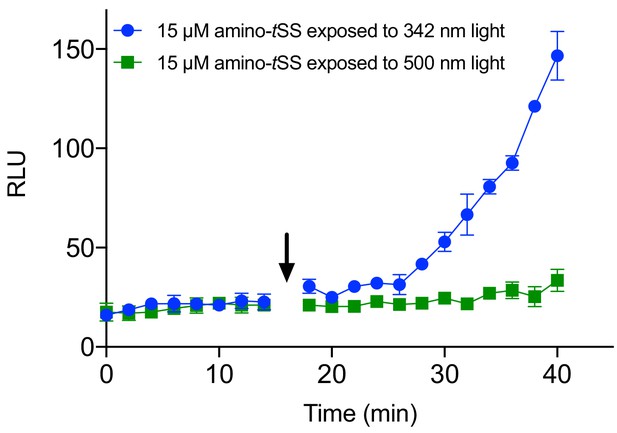
Were-1 photoregulation of protein expression in vivo.
Were-1 E. coli bioluminescence (± SEM) was measured in the presence of amino-tSS. Initial Fluc expression showed identical increases in bioluminescence for all samples. In the presence of 15 µM amino-tSS, samples were either exposed to (arrow) 342 nm light (blue) to isomerize amino-tSS, or 500 nm (green), a wavelength that does not affect amino-tSS isomerization. 342 nm–exposed samples showed significantly higher bioluminescence compared to those exposed to 500 nm light.
-
Figure 3—figure supplement 3—source data 1
Wavelength-dependent photoregualtion of Were-1-regulated protein expression in vivo.
- https://cdn.elifesciences.org/articles/51737/elife-51737-fig3-figsupp3-data1-v3.xlsx
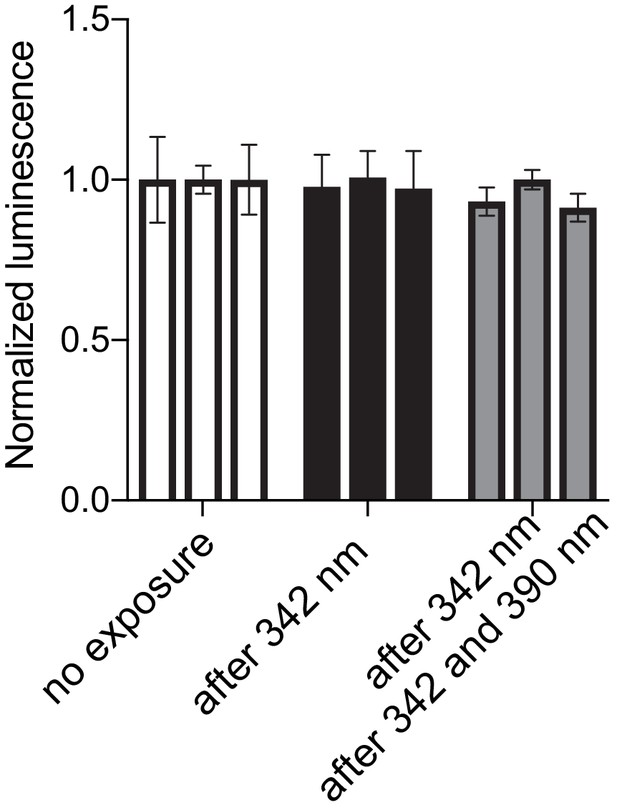
Lack of photoactivity of the Were-1-Fluc G69C variant.
Luciferase expression (± SEM) by the Were-1-Fluc G69C mutant, showing no significant change in expression after exposure to 342 nm (black, second and third bar), or 342 nm followed by 390 nm (second and third gray bar, respectively) compared to unexposed controls (clear).
-
Figure 3—figure supplement 4—source data 1
Lack of photoswitching of the G69C variant of Were-1.
- https://cdn.elifesciences.org/articles/51737/elife-51737-fig3-figsupp4-data1-v3.xlsx
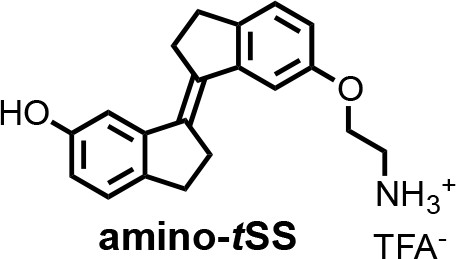
(E)−6'-(2-aminoethoxy)−2,2',3,3'-tetrahydro-[1,1'-biindenylidene]−6-ol (amino-tSS).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | BL21(DE3) | Sigma-Aldrich | CMC0016 | Electrocompetent cells |
| Recombinant DNA reagent | pBV-Luc-c36 (plasmid) | This paper | IPTG inducible Were-1–controlled firefly luciferase gene with T7 RNA polymerase promoter | |
| Recombinant DNA reagent | pBV-Luc (plasmid) | Adgene | RRID: Addgene_16539 | Parent plasmid for Were-1 construct |
| Chemical compound, drug | Amino-tSS | This paper | Were-1 ligand, trans isoform | |
| Chemical compound, drug | Amino-cSS | This paper | Cis isoform of the Were-1 ligand | |
| Chemical compound, drug | tSS | This paper | Were-1 ligand, trans isoform lacking a linker | |
| Chemical compound, drug | tS | Sigma-Aldrich | Cat# 139939 | Trans stilbene |
| Chemical compound, drug | tDHS | Santa Cruz Biotechnology | CAS 659-22-3 | 4,4-trans-dihydroxystilbene |
Additional files
-
Supplementary file 1
Tabulated values shown in graphs of Figures 1–3 and their figure supplements.
- https://cdn.elifesciences.org/articles/51737/elife-51737-supp1-v3.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/51737/elife-51737-transrepform-v3.docx