Yolk-sac-derived macrophages progressively expand in the mouse kidney with age
Figures
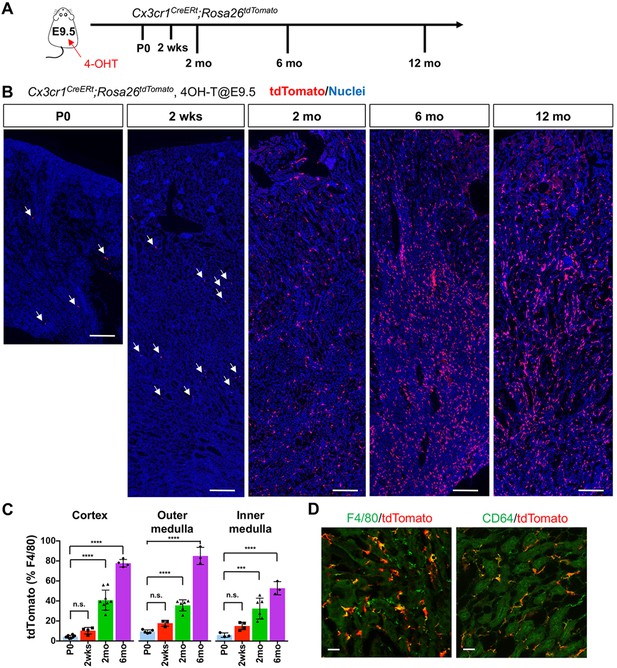
CX3CR1-positive yolk-sac macrophage descendants progressively expand in number in kidneys with age.
(A) Fate-mapping strategies of CX3CR1+ yolk-sac macrophages. 4-hydroxytamoxifen (4-OHT) was injected once into pregnant dams at 9.5 dpc and offspring analyzed at the indicated times (n = 4–6 for P0 to 6-month-old; n = 2 for 12-month-old). Yolk-sac macrophages and their progeny are irreversibly tagged with tdTomato. (B) Distribution of CX3CR1-lineage cells in postnatal kidneys. Arrows: CX3CR1-lineage cells. (C) Percentage of tdTomato+ to F4/80+ cells. Data are represented as means ± S.D. ***, p<0.001; ****, p<0.0001; n.s., not significant. (D) Confocal images of F4/80 and CD64 staining in aged kidneys (six mo) with CX3CR1-lineage tracing (n = 3). Scale bars: 200 μm in B; 20 μm in D.
-
Figure 1—source data 1
Percentage of tdTomato+ to F4/80+ cells.
- https://cdn.elifesciences.org/articles/51756/elife-51756-fig1-data1-v2.xlsx
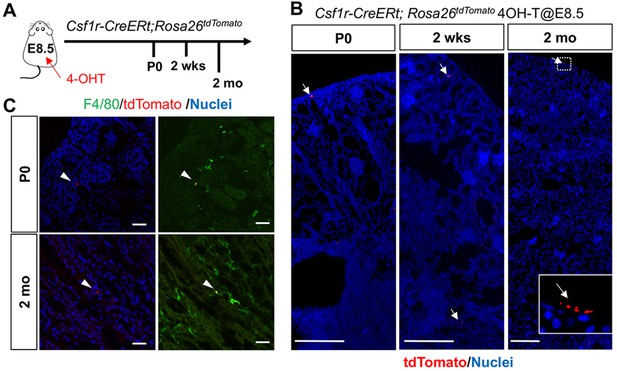
CSF1R-positive yolk-sac macrophage descendants do not expand in number in kidneys.
(A) Fate-mapping strategies of CSF1R+ yolk-sac macrophages. 4-hydroxytamoxifen (4-OHT) was injected once into pregnant dams at 8.5 dpc and offspring analyzed at the indicated times (n = 4–6). Yolk-sac macrophages and their progeny are irreversibly tagged with tdTomato. (B) Distribution of CSF1R-lineage cells in postnatal kidneys (n = 4). Arrows: CSF1R-lineage cells. Inset: higher magnification of dotted box. (C) Confocal images of F4/80 staining with CSF1R-lineage tracing (n = 3). Arrowheads: F4/80+CSFIR-lineage cells. Scale bars: 200 μm in B; and in 20 μm in C.
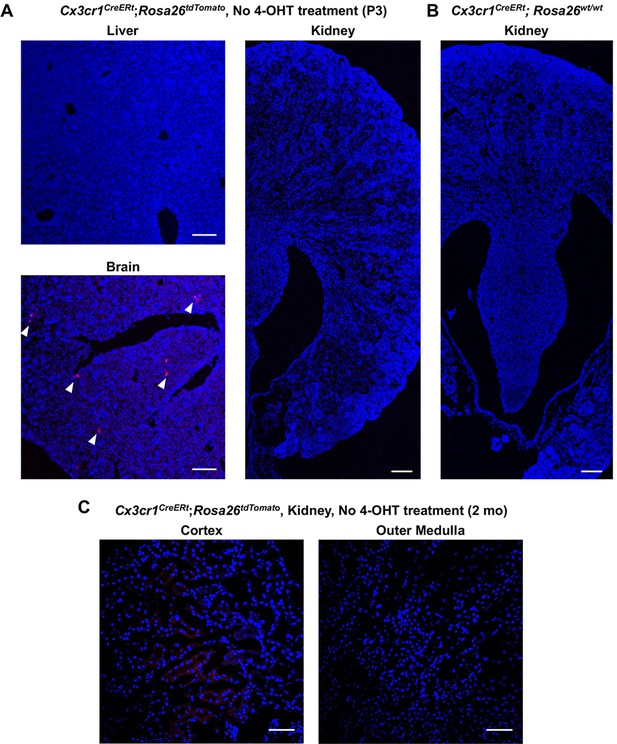
There is no basal Cre activity in kidneys without 4-hydroxytamoxifen (4-OHT) treatment.
Cx3cr1CreERt; Rosa26tdTomato/wt mice were used to determine tdTomato reporter activity in the absence of 4-OHT treatment. (A) While we observed a few tdTomato-expressing cells in the brain, there were no tdTomato-positive cells in the liver and kidney at P3 without 4-OHT treatment. Scale bars: 100 μm. (B) Cx3cr1CreERt; Rosa26wt/wt mice from the same litter were used as a negative control. (C) There were no tdTomato-positive cells in the kidney of 2-month-old Cx3cr1CreERt; Rosa26tdTomato/wt mice without 4-OHT treatment. Arrowheads: tdTomato-expressing cells. Scale bars: 100 μm in A and B; and 50 μm in C.
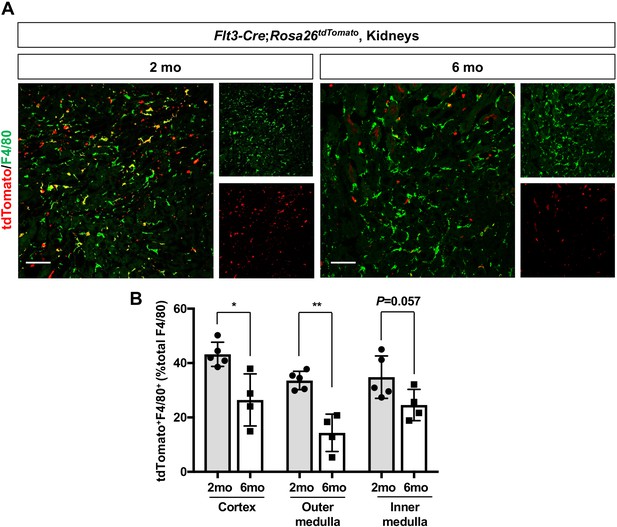
Age-dependent decrease of HSC-derived F4/80+ cells in the kidneys.
The Flt3-Cre; Rosa26tdTomato mouse line was used to examine the contribution of HSC-derived tissue-resident macrophages. (A) Distribution of F4/80+ Flt3-lineage cells in postnatal kidneys. The kidneys were analyzed at the indicated times (n = 4–5). HSC-derived cells and their progeny are irreversibly tagged with tdTomato. (B) Percentage of tdTomato+ F4/80+ to total F4/80+ cells. Note that the number of tdTomato+ F4/80+ cells decreases with age. Data are represented as means ± S.D. *, p<0.05; **, p<0.01.
-
Figure 2—source data 1
Percentage of tdTomato+ F4/80+ cells to total F4/80+ cells.
- https://cdn.elifesciences.org/articles/51756/elife-51756-fig2-data1-v2.xlsx
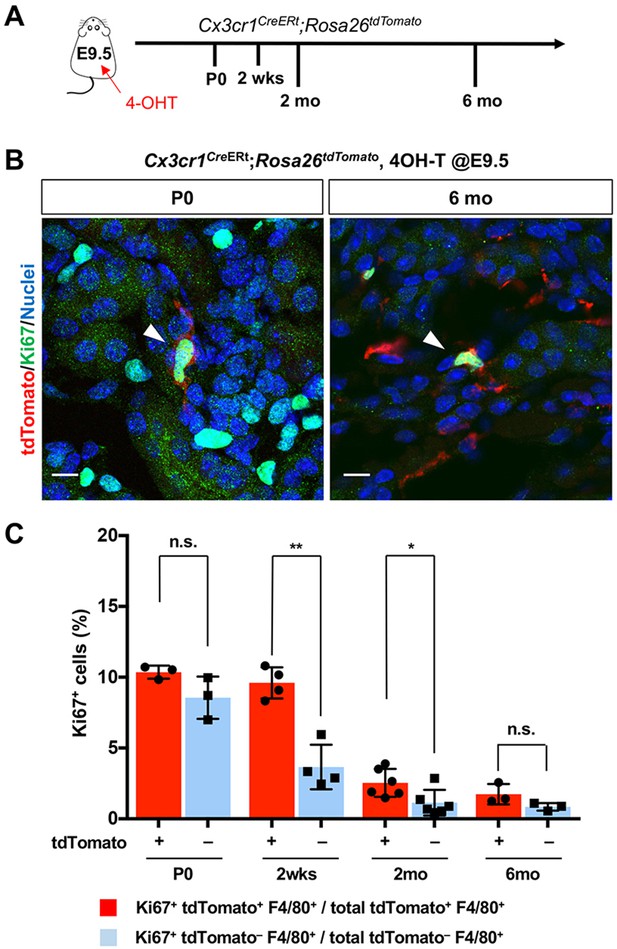
CX3CR1-positive yolk-sac macrophage descendants proliferate locally in the kidneys.
(A) Schematic of fate-mapping strategy. (B and C) CX3CR1-lineage macrophages proliferate in neonatal and aged kidneys. Cx3cr1CreERt; Rosa26tdTomato mice were treated with 4-hydroxytamoxifen (4-OHT) at E9.5 (n = 3–5). Arrowheads: Ki67+ CX3CR1-lineage cells. Percentage of Ki67+ proliferating cells are shown in C. Note that a higher percentage of CX3CR1-lineage F4/80+ cells (tdTomato+) are Ki67-positive compared to tdTomato– F4/80+ cells. Data are represented as means ± S.D. *, p<0.05; **, p<0.01; n.s., not significant. Scale bars: 10 μm.
-
Figure 3—source data 1
Percentage of Ki67+ proliferating F4/80+ cells.
- https://cdn.elifesciences.org/articles/51756/elife-51756-fig3-data1-v2.xlsx
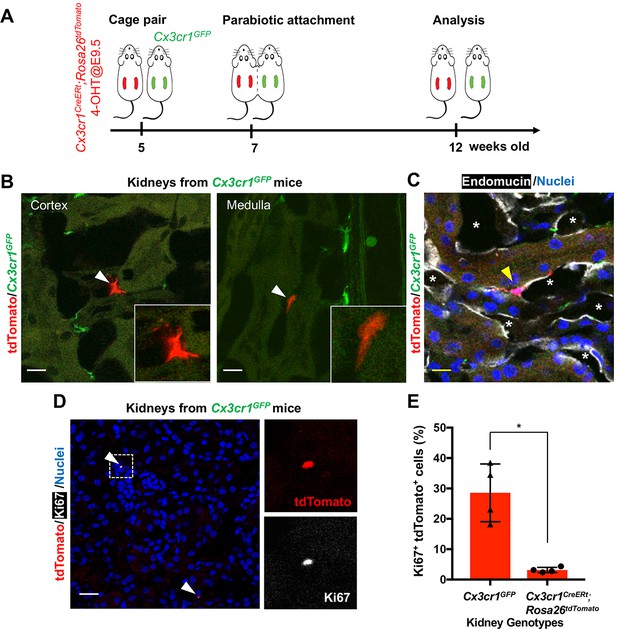
CX3CR1-positive yolk-sac macrophage descendants are recruited into adult kidneys from the circulation.
(A) Schematic of parabiotic experiments. (B and C) Localization of tdTomato+ CX3CR1-lineage cells in Cx3cr1GFP kidneys. The tdTomato-positive cells were lineage-labeled in Cx3cr1CreERt; Rosa26tdTomato mice in utero at E9.5. They migrated into the parabiont Cx3cr1GFP kidneys from circulation. Note that tdTomato+ cells were detected in extravascular interstitium (n = 4 per group). Endomucin; an endothelial cell marker. *, lumen of capillaries. Arrowheads, CX3CR1-lineage cells from circulation. (D and E) Circulation-derived CX3CR1-lineage cells proliferate in adult kidneys. Arrowheads, CX3CR1-lineage cells from circulation. GFP fluorescence was lost during the antigen retrieval process to detect Ki67. Percentages of Ki67+tdTomato+ cells relative to tdTomato+ cells in the kidneys of indicated genotype are shown in E (n = 4 per group). Note that tdTomato+ cells in Cx3cr1GFP kidneys are derived from circulation. Data are represented as means ± S.D. *, p<0.05. Scale bars: 10 μm in B and C; and in 20 μm in D. Legends for the Supplementary Figures.
-
Figure 4—source data 1
Percentages of Ki67+tdTomato+ cells relative to tdTomato+ cells in the kidneys of indicated genotypes.
- https://cdn.elifesciences.org/articles/51756/elife-51756-fig4-data1-v2.xlsx
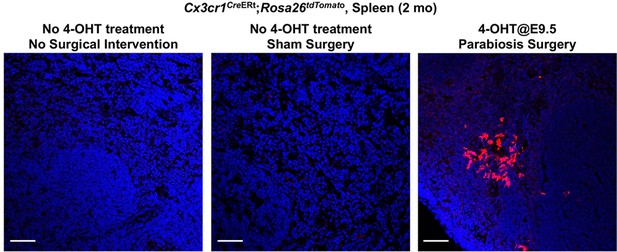
There is no basal Cre activity in kidneys without 4-hydroxytamoxifen (4-OHT) treatment in the spleen of 2-month-old-mice.
Cx3cr1CreERt; Rosa26tdTomato/wt mice were used to determine tdTomato reporter activity in the absence of 4-OHT treatment. Note that 4-OH tamoxifen treatment at E9.5 effectively identifies CX3CR1-lineage cells (red) in the spleen of all animals received the drug (N = 7 out of 7 mice tested). Scale bars: 50 μm.
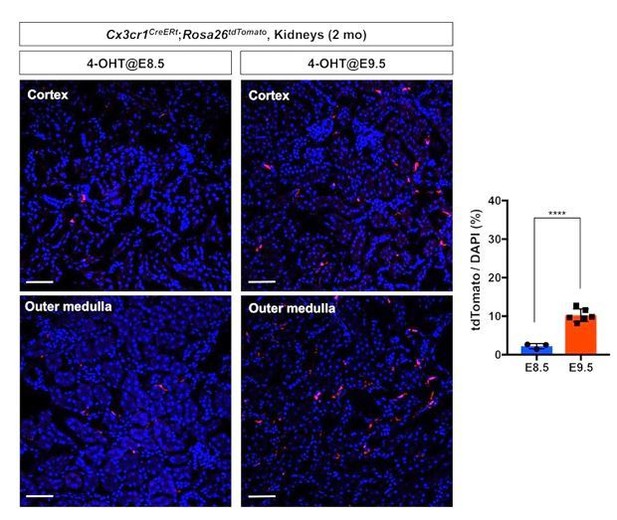
Comparison of fate-mapping strategies using different 4-OH tamoxifen treatment protocols.
4-hydroxytamoxifen (4-OHT) was injected once into pregnant dams at 8.5 dpc or 9.5 dpc, and the offspring were analyzed at 2 months of age. The first wave of pre-macrophages can be lineage traced with 4-OH tamoxifen injection at 8.5 dpc.
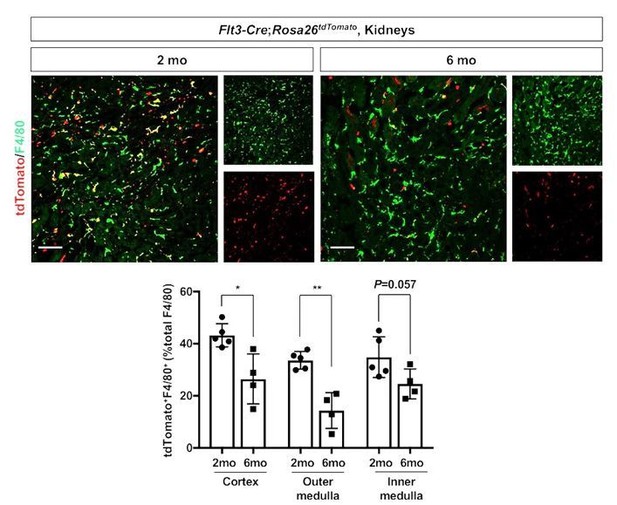
Age-dependent decline of HSC-derived F4/80-positive cells in kidneys.
The Flt3-Cre; Rosa26tdTomato mouse line was used to delineate the contribution of HSC-derived tissue resident macrophages. Please note that the number of tdTomato+ F4/80+ cells (yellow), which indicates HSC origin, decreases with age. *P<0.05, **P<0.01. Image-J was used for quantification. Flt3-Cre mice were kindly provided by Dr. K. Lavine (Washington University, St. Louis MO). Flt3-Cre mice were bred into the C57BL6/J background for 6 generations by the Shinohala lab.
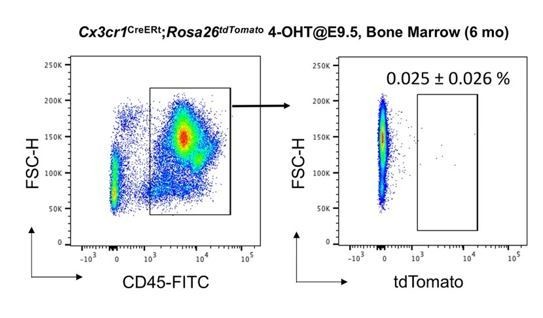
No increase of CD45+ tdTomato+ cells in bone marrow of 6 month-old Cx3cr1CreERt; Rosa26tdTomato mice.
We analyzed thenumber of tdTomato+ CD45+ cells in bone marrow (N=4). Compared to 2-month-old bone marrow (0.02% of CD45+ cells, Yahara et al., 2020), the number of tdTomato+CD45+ cells did not increase with age (0.025%).
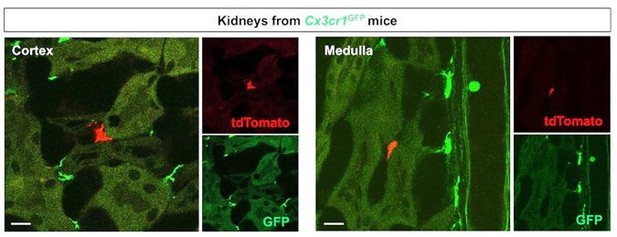
Merged and single color panels of Figure 4B.
The tdTomato-positive cells in the Cx3cr1GFP kidneys are derived from the parabiont animals (Cx3cr1CreERt; Rosa26tdTomato) through circulation. GFP-positive cells are resident Cx3cr1-expressing cells.
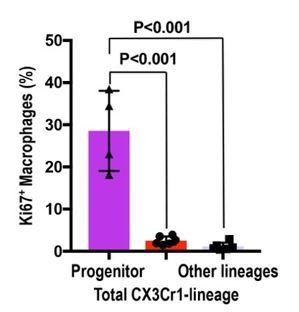
Circulation-derived CX3CR1-lineage cells have a profound proliferative capacity.
The percentage of Ki67+ cells is shown. Data are represented as means ± S.D. The mice were exposed to 4-OH tamoxifen in utero at E9.5. The tdTomato+ CX3CR1-lineage cells are descendants of yolk-sac macrophages. Data are derived and summarized from Figures 3 and 4 (see representative images in these figures). Progenitor, circulation-derived CX3CR1-lineage cells identified in Cx3cr1GFP kidneys; Total CX3CR1-lineage cells, macrophages derived from CX3CR1+ yolk-sac macrophages in the kidneys of Cx3cr1CreERt; Rosa26tdTomato mice; Other lineages, F4/80+tdTomato– macrophages in the kidneys of Cx3cr1CreERt; Rosa26tdTomato mice. Image-J was used for quantification.
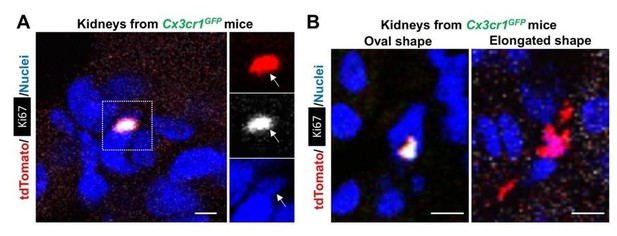
Morphological variation of CX3CR1-lineage cells.
The CX3CR1-lineage tdTomato-positive cells in the Cx3cr1GFP kidneys are derived from the parabiont animals (Cx3cr1CreERt; Rosa26tdTomato) through circulation. The Cx3cr1CreERt; Rosa26tdTomato mice were exposed to 4-OH tamoxifen in utero at E9.5. (A) Ki67+ CX3CR1-lineage cells are positive for DAPI (nuclear staining). (B) The CX3CR1-lineage tdTomato-positive cells exhibit oval and elongated shapes. Scale bars: 2 µm in A and 5 µm in B.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic Reagent (M. musculus) | Cx3cr1CreERt | The Jackson laboratory | RRID:IMSR_JAX:020940 | |
| Genetic Reagent (M. musculus) | Csf1r-CreERt (aka, Csf1r-Mer-iCre-Mer) | The Jackson laboratory | RRID:IMSR_JAX:019098 | |
| Genetic Reagent (M. musculus) | Flt3-Cre | RRID:IMSR_EM:11790 | Flt3-Cre mice were bred into the C57BL6/J background for six generations by the Shinohara lab. | |
| Genetic Reagent (M. musculus) | Rosa26tdTomato | The Jackson laboratory | RRID:IMSR_JAX:007914 | |
| Genetic Reagent (M. musculus) | Cx3cr1GFP | The Jackson laboratory | RRID:IMSR_JAX:005582 | |
| Antibody | Anti-F4/80 (Rat monoclonal) | Bio-Rad (MCA497) | RRID:AB_2098196 | Clone C1:A3-1 IF: 1:100 |
| Antibody | Anti-CD64 (Rat monoclonal) | Bio-Rad (MCA5997) | RRID:AB_2687456 | Clone AT152-9 IF: 1:200 |
| Antibody | Anti-Endomucin (Rat monoclonal) | Abcam (ab106100) | RRID:AB_10859306 | Clone V.7C7.1 IF: 1:100 |
| Antibody | Anti-Ki67 (Rat monoclonal) | eBioscience (14-5698-82) | RRID:AB_10854564 | Clone SolA15 IF: 1:200 |
| Antibody | Anti-Ki67 (Rabbit monoclonal) | Thermo (MA5-14520) | RRID:AB_10979488 | Clone SP6 IF: 1:200 |
| Antibody | Anti-dsRed (Rabbit polyclonal) | Rockland (600-401-379) | RRID:AB_2209751 | IF: 1:200 |
| Software, algorithm | ImageJ | NIH, Bethesda, MD (Version 1.52P) | RRID:SCR_003070 | https://imagej.nih.gov/ij/ |
| Software, algorithm | GraphPad Prism | RRID:SCR_002798 | https://www.graphpad.com/scientific-software/prism/ |





