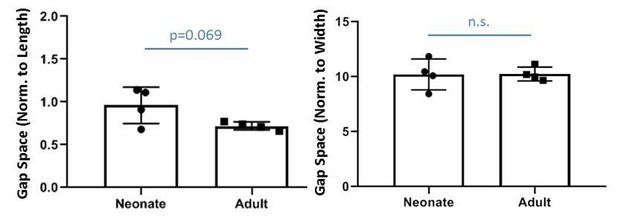
Tgfβ signaling is required for tenocyte recruitment and functional neonatal tendon regeneration
Figures
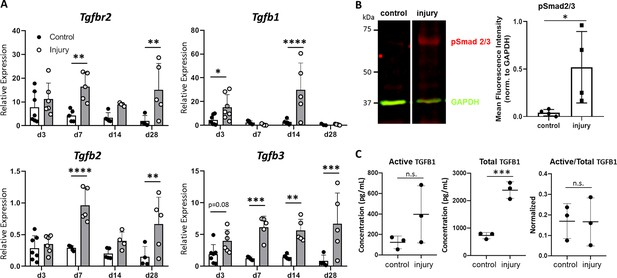
TGFβ signaling is activated after neonatal tendon injury.
(A) Gene expression in control and injured tendons at d3, d7, d14, and d28 post-injury by qPCR showed upregulation of Tgfbr2 receptor and Tgfb1, Tgfb2, Tgfb3 ligands. Expression levels were normalized to Gapdh using a standard curve method (n = 5–7 mice). (B) Western blot of control and injured tendons at d14 showed Smad2/3 phosphorylation after injury indicative of active TGFβ signaling (3 tendons per sample, n = 3 samples). (C) ELISA detection of active and total TGFB1 protein at d14 shows no change in active TGFB1 and increased total TGFB1 with injury (3 tendons per sample, n = 3 samples). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
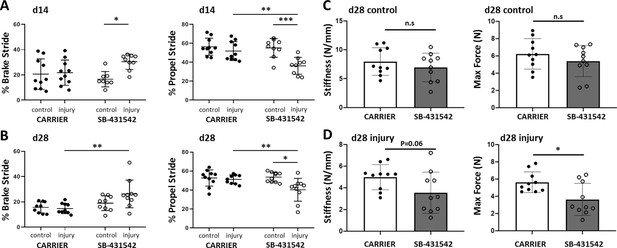
TGFβ signaling is required for functional recovery.
Gait analysis at (A) d14 and (B) d28 showed impaired % brake stride and % propel stride after injury with SB-431542 treatment. (C, D) Tensile testing at d28 revealed reduced stiffness and max force with SB-431542 treatment. *p<0.05, **p<0.01, ***p<0.001 (n = 8–10 mice).
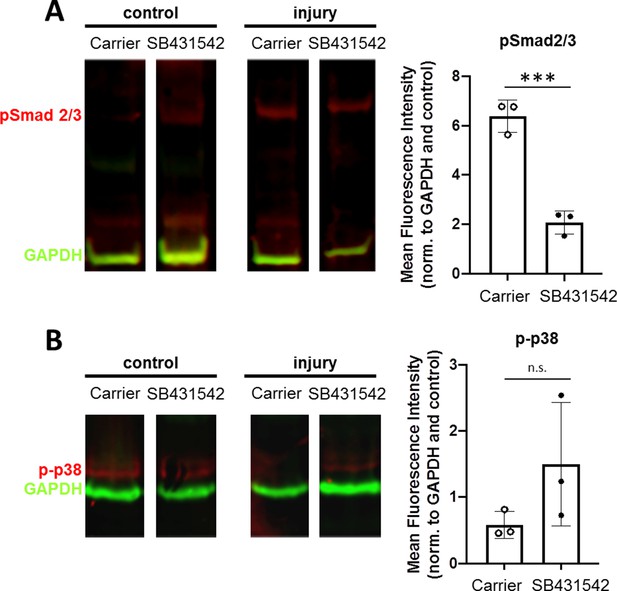
SB-431542 inhibition modulates canonical TGFβ signaling.
Western blotting and quantification of (A) pSmad2/3 and (B) phospho-p38 in injured tendons (normalized to GAPDH and control tendons) treated with carrier or SB-431542. *p<0.05 ***p<0.001, n.s. indicates p>0.1.
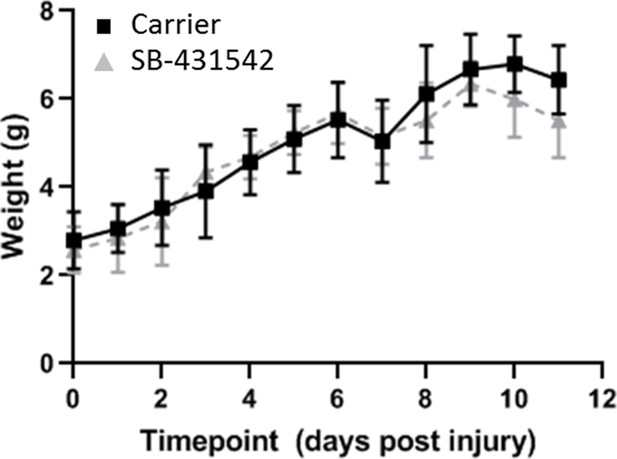
Postnatal growth is not affected by SB-431542 treatment.
Weights are comparable between carrier-treated and SB-431542-treated mice with no significant differences observed at any timepoint.
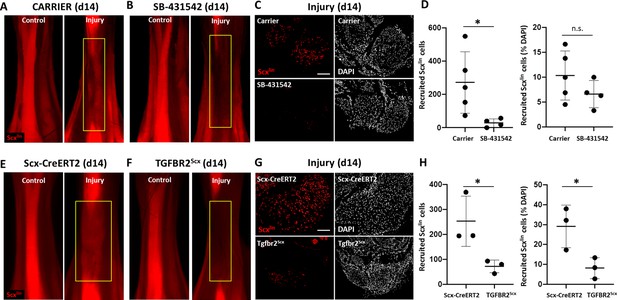
TGFβ signaling is required for tenocyte recruitment at d14.
(A, B) Whole mount images of control and injured limbs in carrier-treated and SB-431542-treated Scx-CreERT2; ROSA-Ai14 mice at d14. (C) Transverse sections through the neotendon (yellow boxes) near the mid-substance and (D) quantification showed reduced Scxlin, TdTomato+ cell recruitment with SB-431542 treatment, but no difference when normalized to total cells (n = 4–5 mice). (E, F) Whole mount images of control and injured limbs in wild type Scx-CreERT2; ROSA-Ai14 and TGFBR2Scx; ROSA-Ai14 mice. (G) Transverse sections through the neotendon near the mid-substance and (H) quantification showed reduced Scxlin, TdTomato+ cell recruitment in TGFBR2Scx mutants (n = 3 mice). *p<0.05, n.s. indicates p>0.05. Scale bars: 100 µm.
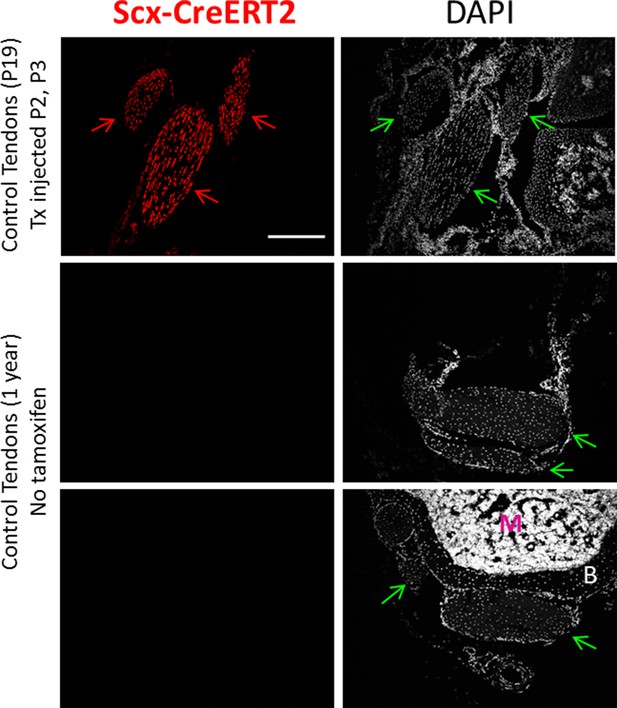
Scx-CreERT2 does not label any cells in the absence of tamoxifen.
One year old mice do not recombine ROSA-Ai14 in any cells in the absence of tamoxifen treatement, indicating no leakiness of Scx-CreERT2. Scale bar: 100 µm.
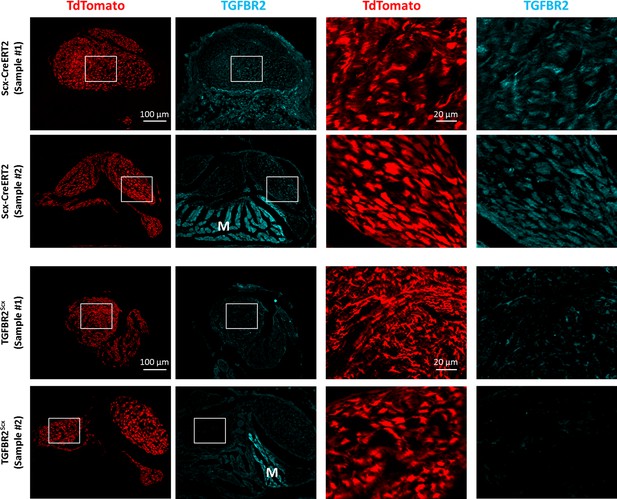
Reduced immunostaining of TGFBR2 in Scxlin cells with TGFBR2Scx deletion.
Transverse section images through injured tendon stubs stained with TGFBR2 showed reduced TGFBR2 staining of TdTomato+ Scxlin cells in TGFBR2Scx mutants compared to wild type. M indicates muscle which was used as internal positive control staining. Scalebars: 100 µm and 20 µm.
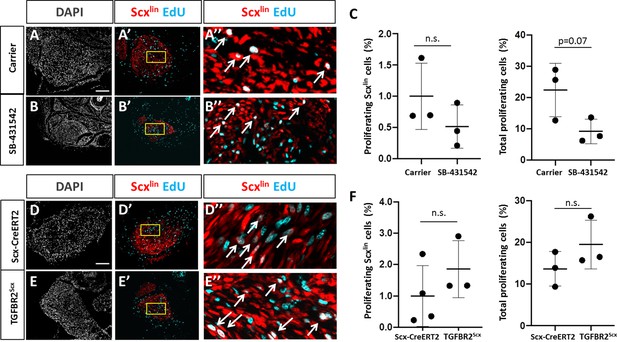
TGFβ signaling is not required for tenocyte proliferation.
Transverse sections through the cut site of (A, A’, A’) carrier-treated injured tendon or (B, B’, B’) SB-431542-treated injured tendon stained for EdU and counterstained with DAPI. (C) Quantification of EdU and Scxlin overlays showed no difference in Scxlin cell proliferation after injury with TGFβ inhibition, while % total proliferating cells was decreased with SB-431542 treatment (n = 3 mice). Transverse sections through the cut site of (D, D’, D’) wild type injured tendon or (E, E’, E’) TGFBR2Scx injured tendon stained for EdU and counterstained with DAPI. (F) Quantification of EdU and Scxlin overlays show no difference in Scxlin cell proliferation or total cell proliferation after injury with TGFBR2Scx deletion (n = 3 mice). A’, B’, D’, E’ are enlarged images from yellow boxed regions shown in A’, B’, C’, D’. White arrows indicate EdU+, Scxlin cells. n.s. indicates p>0.1. Scale bars: 100 µm.
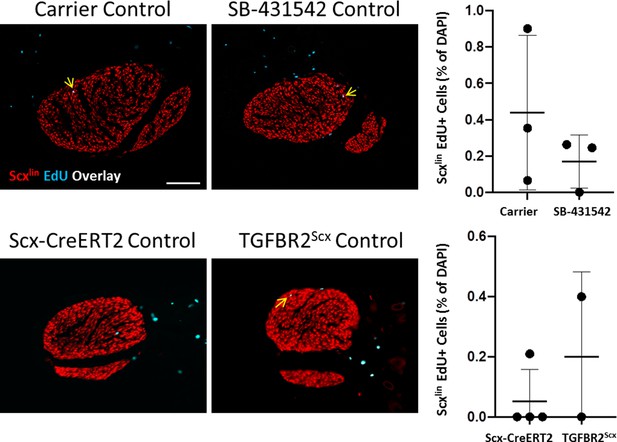
Proliferation in control, uninjured tendons is not affected by SB-431542 treatment or TGFBR2Scx deletion.
Transverse section images through control tendons stained with EdU show no differences between control tendons with SB-431542 treatment or TGFBR2Scx. Arrows indicate EdU+, Scxlin cells. Scalebar: 100 µm.
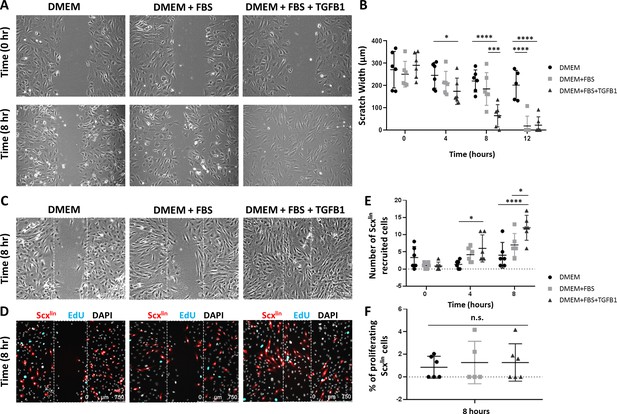
TGFβ enhances neonatal tenocyte migration in vitro.
(A) Phase contrast images and (B) quantification of in vitro wound assay show accelerated closure with TGFB1 supplementation relative to DMEM and DMEM+FBS conditions (n = 6). (C) Phase contrast and (D) fluorescence images show enhanced (E) Scxlin cell migration with TGFB1 treatment. (F) No difference was observed in cell proliferation at 8 hr (n = 5–6). *p<0.05, ***p<0.001, ****p<0.0001. n.s. indicates p>0.1.
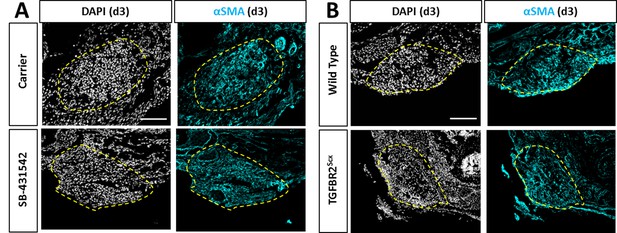
Recruitment of αSMA+ cells is not affected by TGFβ inhibition or TGFBR2Scx deletion.
Transverse sections through the gap space at d3 showed abundant αSMA+ cells with (A) SB-431542 treatment or (B) TGFBR2Scx deletion at levels comparable to carrier-treated or wild type. Yellow dashed outlines highlight gap area formerly occupied by the Achilles tendon.
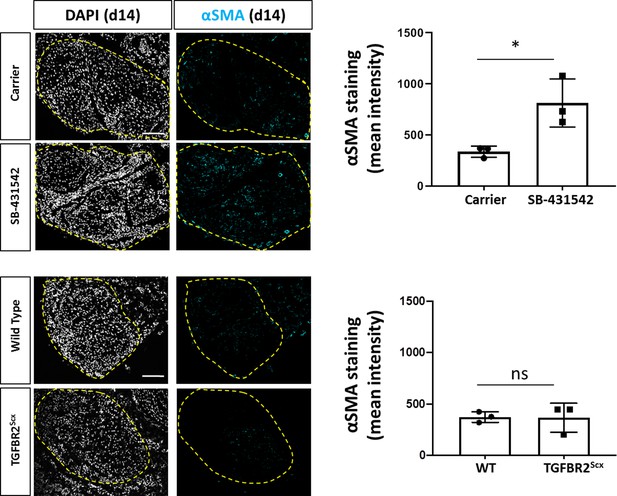
Increased detection of αSMA+ cells with SB-431542 treatment at d14 post-injury.
Transverse sections stained for αSMA show increased staining with SB-431542 treatment after injury. No difference was detected in αSMA staining between WT and TGFBR2Scx mice. Dashed yellow outlines indicate neotendon region. *p<0.05. Scale bar: 100 µm.
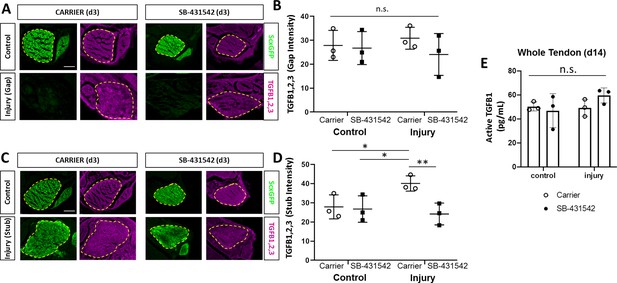
TGFβ ligand synthesis after injury is regulated by TGFβ signaling.
(A) Transverse sections through the midsubstance tendon and neotendon regions at d3 immunostained with antibody against all three TGFβ isoforms. (B) Quantification of intensity levels show no difference in staining between groups. (C) Transverse sections through the tendon cut site regions at d3 immunostained with antibody against all three TGFβ isoforms. (D) Quantification of intensity levels show increased TGFβ ligands after injury in carrier-treated tendons that is no longer observed with SB-431542 treatment. Yellow dashed outlines indicate region of interest quantified. (E) TGFB1 protein quantification by ELISA showed no differences with injury or SB-431542 treatment at d14. *p<0.05 **p<0.01 n.s. indicates p>0.1. Scale bar: 100 µm.
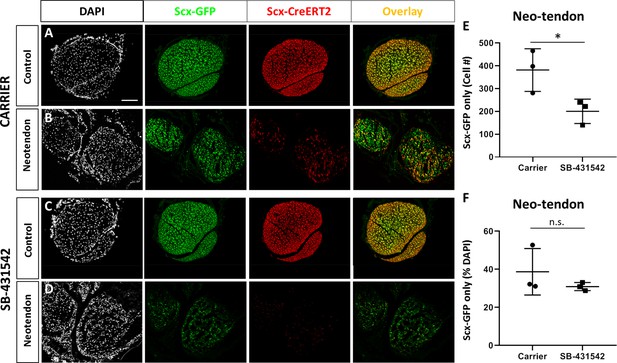
A population of Scx-GFP+, non-Scxlin cells are recruited after neonatal tendon injury.
Transverse sections through the neotendon of control and injured limbs in (A, B) carrier-treated and (C, D) SB-431542-treated Scx-CreERT2; ROSA-Ai14; Scx-GFP mice. (E, F) Quantification of non-Scxlin, Scx-GFP+ cells show reduction in cell number with SB-431542 treatment but not when normalized to total DAPI+ cells (n = 3 mice). *p<0.05. n.s. indicates p>0.1. Scalebar: 100 µm.
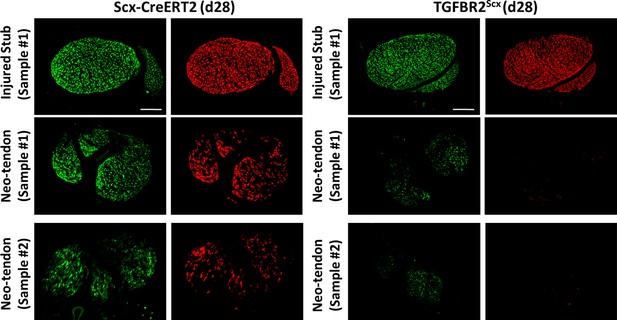
Scx-GFP+ cells are still detected in the absence of Scxlin recruitment in TGFBR2Scx mutants.
At d28, Scxlin cells are minimally detected in the gap space of TGFBR2 mutants, but Scx-GFP-only cells are present. Scale bar: 100 µm.
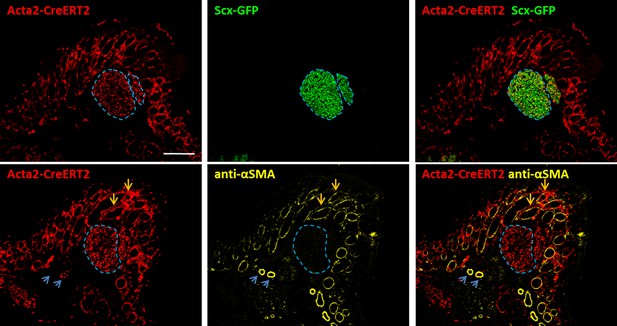
Lineage tracing with Acta2-CreERT2 show unexpected labeling in neonatal tenocytes.
Tamoxifen delivered at P2, P3 with P5 harvest show extensive aSMAlin labeling in Scx-GFP+ tenocytes (blue dashed outlines). Immunostaining for αSMA showed that tenocytes do not express αSMA. Overlays show extensive aSMAlin labeling and immunostaining in hair follicles (orange arrows) while blood vessels are inconsistently labeled (green arrows).
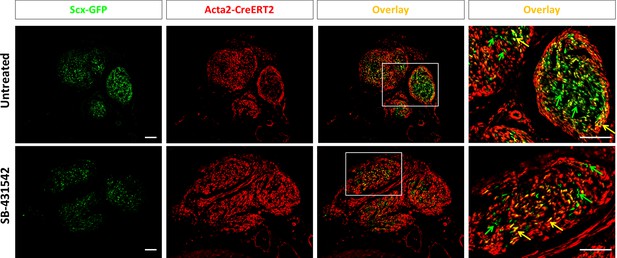
αSMAlin cells are recruited by d14 post-injury but do not account for all Scx-GFP+ cells.
Labeling with Acta2-CreERT2 (P2, P3 with P5 injury) shows the αSMAlin population comprises both Scx-GFP- and Scx-GFP+ cells at d14. In both untreated and SB-431542 treated tendons, a population of Scx-GFP-only cells is observed.
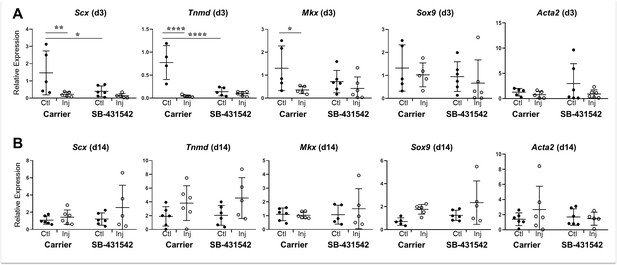
Tendon marker gene expression is not altered by TGFβ signaling inhibition at d14 post-injury.
Real time qPCR analysis of tendons harvested at (A) d3 and (B) d14 from carrier-treated and SB-431542-treated animals (n = 4–6 mice). Tendon genes were decreased after injury in carrier-treated mice but not with SB-431542 treatment at d3. Differences were no longer detected by d14. Sox9 and Acta2 were not significantly different at either time point. *p<0.05, **p<0.01, ****p<0.0001.
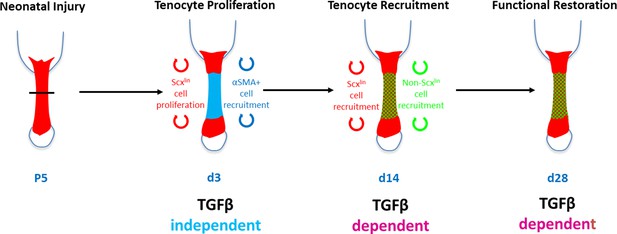
Requirement for TGFβ signaling in neonatal tendon regeneration.
Schematic depiction of conceptual model of the TGFβ-dependent and TGFβ-independent cellular processes during neonatal tendon regeneration. While tenocyte proliferation and αSMA cell recruitment at d3 occur independently of TGFβ signaling, subsequent tenogenic cell recruitment and functional tendon tissue restoration requires TGFβ signaling.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (M. musculus) | Tg(Scx-GFP)1Stzr (ScxGFP) | Ronen Schweitzer (Pryce et al., 2007) | RRID:MGI:3717419 | |
| Genetic reagent (M. musculus) | Scx-CreERT2 | Ronen Schweitzer | N/A | |
| Genetic reagent (M. musculus) | Tg(Acta2-cre/ERT2)1Ikal(Acta2-CreERT2) | Ivo Kalajzic (Grcevic et al., 2012) | RRID:MGI:5461154 | |
| Genetic reagent (M. musculus) | Gt(ROSA)26Sortm14(CAG-tdTomato)Hze (ROSA-Ai14) | Jackson Labs (Madisen et al., 2010) | Stock: 007908 RRID:MGI:3809524 | |
| Genetic reagent (M. musculus) | Tgfbr2tm1.1Hlm (Tgfbr2f/f) | Harold Moses (Chytil et al., 2002) | RRID:MGI:2384512 | |
| Chemical compound, drug | SB-431542 | Stemgent | Cat. # 04-0010-10 | |
| Antibody | Anti-alpha-SMA (mouse monoclonal) | Sigma-Aldrich | Cat. # A5228 RRID:AB_262054 | (1:100) |
| Antibody | Anti-TGFB1,2,3 MAb (Clone 1D11, mouse monoclonal) | R and D Systems | Cat. # MAB1835 RRID:AB_357931 | (1:50) |
| Antibody | Human Anti-TGFBR2 (goat polyclonal) | R and D Systems | Cat. # AF-241 | 5 ug/mL |
| Antibody | Donkey Anti-Rabbit Cy5 (rabbit polyclonal) | Jackson ImmunoResearch | Cat. # 711-175-152 RRID:AB_2340607 | (1:400) |
| Antibody | Strepdavidin Cy5 | Jackson ImmunoResearch | Cat. # 016-170-084 RRID:AB_2337245 | (1:400) |
| Antibody | Anti-phospho- Smad2/3 (rabbit polyclonal) | Cell Signaling | Cat. # 8828 RRID:AB_2631089 | (1:1000) |
| Antibody | Anti-phospho-p38 (rabbit monoclonal) | Cell Signaling | Cat. # 4511 RRID:AB_2139682 | (1:1000) |
| Antibody | Anti-GAPDH (mouse monoclonal) | Millipore Sigma | Cat. # MAB374 | (1:1000) |
| Antibody | Goat anti-rabbit IRDye (rabbit polyclonal) | Licor | Cat. # 926–68071 RRID:AB_2721181 | (1:10,000) |
| Antibody | Goat anti-mouse IRDye (mouse polyclonal) | Licor | Cat. # 926–32210 RRID:AB_621842 | (1:10,000) |
| Software, algorithm | ZEN Digital Imaging for Light Microscopy | Zeiss https://www.zeiss.com/microscopy/int/products/microscope-software/zen-lite.html | RRID:SCR_013672 | |
| Software, algorithm | ImageJ | ImageJ (http://imagej.nih.gov/ij/) | RRID:SCR_003070 | |
| Software, algorithm | Graphpad Prism | GraphPad Prism (https://graphpad.com) | RRID:SCR_015807 |
Additional files
-
Supplementary file 1
Primer sequences for real time qPCR.
- https://cdn.elifesciences.org/articles/51779/elife-51779-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/51779/elife-51779-transrepform-v2.docx