Repression of viral gene expression and replication by the unfolded protein response effector XBP1u
Figures
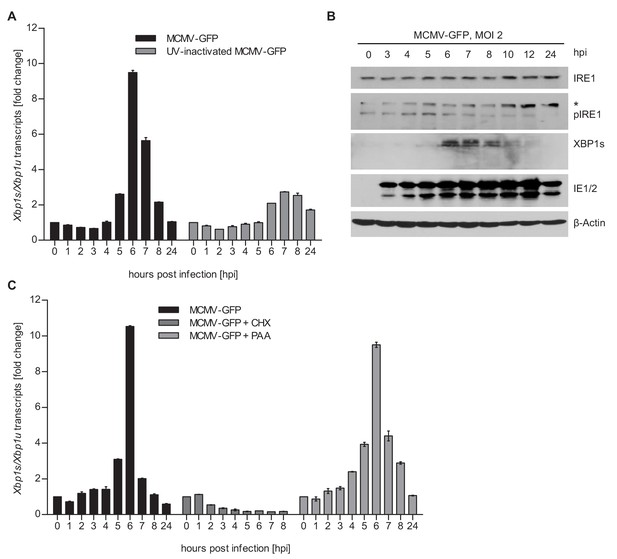
MCMV induces Xbp1s mRNA splicing at early time of infection.
(A) MEFs were infected with MCMV-GFP or UV-inactivated MCMV-GFP (MOI 4). Cells were harvested at the indicated times, total RNA was extracted, and Xbp1s and Xbp1u transcripts were quantified by qPCR. Changes in the Xbp1s/Xbp1u ratio relative to uninfected cells are plotted as bar diagram (means ± SEM of 3 biological replicates). (B) Immunoblot analysis of MEFs infected with MCMV-GFP. Endogenous IRE1, phosphorylated IRE1, and XBP1s were detected using specific antibodies. *, unspecific band. The immunoblot is representative of 2 independent experiments. (C) MEFs were infected with MCMV-GFP as described above and treated with vector, CHX (50 μg/ml) or PAA (250 ng/ml). Changes in the Xbp1s/Xbp1u ratio were determined as described above. Data provided in Figure 1—source data 1.
-
Figure 1—source data 1
Data points of qRT-PCR.
- https://cdn.elifesciences.org/articles/51804/elife-51804-fig1-data1-v2.xlsx
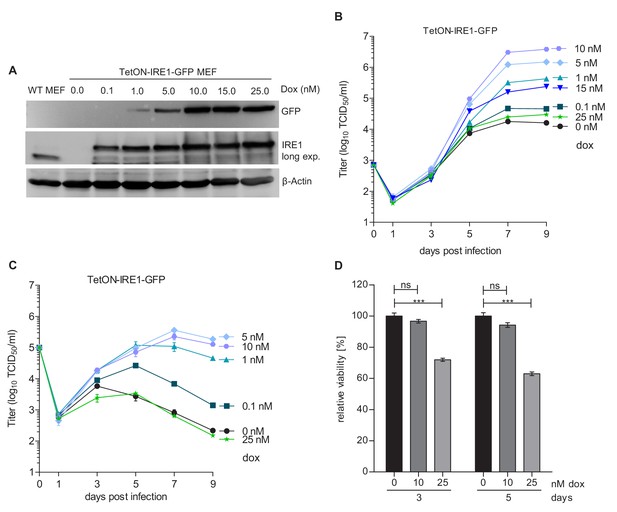
Moderate IRE1 expression is beneficial for MCMV replication.
(A) Immunoblot analysis of IRE1-deficient (Ern1-/-) MEFs expressing IRE1-GFP (TetON-IRE1-GFP) in a doxycycline (dox)-inducible manner. Cells were treated with different dox concentrations for 24 hr. IRE1-GFP expression was detected with GFP or IRE1-specific antibodies. Endogenous IRE1 levels in WT MEFs were detected only with the IRE1-specific antibody. The immunoblot is representative of 3 independent experiments. (B) Multistep MCMV replication kinetics on TetON-IRE1-GFP MEFs induced with different dox concentrations. 24 hr after induction, cells were infected with MCMV-GFP (MOI 0.1). Virus titers in the supernatants were determined by titration and are shown as means ± SEM of 3 biological replicates. (C) Single step MCMV replication kinetics on TetON-IRE1-GFP MEFs induced with dox as above, infected with MCMV-GFP (MOI 3) and are shown as means ± SEM of 3 biological replicates. (D) Cell viability of TetON-IRE1-GFP MEFs treated with different dox concentrations. Cell viability was measured after 3 and 5 days of dox treatment and is shown as relative viability compared to untreated cells (means ± SEM of 6 biological replicates). Data provided in Figure 2—source data 1. Additional data provided in Figure 2—figure supplement 1.
-
Figure 2—source data 1
Data points of growth curves.
- https://cdn.elifesciences.org/articles/51804/elife-51804-fig2-data1-v2.xlsx
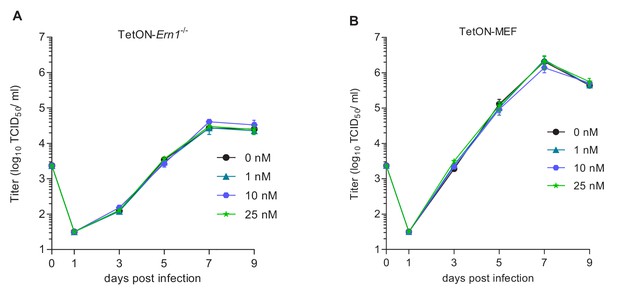
MCMV replication kinetics on TetON expressing cells.
(A) Multistep MCMV replication kinetics on TetON-Ern1-/- MEFs induced with different doxycycline concentrations. 24 hr after induction, cells were infected with MCMV-GFP (MOI 0.1). Virus titers in the supernatants were determined by titration and are shown as means ± SEM of 3 biological replicates. (B) Multistep MCMV replication kinetics on WT MEFs transduced with a TetON-expressing lentiviral vector. 24 hr after doxycycline treatment, cells were infected with MCMV-GFP (MOI 0.1). Virus titers in the supernatants were determined as above. Data provided in Figure 2—source data 1.
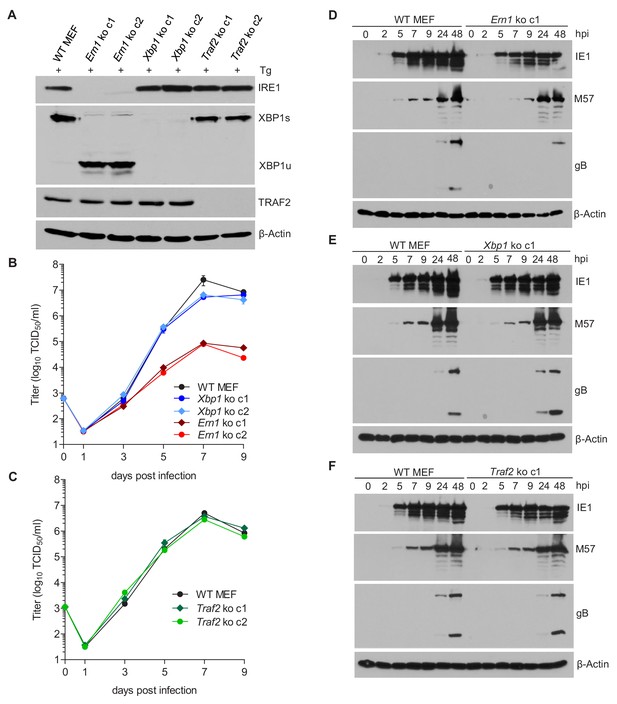
IRE1, but not XBP1 or TRAF2, is required for efficient MCMV replication and viral protein expression.
(A) Immunoblot analysis of IRE1, XBP1, and TRAF2-deficient (Ern1, Xbp1, and Traf2 ko) cell lines. Two ko clones were generated for each gene by CRISPR/Cas9 gene editing using different gRNAs. Cells were treated for 4 hr with Thapsigargin (Tg) to induce Xbp1 mRNA splicing and to increase XBP1 expression. (B,C) Multistep MCMV replication kinetics in Ern1, Xbp1 and Traf2 ko cells, respectively. Cells were infected with MCMV-GFP (MOI 0.1). Virus titers in the supernatants were determined by titration and are shown as means ± SEM of 3 biological replicates. (D–F) Immunoblot analysis of viral protein expression kinetics in Ern1, Xbp1 and Traf2 ko cells, respectively. Cells were infected with MCMV-GFP (MOI 3) and harvested at different times post infection. Expression levels of the viral immediate-early 1 (IE1) protein, the major DNA binding protein (M57; an early protein), and glycoprotein B (gB; a late protein) were detected with specific antibodies, β-Actin served as loading control. Immunoblots are representative of 2 independent experiments. Data provided in Figure 3—source data 1. Additional data provided in Figure 3—figure supplement 1.
-
Figure 3—source data 1
Data points of growth curves and qRT-PCR.
- https://cdn.elifesciences.org/articles/51804/elife-51804-fig3-data1-v2.xlsx
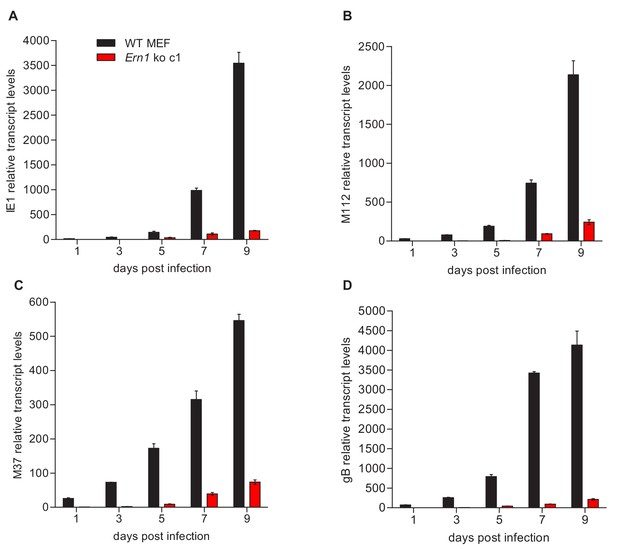
qRT-PCR analysis of viral transcripts in WT and IRE1-deficient cells.
(A–D) WT and Ern1 ko MEFs were infected with MCMV-GFP (MOI 0.1). Cells were harvested at the indicated times, total RNA was extracted, and IE1 (M123), E1 (M112), M37, and gB (M55) transcripts were quantified by qRT-PCR. Transcript levels were normalized to Ern1 ko cells at day one post infection (means ± SEM of 3 biological replicates). Data provided in Figure 3—source data 1.
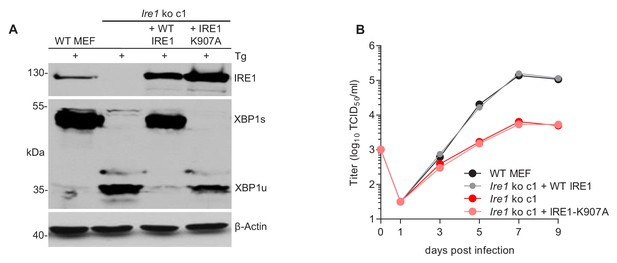
The RNase function of IRE1 is required for efficient MCMV replication.
(A) Immunoblot analysis of IRE1-deficient cells (Ern1 ko c1) transduced with retroviral vectors expressing WT IRE1 or IRE1-K907A (RNase-dead). Cells were treated for 4 hr with Thapsigargin (Tg) to induce Xbp1 mRNA splicing and to increase XBP1 expression. IRE1 and XBP1 protein expression was detected by immunoblot (representative of 2 independent experiments). (B) Multistep MCMV replication kinetics in Ern1 ko cells complemented with WT IRE1 or IRE1-K907A, respectively. Cells were infected with MCMV-GFP (MOI 0.1). Virus titers in the supernatants were determined by titration and are shown as means ± SEM of 3 biological replicates. Data provided in Figure 4—source data 1.
-
Figure 4—source data 1
Data points of growth curves.
- https://cdn.elifesciences.org/articles/51804/elife-51804-fig4-data1-v2.xlsx
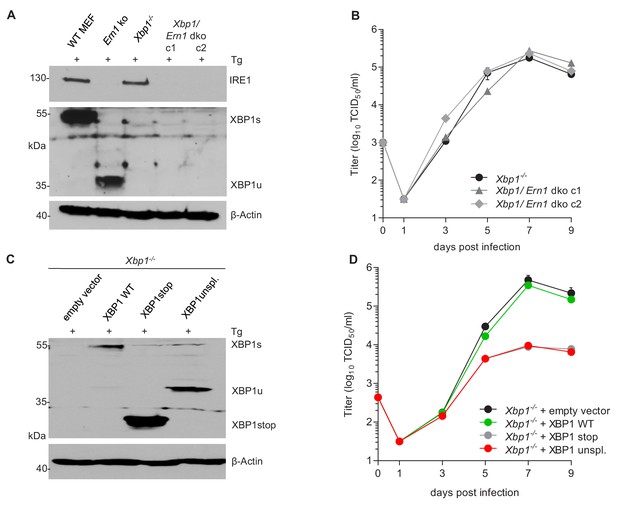
XBP1u acts as a repressor for MCMV replication.
(A) Xbp1-/- MEFs were used to knock out the IRE1-encoding Ern1 gene by CRISPR/Cas9 gene editing. Two double ko (dko) cell clones were generated with different gRNAs. IRE1 and XBP1 protein expression in the two dko clones and control cells was detected by immunoblot analysis. Cells were treated for 4 hr with Thapsigargin (Tg) to induce Xbp1 mRNA splicing and to increase XBP1 expression. (B) Multistep MCMV replication kinetics in Xbp1-/- and Xbp1/Ern1 dko MEFs. Cells were infected with MCMV-GFP (MOI 0.1). Virus titers in the supernatants were determined by titration and are shown as means ± SEM of 3 biological replicates. (C) Immunoblot analysis of Xbp1-/- MEF transduced with retroviral vectors expressing a WT (spliceable) Xbp1 transcript, an unspliceable Xbp1 transcript, or a truncated (Xbp1stop) transcript. Cells were treated with Tg as described in A. (D) Multistep MCMV replication kinetics in cells shown in C. Infection and titration was done as in B. Immunoblots are representative of 2 independent experiments. Data provided in Figure 5—source data 1.
-
Figure 5—source data 1
Data points of growth curves.
- https://cdn.elifesciences.org/articles/51804/elife-51804-fig5-data1-v2.xlsx
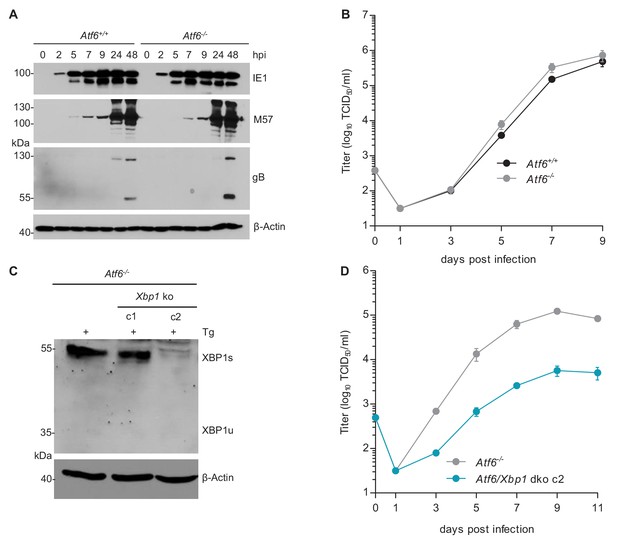
MCMV replication is impaired in cells lacking Atf6 and Xbp1.
(A) Immunoblot analysis of viral gene expression of Atf6+/+ and Atf6-/- cells infected with MCMV-GFP (MOI 3). At the indicated times cells were lysed and stained for the immediate-early one protein (IE1), the major DNA binding protein (M57; early gene) and glycoprotein B (gB; late gene) by immunoblot. β-Actin served as loading control. (B) Multistep replication kinetics. Atf6+/+ and Atf6-/- cells were infected with MCMV-GFP (MOI of 0.1). Virus titers in the supernatants were determined by titration and are shown as means ± SEM of 3 biological replicates. (C) Knockout of Xbp1 in Atf6-/- cells using CRISPR/Cas9 gene editing. Two single cell clones generated by two individual gRNAs (c1 and c2) were analyzed for XBP1s and XBP1u expression by immunoblot. 4 hr prior to harvesting, cells were stimulated with thapsigargin (Tg) to enhance XBP1 expression. The parental Atf6-/- cells are shown as control. (D) Multistep replication kinetics of Atf6/Xbp1 dko cells (clone c2). MCMV infection and titration was done as described in B. Immunoblots are representative of 2 independent experiments. Data provided in Figure 6—source data 1.
-
Figure 6—source data 1
Data points of growth curves.
- https://cdn.elifesciences.org/articles/51804/elife-51804-fig6-data1-v2.xlsx
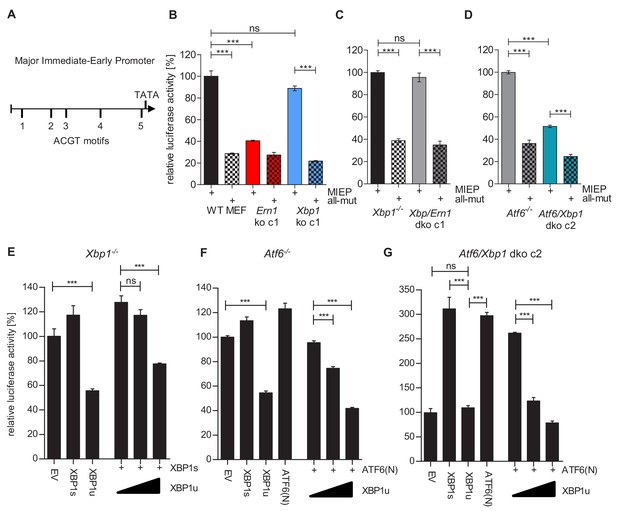
XBP1s and ATF6-mediated activation of the MCMV MIEP is repressed by XBP1u.
(A) Schematic representation of the MCMV major immediate-early promoter (MIEP) with TATA box and 5 ACGT motifs. (B) WT MEFs, Ern1 ko and Xbp1 ko cells were transfected with a firefly luciferase vector containing either the WT MIEP or a MIEP with five mutated ACGT motifs (all-mut). Renilla luciferase was expressed by co-transfection and used for normalization. Relative luciferase activities (firefly: renilla) ± SEM of at least three biological replicates are shown. ***, p<0.001; ns, not significant, p>0.05. (C) Xbp1-/- and Xbp1/Ern1 dko cells were transfected as in B and the relative luciferase activity was determined. (D) Atf6-/- and Atf6/Xbp1 dko cells were transfected as in B and the relative luciferase activity was determined. (E) Xbp1-/- MEFs were co-transfected with firefly and renilla luciferase vectors as in B. Expression vectors for XBP1s, XBP1u, ATF6(N), or empty vector (EV) were co-transfected. Relative luciferase activities (firefly: renilla) ± SEM of 3 biological replicates are shown. (F) Atf6-/- cells were transfected as in E and the relative luciferase activity was determined. (G) Atf6/Xbp1 dko cells were transfected as in E and the relative luciferase activity was determined. Data provided in Figure 7—source data 1.
-
Figure 7—source data 1
Data points of luciferase assays.
- https://cdn.elifesciences.org/articles/51804/elife-51804-fig7-data1-v2.xlsx
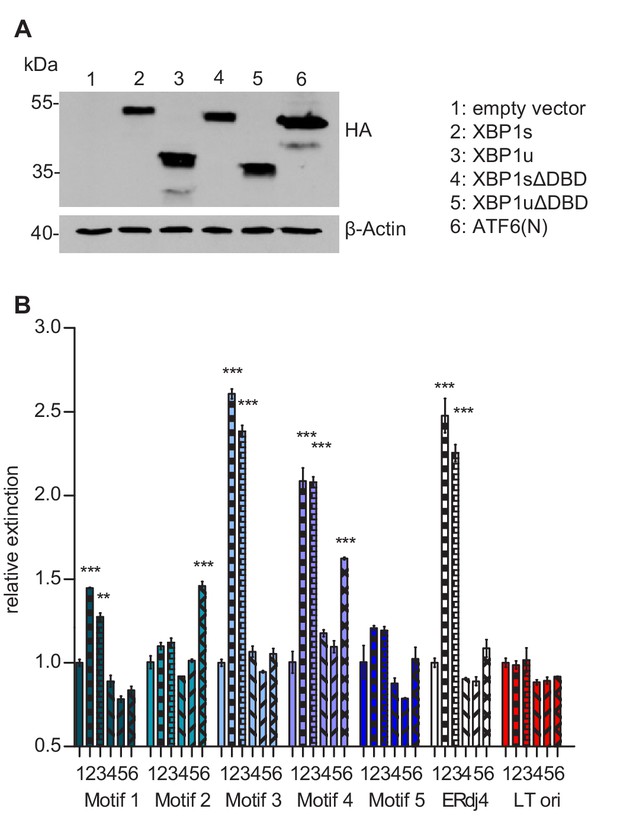
Transcription factor binding the MIE promotor.
(A) HEK 293A cells were transfected with expression vectors encoding HA-tagged WT or mutant XBP1 and ATF6(N) transcription factors. Nuclear extracts were obtained, and transcription factor expression was verified by immunoblot analysis (representative of 2 independent experiments). (B) Microtiter plates coated with dsDNA oligonucleotides containing XBP1 core binding motifs from the MCMV major immediate-early promoter, the ERdj4 promoter (positive control) or an unrelated sequence from the SV40 origin of replication (negative control). Wells were incubated with nuclear extracts 1 to 6 shown in A, and transcription factor binding was measured by DPI-ELISA, and values were normalized to extract 1 (empty vector). Means ± SEM of 3 biological replicates are shown. Significance was determined for all values above the cut-off (1.25). **, p<0.01; ***, p<0.001. Data provided in Figure 8—source data 1.
-
Figure 8—source data 1
Data points of the DPI-ELISA.
- https://cdn.elifesciences.org/articles/51804/elife-51804-fig8-data1-v2.xlsx
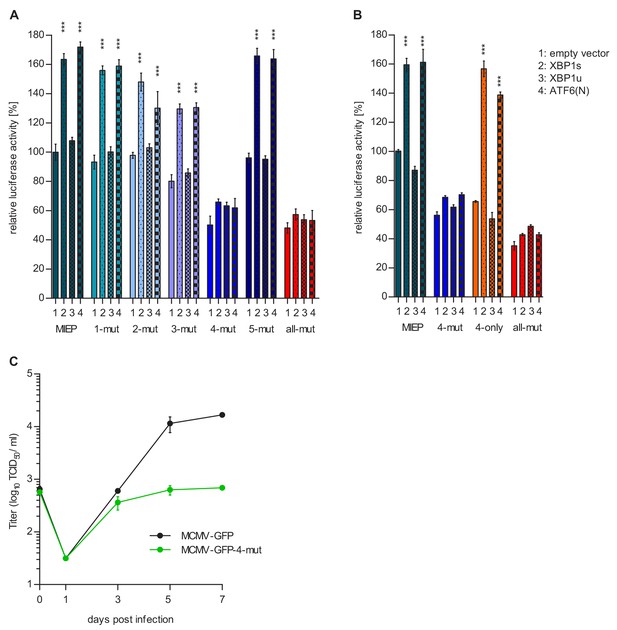
Motif 4 is necessary and sufficient for MIEP activation by XBP1s and ATF6(N).
(A) Atf6/Xbp1 dko MEFs cells were transfected with a firefly luciferase vector containing either the WT major immediate-early promotor (MIEP) or a MIEP with one or all ACGT motifs mutated. Expression vectors for XBP1s, XBP1u, ATF6(N), or empty vector (EV) were co-transfected. Renilla luciferase was expressed by co-transfection and used for normalization. Relative luciferase activities (firefly: renilla) ± SEM of 3 biological replicates are shown. ***, p<0.001; all other differences were not significant (p>0.05). (B) Atf6/Xbp1 dko MEF cells were transfected and analyzed as in A. In addition, a MIEP vector with all ACGT motifs mutated except motif 4 (4-only) was included. (C) Multistep MCMV replication kinetics in WT MEF cells. Cells were infected with MCMV-GFP or MCMV-GFP-4-mut (MOI 0.1), respectively. Virus titers in the supernatants were determined by titration and are shown as means ± SEM of 3 biological replicates. Data provided in Figure 9—source data 1.
-
Figure 9—source data 1
Data points of luciferase assays and growth curve.
- https://cdn.elifesciences.org/articles/51804/elife-51804-fig9-data1-v2.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (E. coli) | GS1783 | PMID:16526409 | BAC host for recombineering | |
| Strain, strain background (murid herpesvirus 1) | MCMV-GFP | PMID:11209080 | BAC-derived MCMV (Smith strain) expressing GFP | |
| Cell line (H. sapiens) | HEK 293A | Invitrogen | R705‐07; RRID:CVCL_6910 | Transformed human embryonic kidney cells |
| Cell line (H. sapiens) | HEK 293T | ATCC | CL-11268 | |
| Cell line (H. sapiens) | Phoenix | ATCC | CRL-3213; RRID:SCR_003163 | Packaging cell line for retrovirus production |
| Cell line (M. musculus) | WT MEF | PMID:19364921 | SV40 TAg immortalized MEF | |
| Cell line (M. musculus) | 10.1 | PMID:1752433 | spontaneously immortalized BALB/c MEF | |
| Cell line (M. musculus) | Ern1-/- MEF | PMID:11780124 | IRE1-deficient MEF | |
| Cell line (M. musculus) | TetON-Ern1-/- | This paper | Ern1-/- MEFs expressing rtTA (TetON) | |
| Cell line (M. musculus) | TetON-IRE1-GFP | This paper | TetON-Ern1-/- cells, inducible expression of IRE1-GFP | |
| Cell line (M. musculus) | Xbp1-/- MEF | PMID:10652269 | ||
| Cell line (M. musculus) | Ern1 ko MEF | This paper | WT MEF, Ern1 knocked out by CRISPR/Cas9 | |
| Cell line (M. musculus) | Xbp1 ko MEF | This paper | WT MEF, Xbp1 knocked out by CRISPR/Cas9 | |
| Cell line (M. musculus) | Traf2 ko MEF | This paper | WT MEF, Traf2 knocked out by CRISPR/Cas9 | |
| Cell line (M. musculus) | Atf6-/- MEF (LT) | This paper PMID:17765679 | SV40 TAg immortalized MEF | |
| Cell line (M. musculus) | Atf6+/+ MEF (LT) | This paper PMID:17765679 | SV40 TAg immortalized MEF | |
| Cell line (M. musculus) | Atf6/Xbp1 dko | This paper | Atf6-/- MEF, Xbp1 knocked out by CRISPR/Cas9 | |
| Recombinant DNA reagent | pMSCVhygro, pMSCVpuro | Clontech | Retroviral vector plasmids | |
| Recombinant DNA reagent | pGL3-Basic | Promega | E1751 | Firefly luciferase reporter plasmid |
| Recombinant DNA reagent | pGL4.73 | Promega | E6911 | Renilla luciferase control plasmid |
| Antibody | anti-HA (mouse monoclonal) | Covance | MMS-101P; RRID:AB_291259 | WB (1:1000) |
| Antibody | anti-IRE1 (rabbit monoclonal) | Cell Signaling | Cat# 3294; RRID:AB_823545 | WB (1:1000) |
| Antibody | anti-pIRE1 (rabbit polyclonal) | Novus Biologicals | NB100-2323; RRID:AB_10145203 | WB (1:500) |
| Antibody | anti-XBP1 (rabbit polyclonal) | Santa Cruz | sc-7160; RRID:AB_794171 | WB (1:500) |
| Antibody | anti-TRAF2 (rabbit monoclonal) | Cell Signaling | Cat# 4724; RRID:AB_2209845 | WB (1:1000) |
| Antibody | anti-IE1 (Chroma101, mouse) | Stipan Jonjic (Univ.of Rijeka) | WB (1:1000) | |
| Antibody | anti-M57 (mouse) | Stipan Jonjic (Univ.of Rijeka) | WB (1:1000) | |
| Antibody | anti-gB (M55.01, mouse) | Stipan Jonjic (Univ.of Rijeka) | WB (1:1000) | |
| Antibody | anti-GFP (mouse monoclonal) | Roche | Cat# 11814460001; RRID:AB_390913 | WB (1:1000) |
| Antibody | anti-β-Actin (mouse monoclonal) | Sigma-Aldrich | A2228; RRID:AB_476697 | WB (1:1000) |
| Chemical compound, drug | GenJet | SignaGen Laboratories | SL100489-MEF | Transfection reagent |
| Chemical compound, drug | Polyethylenimine (PEI) | Sigma-Aldrich | 764604 | Transfection reagent |
| Commercial assay or kit | CellTiter-Glo Luminescent Cell Viability Assay | Promega | G7570 | |
| Commercial assay or kit | Dual-Luciferase Reporter Assay System | Promega | E1910 |
Additional files
-
Supplementary file 1
Wildtyp and mutated Major Immediate-Early Promoter (MIEP) sequences.
Minimal XBP1 binding motifs (ACGT) are underlined (black), mutated motifs are in red. The TATA box is in bold.
- https://cdn.elifesciences.org/articles/51804/elife-51804-supp1-v2.xlsx
-
Supplementary file 2
Oligonucleotides used for the DNA-Protein Interaction (DPI) ELISA.
- https://cdn.elifesciences.org/articles/51804/elife-51804-supp2-v2.xlsx
-
Supplementary file 3
Guide RNAs (gRNAs) used for CRISPR/Cas9 gene editing.
- https://cdn.elifesciences.org/articles/51804/elife-51804-supp3-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/51804/elife-51804-transrepform-v2.pdf





