Paradoxical network excitation by glutamate release from VGluT3+ GABAergic interneurons
Figures
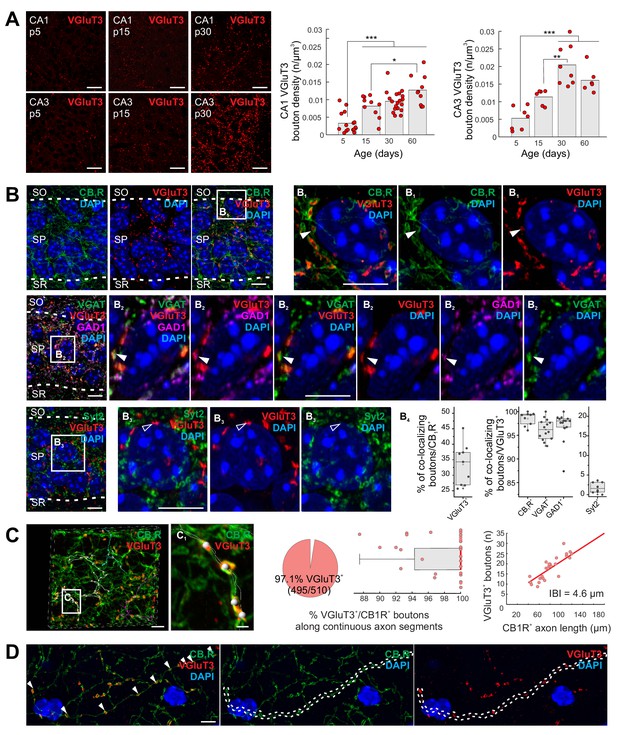
Delayed onset of VGluT3 expression in hippocampal CCK+VGluT3+INTS.
(A) Representative images illustrating native VGluT3 expression within CA1/3 stratum pyramidale (SP) at postnatal day 5, 15, and 30 (p5,15,30) as indicated. Group data for terminal density counts in CA1 and CA3 throughout development are plotted at right (n = 6–19 sections from three to six animals per age were examined; individual observations (n) along with means are provided). (B) Representative high resolution Airyscan images illustrating colocalization of VGluT3 signal in CA1 perisomatic axon terminals with CB1R signal (upper row, (B1) as well as GAD1 and VGAT signals (second row, (B2) but not Syt2 labelled terminals (third row, (B3); SO, stratum oriens; SR, stratum radiatum). Widefield as well as digitally magnified images from boxed regions are provided showing merged and split channels for each set of markers, with DAPI labeling, as indicated. Filled arrowheads highlight terminals showing colocalization while open arrowheads highlight terminals lacking colocalization. Colocalization of VGluT3 with each marker is quantified in the box plots (B4) with data normalized to total CB1R+ terminal counts (left plot) or total VGluT3+ terminal counts (middle and right plots; 1200–6580 terminals from n = 8–14 sections in each staining combination, from three mice were evaluated). (C) Representative Airyscan images and group data illustrating conserved VGluT3 expression at all putative release sites along continuous individual axon segments. Three individual axon segments are highlighted in the CA1 SP widefield image and a segment from one of these axons (boxed region, (C1) is digitally magnified. Group data plotted at right summarize the percentage of putative release sites along individual axons from CB1R+/VGluT3+ cells that are VGluT3+ (pie chart and box plot, normalized to number of boutons in each axon segment) as well as the correlation between total number of boutons scored per length of each continuous axon examined (IBI, interbouton interval; 510 boutons were evaluated from n = 28 axon segments across three mice). (D) An additional example of conserved VGluT3 expression across all putative release sites of a continuous axon segment within SO with merged and split channels as indicated. For statistical analysis *p<0.05, **p<0.01, ***p<0.001. Scalebar in A = 10 µm; in B, C, D = 5 µm; in C1 = 1 µm.
-
Figure 1—source data 1
Data plotted in Figure 1.
- https://cdn.elifesciences.org/articles/51996/elife-51996-fig1-data1-v2.xlsx
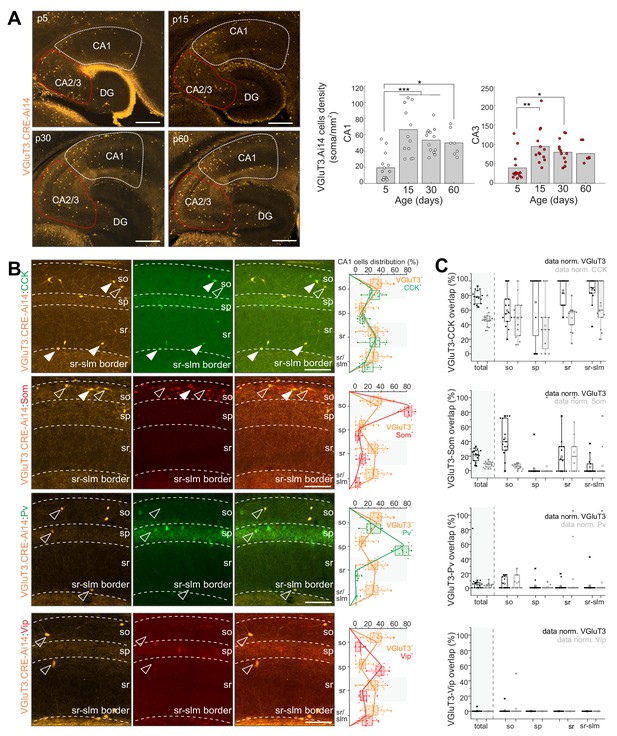
Developmental and IHC evaluation of hippocampal VGluT3-Cre:Ai14 reported cells.
(A) Representative images (left) and group data summary (right) illustrating the developmental profile of RFP+ cell densities in CA1 and CA2/CA3 regions of the hippocampus as indicated (n = 7–14 sections from three to six mice for each time point were evaluated; DG, dentate gyrus). (B) Representative images of CA1 used to evaluate colocalization of RFP+ cells with CCK, Som, PV, and VIP as indicated (left). Merged and split channels are shown for each marker pair with filled arrowheads highlighting sample cells displaying colocalized signal and open arrowheads highlighting sample cells without colocalization. Distributions of labelled cells for each interneuron marker within CA1 from s.o. to s.l.m. (stratum lacunosum moleculare) are plotted for comparison to RFP+ cell distributions (right). (C) The degree of overlap between cells expressing each interneuron marker with those expressing RFP is provided for all of CA1 as well as for each sublayer. Data are plotted normalized to either the RFP+ cell population (black) or the given interneuron marker population (gray; 336–619 cells from n = 9–15 sections from three mice were evaluated per staining combination). Scalebar in A = 500 µm; in B = 100 µm.
-
Figure 2—source data 1
Data plotted in Figure 2.
- https://cdn.elifesciences.org/articles/51996/elife-51996-fig2-data1-v2.xlsx
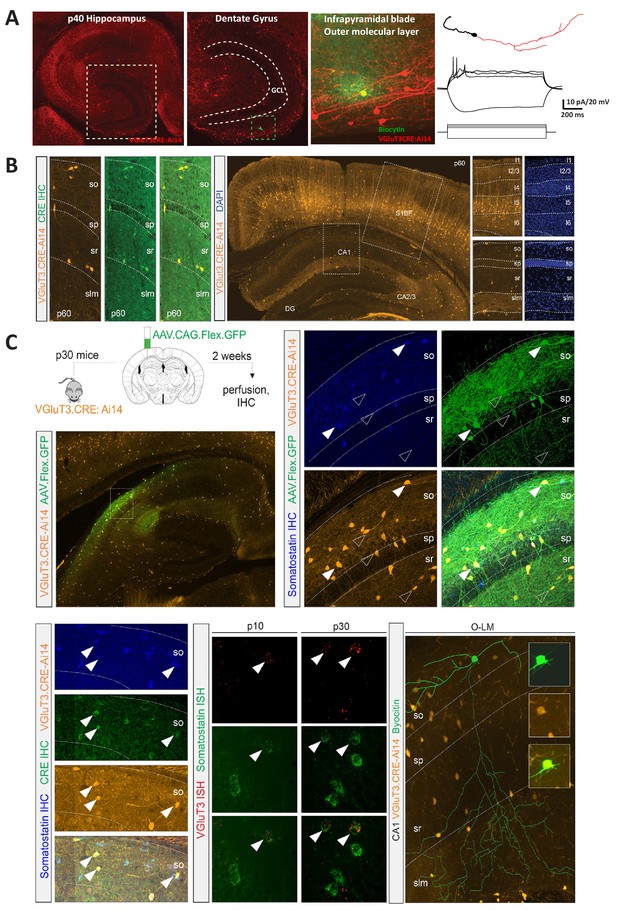
Further characterization of RFP+ cell populations reported in VGluT3-Cre:Ai14 mice.
(A) Representative images highlighting RFP+ Cajal Retzius cells in the hippocampal dentate gyrus outer molecular layer reported in a VGluT3-Cre:Ai14 mouse (GCL, granule cell layer). The highest magnification (right) illustrates a single RFP+ Cajal Retzius that was electrophysiologically recorded and labeled with biocytin (green). The neurolucida reconstruction of this cell (far right) reveals a stereotypical tadpole-like shape with a unipolar dendrite (black) and sparse axon (red) emanating from opposite poles of the soma. Also shown are the membrane voltage responses of this cell to hyperpolarizing and depolarizing current injections from a resting potential of −60 mV illustrating a characteristic strongly accommodating spiking profile with action potentials of small amplitude. (B) Representative images illustrating RFP+ cell distributions throughout hippocampus and neocortex (middle panel with digitally magnified boxed regions at right showing RFP and DAPI signals) as well as Cre expression within RFP+ CA1 interneurons (left panels) in p60 VGluT3-Cre:Ai14 mice. (C) Confirmation of VGluT3 signal in a small subset of hippocampal SOM+ interneurons. Upper panels illustrate that delivery of AAV-CAG-FLEX-GFP into stratum oriens of a P30 VGluT3-Cre:Ai14 mouse (upper left schematic) leads to GFP/RFP coexpression within a subset of SOM+ interneurons (filled arrowheads; open arrowheads highlight SOM- RFP+GFP+, likely CCK+, populations). The bottom left column of images illustrates Cre expression within a subset of SOM+ interneurons in an adult VGluT3-Cre:Ai14 mouse. The bottom middle series of images illustrates native VGluT3 mRNA expression within a subset of SOM+ interneurons (filled arrowheads). The bottom right panel ill1ustrates a post-hoc recovered O-LM interneuron, the prototypical SOM+ interneuron, that was targeted for recording in a VGluT3-Cre:Ai14 slice.
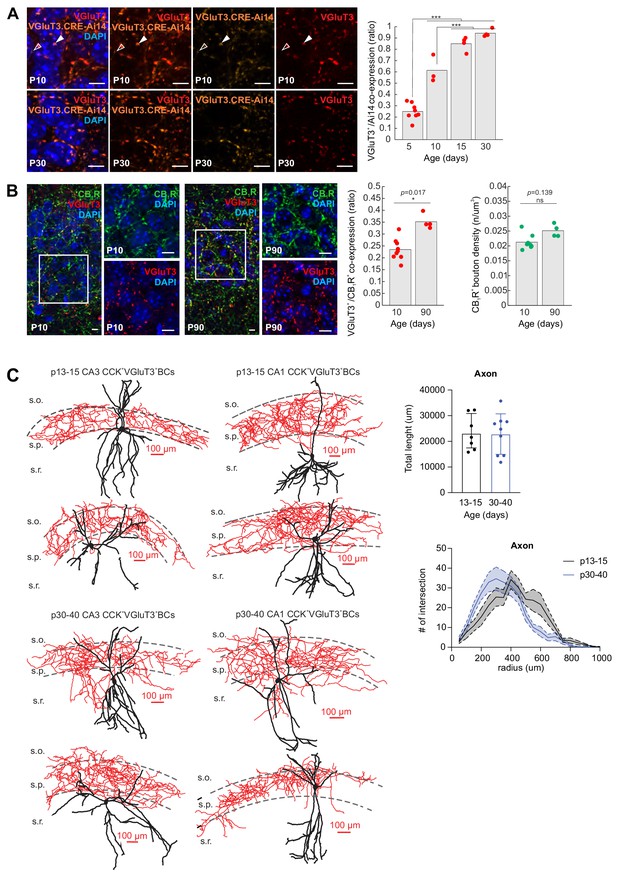
Further developmental characterization of VGluT3+ terminals and VGluT3+ BC anatomies.
(A) Representative high-resolution images illustrating colocalization of VGluT3 and RFP signal in CA1 perisomatic axon terminals of P10 and P30 VGluT3-Cre:Ai14 mice as indicated (scale bars = 5 µm). Colocalization of VGluT3 with RFP across development is quantified in the bar chart at right with data normalized to the total number of RFP+ terminals examined at each age (237–1447 terminals from n = 3–8 sections in each staining combination, from 2 to 4 mice were evaluated at each age; ***p<0.001). (B) Representative high-resolution images illustrating colocalization of VGluT3 and CB1R signal in CA1 perisomatic axon terminals at P10 and P90 as indicated. Widefield as well as digitally magnified images from boxed regions are provided showing merged and split channels for each marker, with DAPI labelling, as indicated (scale bars = 5 µm). The group data bar charts at right illustrate the the colocalization of VGluT3 to CB1R+ terminals at P10 and P90 (normalized to total number of CB1R+ terminals examined) as well as overall CB1R+ terminal density at these ages (1265–2205 terminals from n = 4–9 sections from three mice were evaluated at each age; *p<0.05). (C) Anatomical reconstructions of representative biocytin filled CA1 and CA3 VGluT3+BCs recorded in P13-15 and P30-40 VGluT3-Cre:Ai14 mice as indicated (red axon, black dendrite). Group data summary plots at right illustrate similar overall axon length (upper bar chart) and distribution (lower Sholl plot) at the two developmental stages (n = 7–9 VGluT3+BCs, CA1 and CA3 combined, were recovered from recordings in 2 VGluT3-Cre:Ai14 mice for each time point).
-
Figure 2—figure supplement 2—source data 1
Data plotted in Figure 2—figure supplement 2.
- https://cdn.elifesciences.org/articles/51996/elife-51996-fig2-figsupp2-data1-v2.xlsx
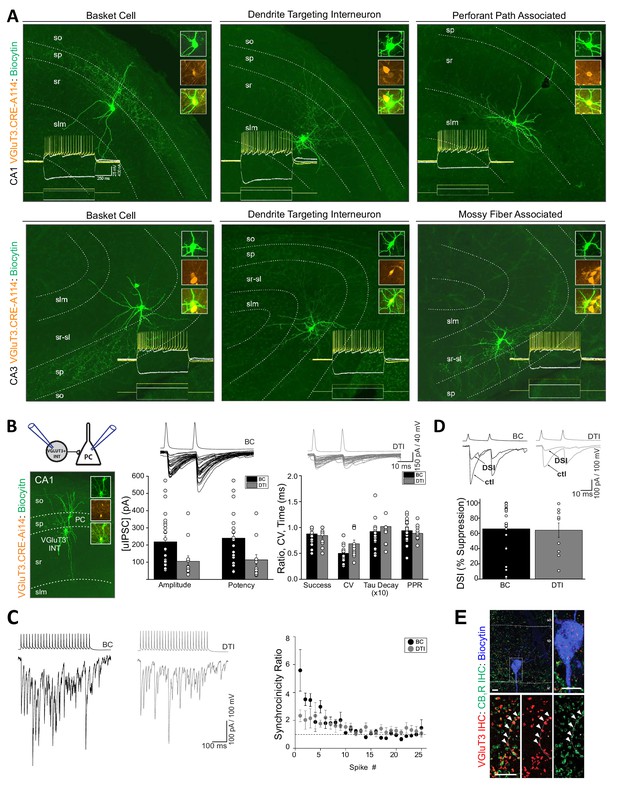
Anatomical, electrophysiological, and basic synaptic properties of hippocampal CCK+VGluT3+INTs.
(A) Images of representative RFP+ cells recorded in VGluT3-Cre:Ai14 mice from CA1 (upper row) and CA3 (lower row) hippocampus illustrating prototypical BCs and DTIs. Inset traces illustrate membrane responses of each representative cell (upper series of traces) to the current injection waveforms shown (lower series of traces). Image insets highlight colocalization of the somatic dye fill (biocytin) with RFP. (B) Basic properties of unitary GABAergic synaptic transmission between VGluT3+BCs (black) or VGluT3+DTIs (gray) with CA1 PCs. At left, the recording configuration is schematized above an image of a sample RFP+BC-CA1PC pair. At right, sample traces show individual trial synaptic events in postsynaptic CA1 PCs (lower traces) following presynaptic spiking (upper traces) in either RFP+ BCs or DTIs as indicated. The basic properties of transmission are summarized in the group data bar charts below (n = 25 and 12 pairs, from 17 and 10 mice for BC and DTI pairs, respectively). Note that for these recordings postsynaptic CA1PC cells were filled with high [Cl-] and held at −70 mV yielding GABAergic dominated inward currents. (C) Sample traces (left) from representative recordings of VGluT3+BC-CA1PC and VGluT3+DTI-CA1PC pairs as indicated illustrating asynchronous release of GABA during presynaptic trains of stimuli. Note the postsynaptic events are tightly synchronized with presynaptic APs early in the train and then fail to remain time locked to presynaptic APs as the train continues resulting in a progressive reduction in the synchronicity ratio plotted at right for all recordings (n = 6 and 5 pairs from 4 and 4 mice for BC and DTI pairs, respectively). (D) Sample traces (above) and group data bar chart summary (below) illustrating DSI of VGluT3+BC/DTI-CA1PC pairs. For each set of traces an averaged uIPSC obtained under control conditions (ctl) is shown in comparison with an averaged uIPSC obtained following depolarization of postsynaptic CA1PCs to 0 mV for 5 s (DSI, 5–10 individual uIPSCs were averaged for each condition). The group data bar chart quantifies the magnitude of uIPSC suppression induced by DSI (n = 22 and 10 pairs from 17 and 9 mice for BC and DTI pairs respectively). (E) Representative sample image of a biocytin filled postsynaptic CA1PC soma decorated with VGluT3+CB1R+ presynaptic terminals consistent with DSI of VGluT3+BC /-CA1 PC pairs. Throughout the figure summary plots provide individual observations as well as group means ± SEM.
-
Figure 3—source data 1
Data plotted in Figure 3.
- https://cdn.elifesciences.org/articles/51996/elife-51996-fig3-data1-v2.xlsx
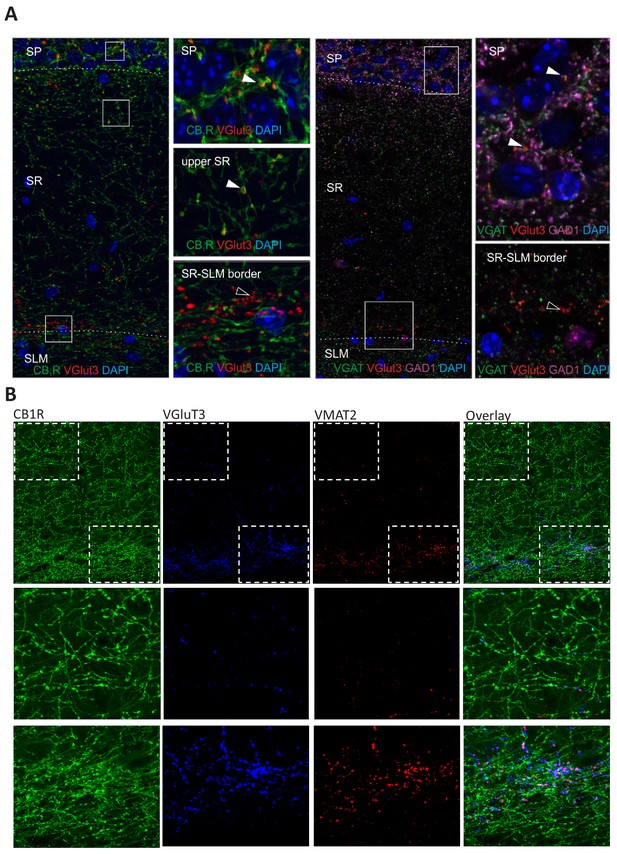
Evaluation of dendrite targeting VGluT3+ terminals.
(A) Representative Airyscan images examining colocalization of VGluT3 with CB1R (left panels), VGAT and GAD1 (right panels) in terminals located from SP to the SR/SLM border. While SP and upper SR VGluT3+ terminals typically coexpress CB1R, VGAT, and GAD (filled arrowheads) a population of large VGluT3+ terminals at the SR/SLM border do not express these markers (open arrowheads). (B) Representative Airyscan images illustrating VMAT2 expression within many of the large VGluT3+ terminals located at the SR/SLM border. Also shown is the coexpression of VGluT3 and CB1R within a subset of terminals located within SR.
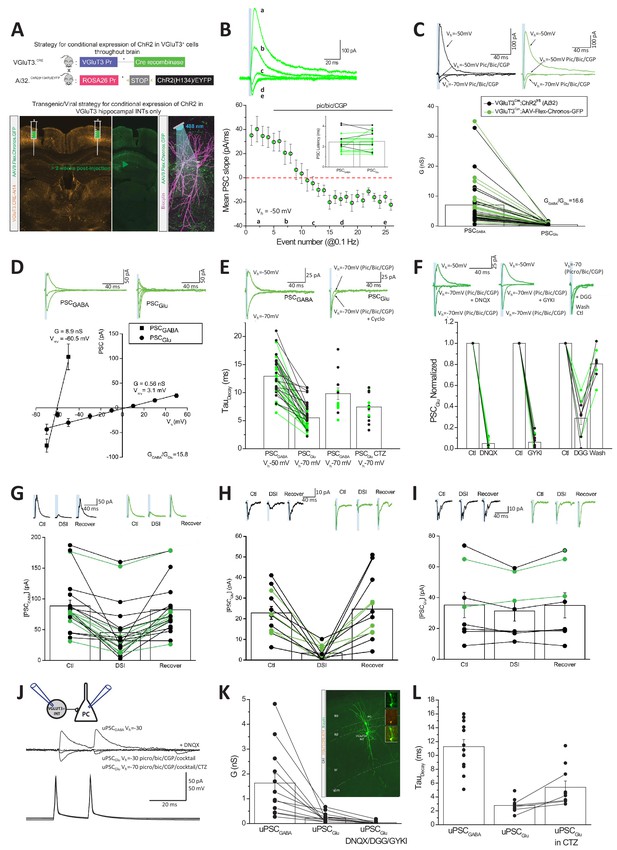
Glutamate and GABA corelease from CCK+VGluT3+INTs.
(A) Schematics depicting two strategies to genetically introduce ChR2 or the Chronos variant within CCK+VGluT3+INTs by either breeding VGluT3-Cre:Ai32 mice (above) or introducing AAV-CAG-Flex-Chronos-GFP selectively into the hippocampus of VGluT3-Cre mice (below) respectively. In both cases, the optogenetic actuator is driven by illumination with blue light while recording CA1PCs as illustrated in the bottom right subpanel. Here and throughout data from both strategies have been pooled and individual recordings from the VGluT3-Cre:Ai32 and VGluT3:Chronos strategies are depicted in black and green respectively. (B) Traces from a representative recording (above) and group data summary (below) for a subset of recordings illustrating the conversion of an outward GABAergic dominated postsynaptic current (PSCGABA) to an inward glutamatergic mediated synaptic event (PSCGlu) following introduction of GABAA/B receptor antagonists as indicated (n = 16 recordings from nine mice). Traces are average events (3–5 individual sweeps) obtained at the times indicated in the group plot below. Note that Cl- reversal potential is set to ~60 mV and postsynaptic cells are initially held positive to this so that GABAergic events are outward (positive inititial slope) while glutamatergic events are inward (negative initial slope). Bar chart inset summarizes the latencies of GABAergic and pharmacologically isolated glutamatergic events for each recording making up the time course plot. (C) Averaged traces from representative recordings (above) and group data summary (below) comparing GABAergic (PSCGABA) and glutamatergic (PSCGlu) synaptic conductances (G, here and throughout corresponding conductances measured for each individual recording are connected in the group data plot, n = 49 recordings from 22 mice). (D) Traces from representative recordings (above) and group data plots illustrating the current-voltage relations of GABAergic and glutamatergic synaptic events as indicated (PSCGABA, n = 12 cells from four mice; PSCGlu, n = 10 cells from four mice). (E) Traces from representative recordings (above) and group data (plotted below) highlighting the distinct kinetics of PSCGABA and PSCGlu (n = 35 cells from 15 mice with corresponding GABAergic and glutamatergic kinetics, connected points). Also shown are PSCGABA kinetics observed at −70 mV for inward GABAergic events (n = 12 cells from three mice) and PSCGlu kinetics obtained in CTZ (n = 13 cells from seven mice). (F) Traces from representative recordings (above) and group data summaries (below) illustrating antagonism of PSCGlu by DNQX (n = 12 cells from four mice), GYKI 53655 (n = 12 cells from seven mice) or γ-DGG (n = 9 cells from six mice, note reversible block following wash). Group data are expressed normalized to control responses obtained prior to antagonist application. (G–H) Traces from representative recordings (above) and group data summary plots (below) illustrating DSI sensitivity of PSCGABA (G, n = 21 cells from seven mice), and PSCGlu (H, n = 12 cells from seven mice). Absolute amplitudes of averaged PSCs before (Ctl) during (DSI) and after DSI (Recover) are plotted. (I) As for H but with BAPTA included in the recording pipette to block endocannabinoid production (n = 8 cells from three mice). (J) Averaged traces from a representative CCK+VGluT3+BC-PC pair recording (schematic inset) illustrating GABA and glutamate cotransmission. Antagonism of the outward unitary GABAergic event (uPSCGABA) unmasks a smaller inward unitary glutamatergic event (uPSCGlu) that is enhanced by CTZ and increased driving force and also sensitive to DNQX. (K) Group data summary of uPSCGABA and uPSCGlu conductances for all pairs exhibiting glutamatergic transmission (n = 12 and 17 pairs in 10 mice for uPSCGABA and uPSCGlu respectively; 12 recordings with corresponding uPSCGABA and uPSCGlu are connected by lines, in the remainder only uPSCGlu was evaluated as pair was obtained in partial or full GABAAR antagonism). Also plotted is antagonism of uPSCGlu by DNQX or GYKI or DGG in a subset of recordings (n = 10 pairs in five mice). Inset image shows sample pair anatomical recovery. (L) Group data summary of the kinetic properties of uPSCGABA (n = 12 pairs) and uPSCGlu (n = 11) as well as uPSCGlu in CTZ (n = 10; in five cases with corresponding control and CTZ conditions as illustrated by the connected points). Throughout the figure summary plots provide individual observations as well as group means ± SEM.
-
Figure 4—source data 1
Data plotted in Figure 4.
- https://cdn.elifesciences.org/articles/51996/elife-51996-fig4-data1-v2.xlsx
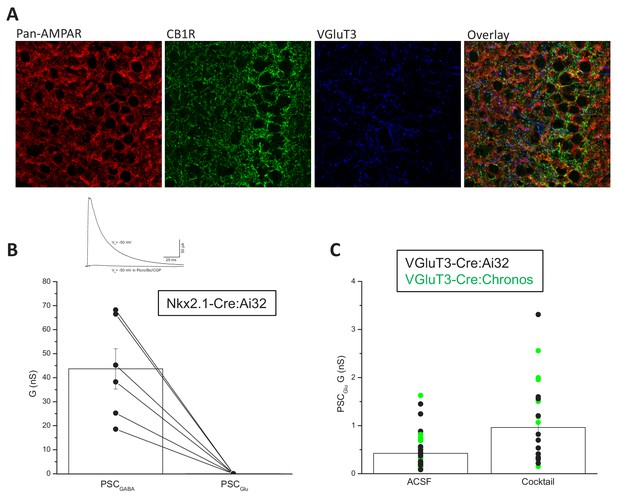
Additional data in support of glutamatergic output from VGluT3+ INTs.
(A) Representative high-resolution Airyscan images highlighting prominent somatic AMPAR signal within CA1 SP in close proximity to perisomatic targeting CB1R+ and VGluT3+ CCKBC terminals. (B) Traces from a representative recording (above) and group data summary (below) illustrating a lack of light driven glutamatergic output onto CA1PCs in Nkx2.1-Cre:Ai32 mice (n = 6 cells from one mouse). (C) Group data summary comparing PSCGlu conductance recorded in CA1 PCs by light evoked activation of VGluT3+INTs the absence (n = 45 cells from 22 mice) or presence (n = 21 cells from three mice) of a cocktail (AM251/CGP 55845/UBP 302/LY 341495/CTZ) designed to minimize presynaptic depression and AMPAR desensitization. Throughout the figure summary plots provide individual observations as well as group means ± SEM.
-
Figure 4—figure supplement 1—source data 1
Data plotted in Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/51996/elife-51996-fig4-figsupp1-data1-v2.xlsx
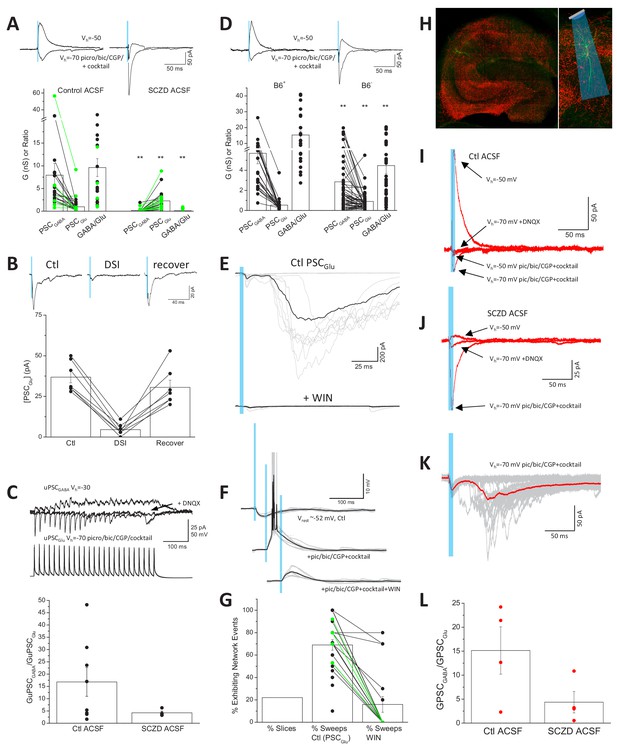
Enhanced glutamate release from CCK+VGluT3+INTs following GAD inhibition.
(A) Traces from representative recordings (above) and group data summary (below) comparing light-evoked GABA and glutamate synaptic conductances observed in CA1 PCs of VGluT3-Cre:Ai32 or VGluT3-Cre:Chronos slices maintained in control (left, n = 22 cells from four mice) or SCZD supplemented ACSF (right, n = 23 cells from four mice; **p<0.01 vs. Control ACSF). (B) Traces from a representative recording (above) and group data plot (below) illustrating DSI of PSCGlu evaluated in a subset SCZD treated slices (n = 7 cells from two mice). (C) Traces from a representative VGluT3+BC-CA1PC pair recording (above) and group data summary (below) illustrating enhanced glutamatergic, relative to GABAergic, transmission in slices maintained in SCZD (n = 4 pairs from two mice) compared to slices maintained in control ACSF (n = 8 pairs from six mice). Traces are averages of 3–5 sweeps per condition obtained by presynaptic train stimulation. (D) Traces from representative recordings (above) and group data plots (below) comparing light evoked GABA and glutamate synaptic conductances in CA1PCs of VGluT3-Cre:Ai32 mice maintained on B6+ diet (left, n = 24 cells from four mice) or B6- diet (right n = 56 cells from nine mice **p<0.01 vs. B6+). (E) Example recording with GABAergic transmission blocked that displayed large polysynaptic network barrages following light evoked monosynaptic glutamatergic events (upper traces) that could be eliminated by the CB1R agonist WIN-55212–2 (lower traces). Individual trials/sweeps are shown overlayed in gray with the averaged events shown in black. (F) Example recording of a VGluT3-Cre:Ai32 CA1PC held in current clamp illustrating the transition from light-evoked GABAergic inhibition (upper traces) to suprathreshold glutamatergic excitation (middle traces) that is suppressed below threshold by WIN-55212–2 (lower traces). (G) Group data summary plot of the proportion of slices exhibiting light-evoked polysynaptic network barrages similar to those illustrated in E as well as the percentage of trials/sweeps in such recordings displaying network events in the absence (n = 25 slices from 11 mice) or presence of WIN-55212–2 (n = 17 slices, from six mice). (H) Images of a representative hippocampal section from a VGluT3-Cre mouse infected with AAV-mDlx-Flex-ChR2-mCherry (VGluT3-Cre:mDlxChR2) with a filled PC. (I–J) Averaged traces from representative PC recordings in slices from VGluT3-Cre:mDlxChR2 mice illustrating light evoked GABA and glutamate cotransmission under control (I) or SCZD (J) conditions. (K) Traces from a sample VGluT3-Cre:mDlxChR2 recording that exhibited light evoked polysynaptic network barrages. (L) Group data summary comparing PSCGABA and PSCGlu conductance ratios for VGluT3-Cre:mDlxChR2 slices maintained in control (n = 4 cells from one mice) or SCZD ACSF (n = 4 cells from one mice). Throughout the figure summary plots provide individual observations as well as group means ± SEM.
-
Figure 5—source data 1
Data plotted in Figure 5.
- https://cdn.elifesciences.org/articles/51996/elife-51996-fig5-data1-v2.xlsx
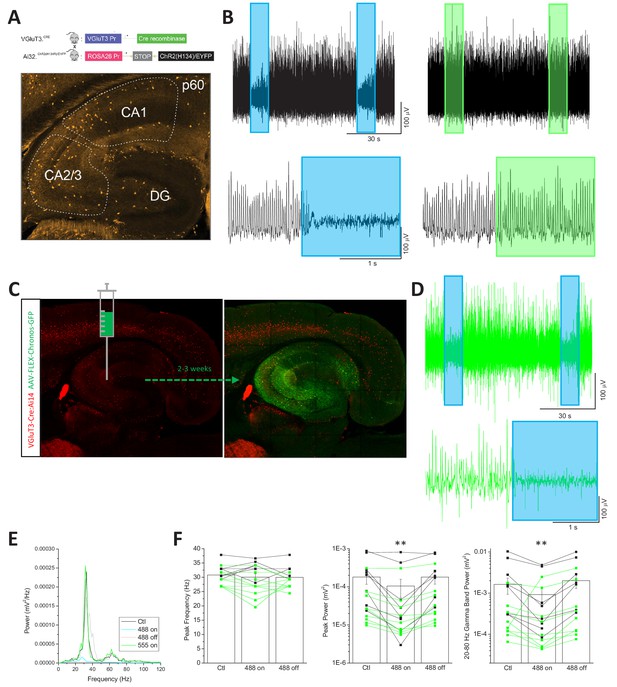
Supression of network activity by GABAergic output from CCK+VGluT3+ INTs.
(A) Schematic depicting breeding strategy to genetically introduce ChR2 within CCK+VGluT3+INTs by breeding VGluT3-Cre:Ai32 mice (above) and representative image of VGluT3+ cells (from VGluT3-Cre:Ai14 reporter) distributed throughout the mature hippocampus that can contribute to controlling network activity with light stimulation (below). (B) Traces from a representative local field potential recording in CA3 SP illustrating the sensitivity of carbachol induced gamma oscillations to blue light mediated activation of CCK+VGluT3+INTs in slices from VGluT3-Cre:Ai32 mice. The upper traces illustrate two epochs of either blue or green light stimulation during ongoing gamma activity while the lower traces illustrate the effect of light onset at higher temporal resolution. (C) Images, with schematics overlayed, illustrating a representative slice with selective hippocampal expression of Chronos that was evaluated for gamma activity. (D) Traces of the local field potential recording in CA3 from the slice displayed in C illustrating the sensitivity of carbachol induced gamma oscillations to blue-light-mediated activation of CCK+VGluT3+INTs infected with Chronos. The upper traces illustrate two epochs of blue light stimulation during ongoing gamma activity while the lower traces illustrate the effect of light onset at higher temporal resolution. (E) Power spectra density plots for the sample traces in B under the conditions indicated. Plots were generated from 10 s epochs before (Ctl) during (488 on) of after (488 off) blue light stimulation as well as during an epoch of green light stimulation (555 on). (F) Group data summary plots illustrating the effects of light evoked CCK+VGluT3+INT activation on peak gamma frequency (left) and power (middle) as well as gamma band power (right) as indicated (n = 8 slices from 3 VGluT3-Cre:Ai32 mice; n = 9 slices from 4 VGluT3-Cre:Chronos mice). **p<0.01 vs. Ctl.
-
Figure 5—figure supplement 1—source data 1
Data plotted in Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/51996/elife-51996-fig5-figsupp1-data1-v2.xlsx
Tables
CCK+VGluT3+INTs membrane properties.
| Active/Passive Membrane Properties | VGluT3+ BCs (mean ± SEM, n = 30–37 observations/parameter, from 37 cells) | VGluT3+ DTIs (mean ± SEM, n = 24–26 observations/parameter, from 26 cells) |
|---|---|---|
| Resting potential (mV) | −56.9 ± 0.8 | −59.0 ± 1.1 |
| Input resistance (MΩ) | 158.5 ± 9.9 | 149.7 ± 10.1 |
| Time constant (ms) | 20.8 ± 1.1 | 22.0 ± 1.0 |
| Capacitance (pF) | 135.7 ± 5.8 | 150.1 ± 6.3 |
| Sag index | 0.83 ± 0.01 | 0.82 ± 0.02 |
| Spike threshold (mV) | −41.9 ± 0.8 | −39.86 ± 1.1 |
| Spike amplitude (mV) | 57.4 ± 1.7 | 55.7 ± 1.9 |
| Spike half-width (ms) | 0.74 ± 0.03 | 0.80 ± 0.05 |
| Maximal rise slope (mV/ms) | 197.7 ± 8.5 | 186.4 ± 13.9 |
| Maximal decay slope (mV/ms) | −87.8 ± 4.4 | −81.1 ± 7.2 |
| Frequency at 2x Threshold (Hz) | 34.9 ± 2.4 | 31.3 ± 1.8 |
| Adaptation ratio at 2x threshold | 0.43 ± 0.04 | 0.43 ± 0.05 |
| AHP amplitude (mV) | −10.3 ± 0.6 | −11.8 ± 0.9 |
-
Table 1—source data 1
Individual neuron properties.
- https://cdn.elifesciences.org/articles/51996/elife-51996-table1-data1-v2.xlsx





