A processive rotary mechanism couples substrate unfolding and proteolysis in the ClpXP degradation machinery
Figures
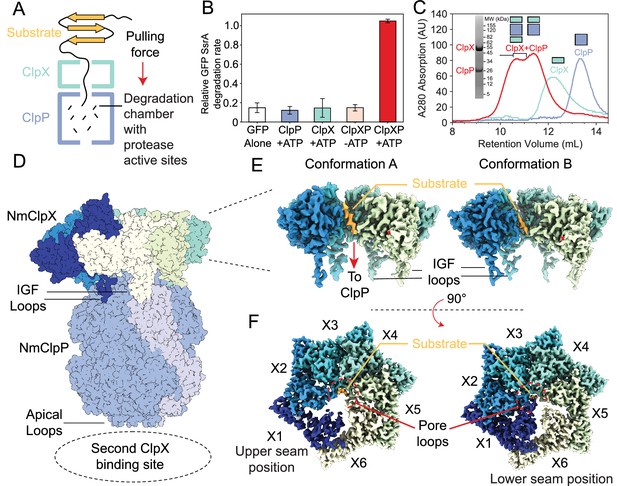
Functional and structural characterization of ClpXP from N. meningitidis.
(A) Schematic representation of the ClpXP degradation machinery. The overall positions of the substrate (orange), ClpX unfoldase (green), and the ClpP protease (blue) are shown; (B) GFP-SsrA degradation by ClpXP is ATP-dependent. The degradation rate of GFP-SsrA is monitored by measuring loss of fluorescence. The components (ClpX, ClpP, ATP) included for each measurement are denoted on the plot. All measurements included GFP-SsrA and were performed in triplicate on WT ClpXP; (C) ClpXP complex formation monitored by size exclusion chromatography (SEC). SEC profiles of isolated ClpP (blue trace) and ClpX (green trace) are consistent with their expected molecular weights. SEC profile of a 2:1 ClpX6:ClpP14 mixture (red trace) incubated for 10 min in the presence of MgATP shows the formation of doubly- and singly-capped ClpXP complexes. The running buffer for all traces contained 2 mM MgATP. SDS-PAGE gel of SEC fractions (fractions within the region delineated by black bracket) shows that they contain both ClpP and ClpX; (D) Overall architecture of the ClpXP complex as established by cryo-EM. The ClpP double ring component is shown in dark blue, with the exception of a pair of opposing protomers shown in light blue shade to delineate the IGF loop binding site that is located at the interface between protomers. Cutaway side (E) and top (F) views of Conformations A and B of ClpX resulting from focused classification and local refinement. The IGF loops that bind to ClpP, and the presence of substrate (orange) in the axial channel of ClpX, are highlighted. Each of the ClpX protomers is labeled. In panel (F), the engagement of the substrate (orange) with five of the six pore loops is indicated (red semi-circle).
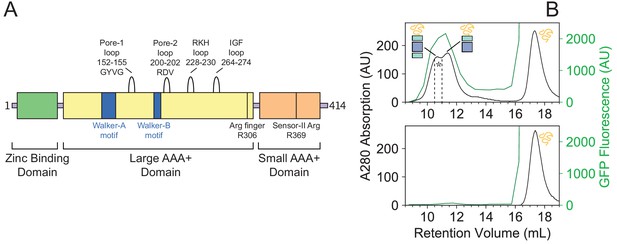
Sequence map of NmClpX and SEC analysis of substrate binding by NmClpXP.
(A) NmClpX primary sequence domain map. Important structural and functional motifs are denoted. (B) NmClpXP binds a SsrA-tagged GFP. SEC profiles for ClpXP incubated with GFP-SsrA (top panel) and GFP-SsrA alone (bottom panel) measured using absorbance at 280 nm (black trace) and GFP fluorescence (green trace - λex:480 nm, λem:508 nm). The region used for SDS-PAGE analysis is highlighted by * (Figure 1C).
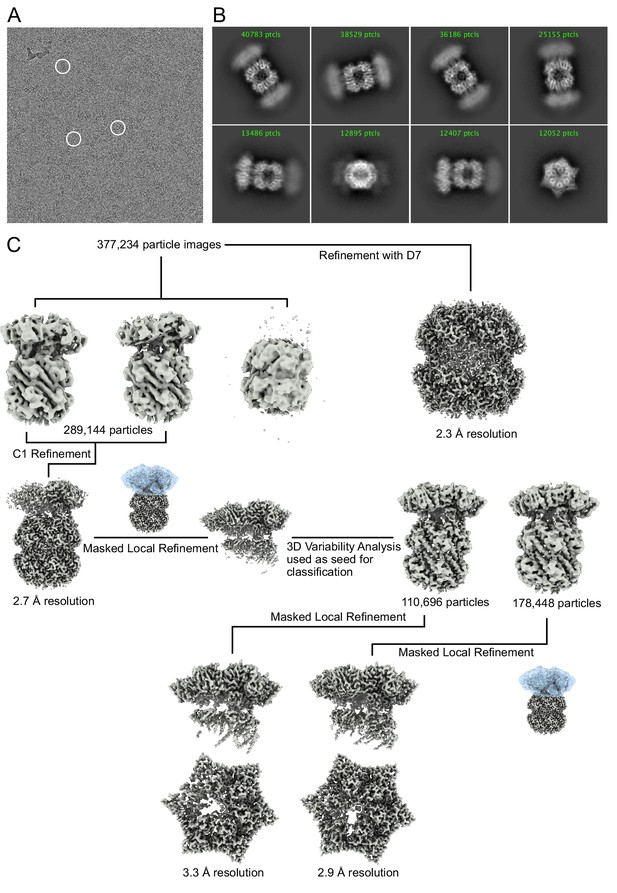
Cryo-EM image processing.
(A) Representative cryo-EM micrograph of NmClpXP bound to GFP-SsrA. Example complexes are circled in white. (B) Representative 2D class averages of NmClpXP with GFP-SsrA. (C) Image processing workflow used to obtain final maps.
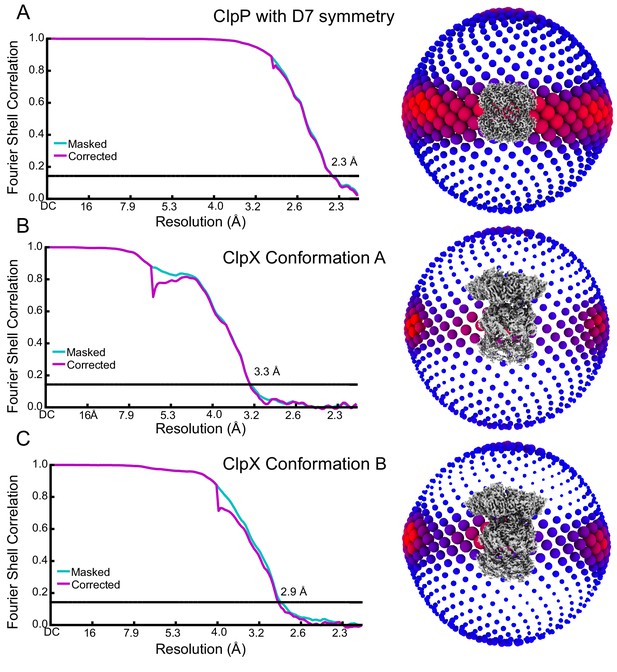
Cryo-EM map validation.
(A to C) Fourier Shell Correlation (FSC) plots (left) and orientation distribution (right). Resolution values reported are for FSC = 0.143.
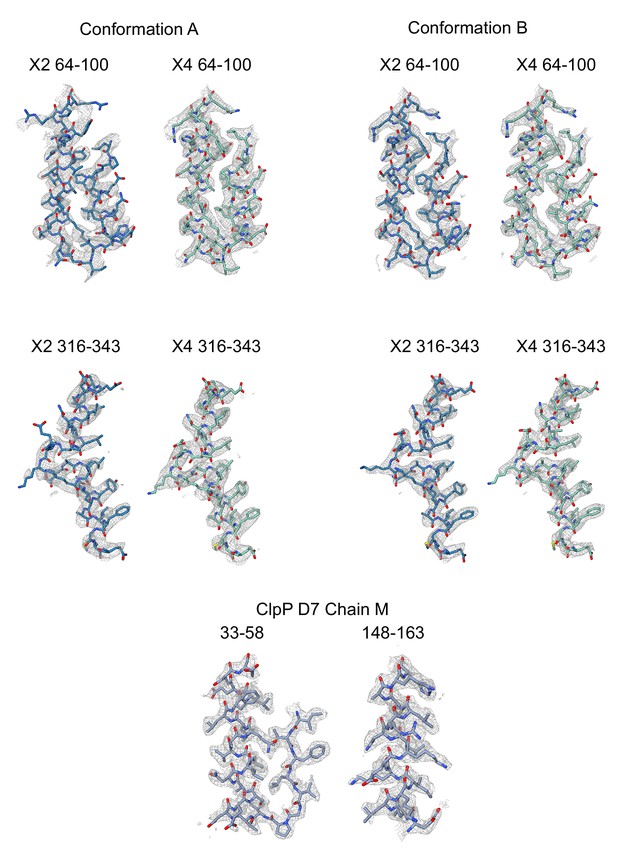
Examples of regions of the atomic models built into the experimental cryo-EM maps.
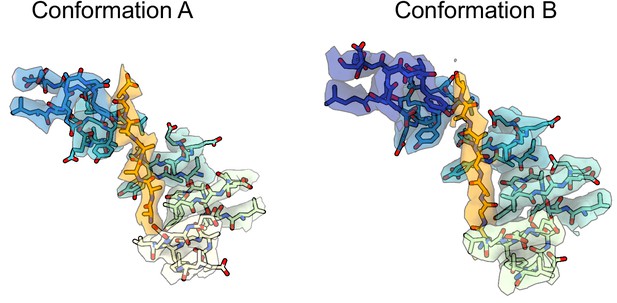
Experimental density maps and models for the substrate pore-1 loop interaction interface.
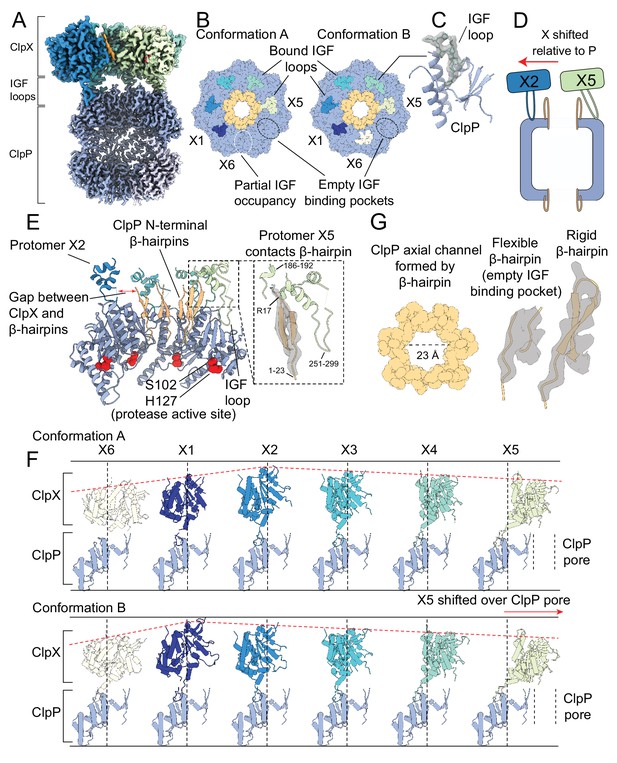
The interaction interface between ClpX and ClpP.
Protomer X1 occupies the US position in Conformation A and protomer X6 the LS position in Conformation B. (A) Cutaway density map of the overall architecture of the ClpXP interaction interface. (B) Models for the ClpP apical surface and the ClpX IGF loops (residues 265–275). The empty IGF-binding pocket (dotted oval) resides clockwise to the X5 protomer in both conformations. In Conformation A, an IGF loop was not built into the map for protomer X6 due to weak loop density (yellow dotted line). (C) Model in map fit for an IGF loop (grey), surrounded by regions of ClpP comprising the IGF binding site (blue). (D) Interaction of the ClpP N-terminal gates with ClpX. The X2 protomer moves away from the gates, while the X5 protomer directly contacts the gate of the ClpP protomer to which its IGF loop is bound, shown in (E). Inset shows the details of this interaction, with model in map fit for the ClpP β-hairpin. (F) Positions of ClpX protomers relative to ClpP. Images were generated by fitting all ClpP protomers to a common protomer and displaying the corresponding ClpX protomer. In both conformations, ClpX adopts a spiral arrangement relative to ClpP (dotted red line). In Conformation A, the X2 protomer is located at the top of the spiral, while in Conformation B the X1 protomer occupies the top position, ~7 Å higher than its position in Conformation A. The ClpX protomers rotate and translate relative to ClpP; protomers X2 and X3 sit nearly atop their ClpP protomers, while X4 and especially X5 show large deviations from this position (vertical lines), with X5 sitting overtop the ClpP axial pore. (G) View down the channel formed by the N-terminal ClpP β-hairpins (left); density and models for the most flexible and rigid β-hairpins (center and right, respectively).
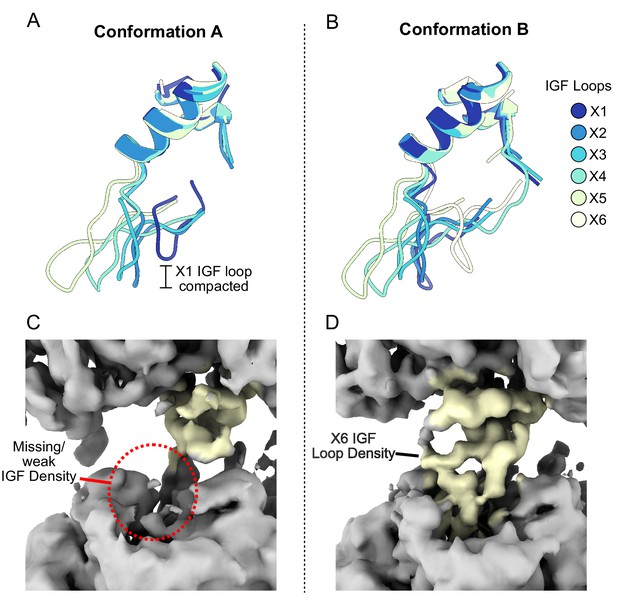
IGF loop flexibility.
(A and B) Large domains of ClpX were aligned and models for the IGF loops are shown. (C and D) Unsharpened density map for Conformations A and B looking at protomer X6.
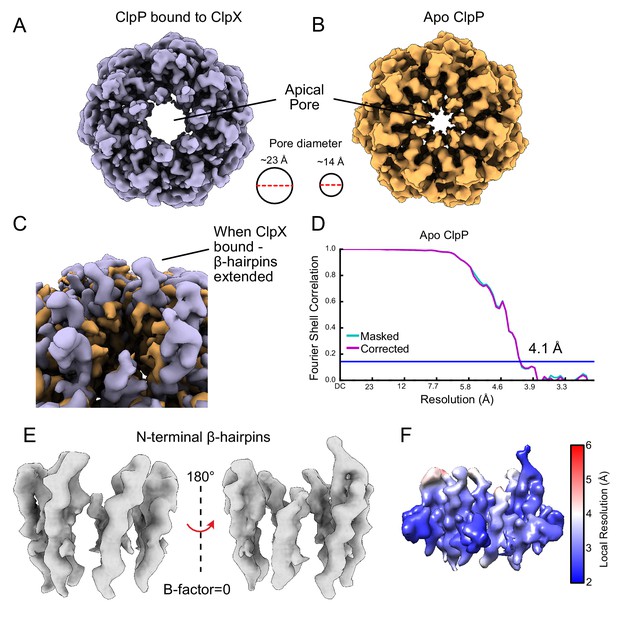
The ClpP apical loops extend upwards from the ring surface.
(A) Unsharpened density map of the ClpP N-terminal β-hairpins when bound to ClpX. (B) Cryo-EM map of apo ClpP from N. meningitidis showing that the apical loops no longer extend ‘up’. (C) ClpX-bound and apo forms of ClpP overlaid. There is no noticeable density for the turn of the ClpP N-terminal β-hairpin in the apo form, and the β-hairpins point inwards, constricting the axial pore of ClpP. (D) Fourier Shell Correlation (FSC) plot for Apo-NmClpP. (E) Closeup view of the N-terminal β-hairpins when bound to ClpX. (F) Density map of the β-hairpins in the ClpXP complex, colored by local resolution.
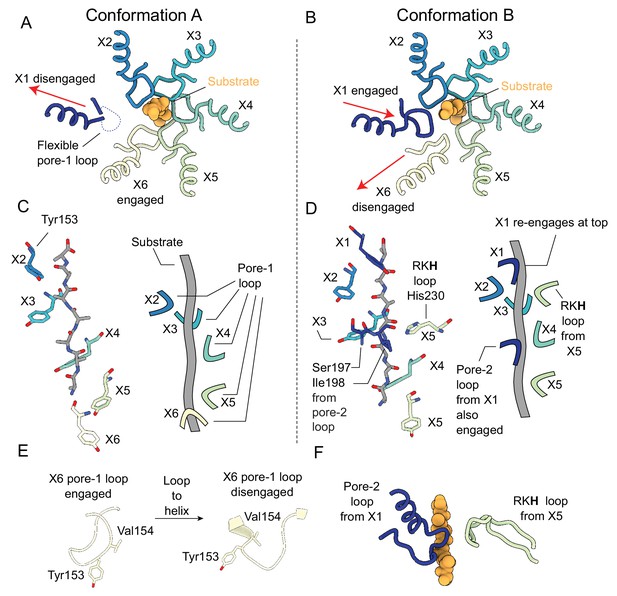
Substrate engagement by the ClpX pore loops.
(A and B) Pore-1 loop residues grip the substrate, as observed in this view looking down the axial channel. The substrate is modeled as poly-Ala. (A) In Conformation A, the X1 protomer is disengaged from the substrate in the US position (red arrow) and shows noticeable flexibility in its pore-1 loop. (B) In Conformation B, the X1 protomer contacts the substrate but the X6 protomer has disengaged into the LS position (red arrows). (C and D) Model of substrate and interacting residues, along with schematic, viewed perpendicular to the axial channel. (C) In Conformation A, only the Tyr153 residue of the pore-1 loop makes significant contacts with the substrate; the five protomers (X2, X3, X4, X5, X6) form a downward spiral surrounding the backbone of the substrate chain, extending to the apical surface of ClpP. (D) In Conformation B, in addition to the five Tyr153 residues from the five gripping protomers (X1, X2, X3, X4, X5), more contacts are formed: Ser197 and Ile198 from the pore-2 loop of the X1 protomer and His230 from the RKH loop of the X5 protomer. (E) Substrate-induced conformational changes of the X6 pore-1 loop upon releasing client. When the X6 protomer is substrate engaged its pore-1 loop forms a lasso like conformation, with Tyr153 and Val154 both pointing toward the axial channel (left). When disengaged, a part of the pore-1 loop becomes an α-helix, with Val154 and Tyr153 no longer pointing outwards (right). (F) Architecture of the additional contacts made by the pore-2 and RKH loops from the X1 and X5 protomers, respectively.
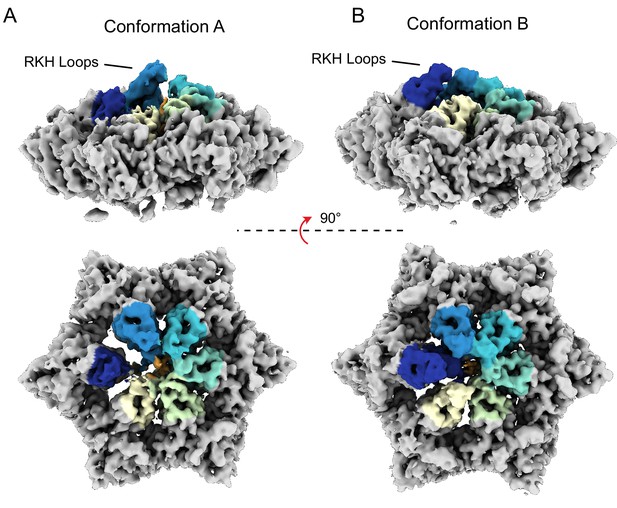
RKH loop positions.
(A and B) Unsharpened maps for both conformations A and B of ClpX are shown, with RKH loops highlighted in color (coloring done by a radius of 10 Å around residues 220–240).
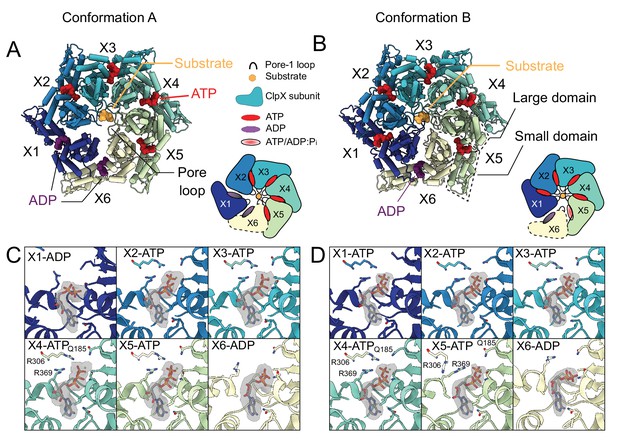
Nucleotide occupancy and interactions with ClpX.
(A and B) Model of ClpX in conformations A and B, looking into the ClpX pore. Nucleotides are shown and color-coded, with ATP red and ADP purple, and bind between the large and small domains (boxed). Inset shows a schematic of the protomer positions and nucleotide occupancies. Conformation A has two bound ADP, in the X1 (dark blue, US in Conformation A in this representation) and X6 (yellow, LS in Conformation B in this representation) protomers, with the X1 protomer disengaged from the substrate and away from the axial pore (US position). In Conformation B only a single ADP is bound, protomer X1 is reengaged with substrate and X6 has disengaged. (C) Nucleotide-binding sites of Conformation A, with key interacting residues shown along with the experimental density maps corresponding to bound nucleotide. In the ADP-bound sites, both the arginine finger (R306) and the sensor-II arginine (R369) have moved away, while in the ATP bound sites they form close contacts with the β and γ-phosphates. (D) Nucleotide binding in Conformation B. As in Conformation A, R306 and R369 have moved away from the ADP, while in the X5 protomer only the arginine finger has moved away, while the sensor-II transitions closer to the γ-phosphate.
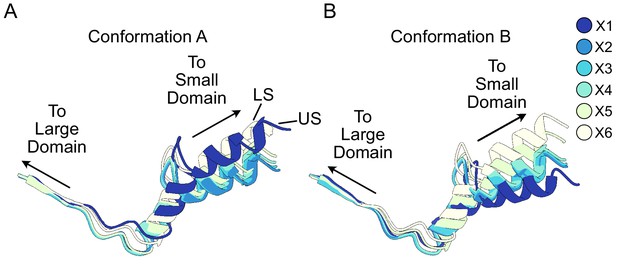
Relative orientation between large and small domains of ClpX.
(A and B) Residues 308–343 are shown. Note that protomers X1 and X6 in Conformations A and B, respectively, are displaced from substrate. These show among the largest differences in domain orientations.
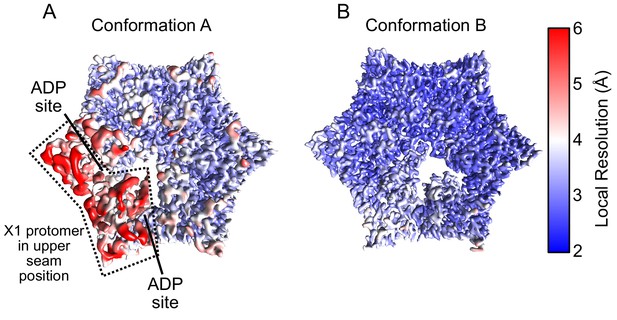
Local resolution maps of ClpX.
(A and B) Density maps colored by local resolution for Conformations A and B. In Conformation A the US protomer is highlighted with a dashed line.
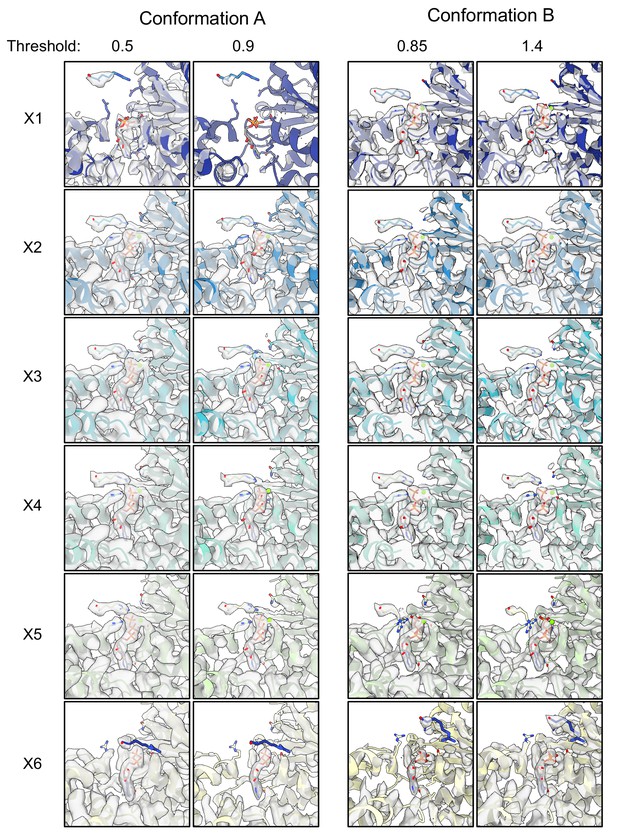
ATP-binding pocket densities.
Experimental density maps and models are shown for the ATP-binding pockets of all protomers. Two numerical thresholds are shown to highlight differences in density.
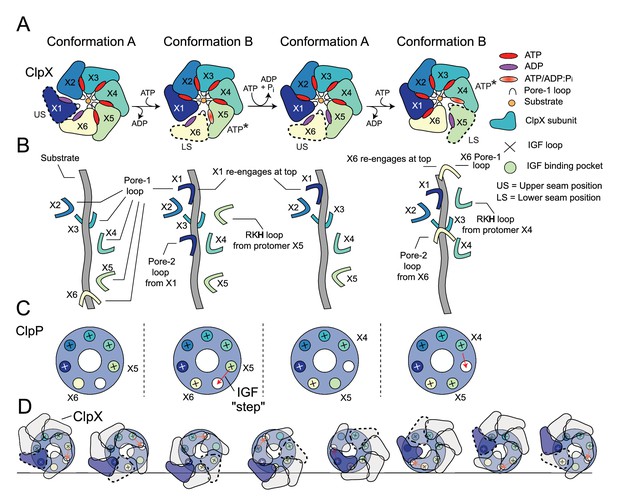
Translocation model for ClpXP.
(A) Schematic of the ClpX-nucleotide-dependent conformational cycle. The transition from Conformation A to Conformation B is mediated through nucleotide exchange in the US position (1st to 2nd ring), with ATP hydrolysis and phosphate release in the ATP* position restoring the A conformation from the B state (2nd to 3rd ring) so that the cycle can repeat. In the new Conformation A (3rd ring), protomer X6 (yellow) adopts the US conformation and the X1 protomer (previous US) moves to the top of the spiral. In this way each protomer of the complex cycles through all positions as ATP is hydrolyzed in successively new ATP* protomer states, requiring the hydrolysis of seven ATPs. (B) Hand-over-hand mechanism of pore-loop mediated translocation. In the first A to B transition depicted in (A) X1 (US protomer) reengages at the top of the spiral and the LS pore one loop (X6 in Conformation B) disengages from the bottom of the spiral. As this is repeated the pore loops move ‘up’ the substrate and pull it into the degradation chamber. (C) IGF loop movements accompanying the hand-over-hand movements of ClpX. Shown in the small circles are the seven IGF-loop-binding sites in ClpP with a ‘+’ sign indicating engagement with an IGF loop of a given ClpX protomer. The binding sites are color coded according to the color of the protomer that interacts with the corresponding site. In conformation A, the IGF loop of X6 is unbound or only weakly bound, while in Conformation B it is bound. A new Conformation A is generated when the IGF loop from the ATP* protomer unbinds. It subsequently reengages with ClpP in a new Conformation B (third and fourth rings in (A)). In this way, the IGF loops ‘step’ around the ClpP ring, with each complete rotation requiring the hydrolysis of seven ATPs. (D) Schematic of the ClpX-ClpP interactions during seven ATP hydrolysis steps that lead to a net rotation of ClpX protomers by 60o. Because of the six- to seven-fold symmetry mismatch a total of forty-two ATPs must be hydrolyzed to return the protomers back to their original starting positions in the cycle (i.e. ring 1). Each of the seven ring structures corresponds to Conformation A, with the red arrows indicating the subsequent step of the IGF loop of the ATP* protomer along the cycle. The US protomer in each step is highlighted by the dashed trace.
Videos
Overview of the substrate-bound ClpXP complex (Conformation B).
Hand-over-hand translocation cycle of the ClpXP complex.
Interpolation between states A → B → A → B. Side and top views of the complex are shown (top), along with the pore-1 loop Tyr153 and substrate (bottom left) as well as IGF loops (bottom right).
ClpX precession on the ClpP apical surface.
Repeated interpolation between states reveals a precession of the ClpX ring as its IGF loops are placed successively in different ClpP binding sites. The position of the empty binding pocket on ClpP undergoes a complete cycle around the complex for every 60o rotation of the ClpX ring relative to the ClpP ring.
Tables
Cryo-EM data acquisition, processing, atomic model statistics, and map/model depositions.
| A. Cryo-EM data acquisition and image processing. | |||||
|---|---|---|---|---|---|
| Data collection | |||||
| NmClpXP + GFP-SsrA | apo-NmClpP | ||||
| Electron Microscope | Titan Krios | Tecnai F20 | |||
| Camera | Falcon 3EC | K2 Summit | |||
| Voltage (kV) | 300 | 200 | |||
| Nominal Magnification | 75,000 | 25,000 | |||
| Calibrated physical pixel size (Å) | 1.06 | 1.45 | |||
| Total exposure (e/Å2) | 42.7 | 35 | |||
| Exposure rate (e/pixel/s) | 0.8 | 5 | |||
| Number of frames | 30 | 30 | |||
| Defocus range (μm) | 0.9 to 1.7 | 0.8 to 3.3 | |||
| Image processing | |||||
| Motion correction software | cryoSPARC v2 | cryoSPARC v2 | |||
| CTF estimation software | cryoSPARC v2 | cryoSPARC v2 | |||
| Particle selection software | cryoSPARC v2 | cryoSPARC v2 | |||
| Micrographs used | 2680 | 122 | |||
| Particle images selected | 466,549 | 100,132 | |||
| 3D map classification and refinement software | cryoSPARC v2 | cryoSPARC v2 | |||
| B. Map and model statistics. | |||||
| EM maps | NmClpXP with Symmetry | Focused NmClpX Conformation A | Focused NmClpX Conformation B | Apo-NmClpP | |
| Particle images contributing to maps | 377,234 | 110,696 | 178,448 | 50,403 | |
| Applied symmetry | D7 | C1 | C1 | D7 | |
| Applied B-factor (Å2) | −116.2 | −84.1 | −88.1 | −229.7 | |
| Global resolution (FSC = 0.143, Å) | 2.3 | 3.3 | 2.9 | 4.1 | |
| Model Building | NmClpX Conformation A focused | NmClpX Conformation B focused | NmClpP with Symmetry | NmClpXP Conformation A combined | NmClpXP Conformation B combined |
| Modeling software | Coot, Phenix, Rosetta | ||||
| Number of residues | 1912 | 2018 | 2671 | 4583 | 4689 |
| RMS bond length (Å) | 0.0043 | 0.0037 | 0.0039 | 0.0044 | 0.0038 |
| RMS bond angle (°) | 1.04 | 0.93 | 0.91 | 1.00 | 0.92 |
| Ramachandaran outliers (%) | 0.00 | 0.05 | 0.00 | 0.00 | 0.02 |
| Ramachandran favored (%) | 98.33 | 97.32 | 95.68 | 97.07 | 96.39 |
| Rotamer outliers | 0.25 | 0.24 | 0.00 | 0.13 | 0.10 |
| C-beta deviations | 0 | 0 | 0 | 0 | 0 |
| Clashscore | 1.45 | 2.8 | 0.67 | 1.84 | 2.11 |
| MolProbity score | 0.88 | 1.20 | 1.02 | 1.11 | 1.22 |
| EMRinger score | 2.78 | 3.42 | 5.6 | ||
| Map-Model CC_mask | 0.75 | 0.78 | 0.82 | ||
| Ligand | 2 ADP 4 Mg-ATP | 1 ADP 5 Mg-ATP | 2 ADP 4 Mg-ATP | 1 ADP 5 Mg-ATP | |
| C. Residues excluded in atomic models*. | |||||
| ClpX protomer | NmClpXP Conformation A combined | NmClpXP Conformation B combined | |||
| X1 (Chain A) | 1–62, 102–109, 149–156, 191–204, 224–236, 263–264, 273–282, 413–414 | 1–62, 228–233, 275–278, 413–414 | |||
| X2 (Chain B) | 1–62, 102–111, 192–198, 226–235, 275–279, 413–414 | 1–62, 226–233, 275–279, 413–414 | |||
| X3 (Chain C) | 1–62, 192–198, 225–232, 275–279, 413–414 | 1–62, 192–198, 228–233, 274–278, 413–414 | |||
| X4 (Chain D) | 1–62, 103–106, 193–198, 226–234, 276–280, 413–414 | 1–62, 193–198, 229–233, 413–414 | |||
| X5 (Chain E) | 1–62, 193–198, 229–233, 277–281, 413–414 | 1–62, 193–198, 277–279, 413–414 | |||
| X6 (Chain F) | 1–62, 193–201, 225–234, 259–287, 413–414 | 1–62, 191–204, 227–233, 275–282, 413–414 | |||
| ClpP subunits | NmClpXP Conformation A combined | NmClpXP Conformation B combined | |||
| Chain H | 1–5, 16–18, 199–204 | 1–5, 16–18, 199–204 | |||
| Chain I | 1–5, 16–17, 199–204 | 1–5, 16–17, 199–204 | |||
| Chain J | 1–5, 17, 199–204 | 1–5, 17, 199–204 | |||
| Chain K | 1–5, 199–204 | 1–5, 199–204 | |||
| Chain L | 1–5, 16–17, 199–204 | 1–5, 16–17, 199–204 | |||
| Chain M | 1–5, 13–19, 199–204 | 1–5, 13–19, 199–204 | |||
| Chain N | 1–5, 16–17, 199–204 | 1–5, 16–17, 199–204 | |||
| Chain O | 1–5, 16–17, 199–204 | 1–5, 16–17, 199–204 | |||
| Chain P | 1–5, 16–17, 199–204 | 1–5, 16–17, 199–204 | |||
| Chain Q | 1–5, 16–17, 199–204 | 1–5, 16–17, 199–204 | |||
| Chain R | 1–5, 16–17, 199–204 | 1–5, 16–17, 199–204 | |||
| Chain S | 1–5, 16–17, 199–204 | 1–5, 16–17, 199–204 | |||
| Chain T | 1–5, 16–17, 199–204 | 1–5, 16–17, 199–204 | |||
| Chain U | 1–5, 16–17, 199–204 | 1–5, 16–17, 199–204 | |||
| D. Deposited maps and associated coordinate files. | |||||
| Maps | EMDB code | Associated PDB ID | |||
| NmClpXP Conformation A | EMD-21187 | 6VFS | |||
| NmClpXP Conformation B | EMD-21194 | 6VFX | |||
| NmClpXP D7 | EMD-21195 | ||||
| Apo-NmClpP | EMD-21196 | ||||
-
*Protomer X1 is US in Conformation A and protomer X2 is LS in Conformation B.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Neisseria meningitidis) | clpX | UniProtKB - Q9JYY3 | ||
| Gene (Neisseria meningitidis) | clpP | UniProtKB - Q9JZ38 | ||
| Strain, strain background (Escherichia coli) | BL21(DE3) | Sigma-Aldrich | CMC0016 | Chemically competent cells |
| Recombinant DNA reagent | pET28-NmClpP | Synthetic (GenScript) | UniProtKB - Q9JYY3 | Plasmid containing ClpP |
| Recombinant DNA reagent | pET28a-NmClpX | Synthetic (GenScript) | UniProtKB - Q9JZ38 | Plasmid containing ClpX |
| Recombinant DNA reagent | pET28a- GFP-SsrA | (Ripstein et al., 2017) | Plasmid containing GFP-SsrA | |
| Chemical compound, drug | PKM-AMC | GenScript | Fluorogenic peptide for protease assays | |
| Software, algorithm | EPU | Thermo Fischer Scientific | EM imaging software | |
| Software, algorithm | cryoSPARC v2 | (Punjani et al., 2017) | RRID:SCR_016501 | EM reconstruction software |
| Software, algorithm | UCSF Chimera | (Pettersen et al., 2004) | RRID:SCR_004097 | Molecular Visualization Software |
| Software, algorithm | UCSF ChimeraX | (Goddard et al., 2018) | RRID:SCR_015872 | Molecular Visualization Software |
| Software, algorithm | Coot | (Emsley and Cowtan, 2004) | RRID:SCR_014222 | Protein Model Building Software |
| Software, algorithm | Phyre2 | (Kelley et al., 2015) | RRID:SCR_010270 | Protein Model Building Software |
| Software, algorithm | Rosetta | (Wang et al., 2015) | RRID:SCR_015701 | Protein Model Building Software |
| Software, algorithm | Phenix | (Adams et al., 2010) | RRID:SCR_014224 | Protein Model Building Software |
| Software, algorithm | Molprobity | (Arendall III et al., 2010) | RRID:SCR_014226 | Protein Model Evaluation Software |
| Software, algorithm | EMRinger | (Barad et al., 2015) | Protein Model Evaluation Software |