Hepatitis C virus exploits cyclophilin A to evade PKR
Figures
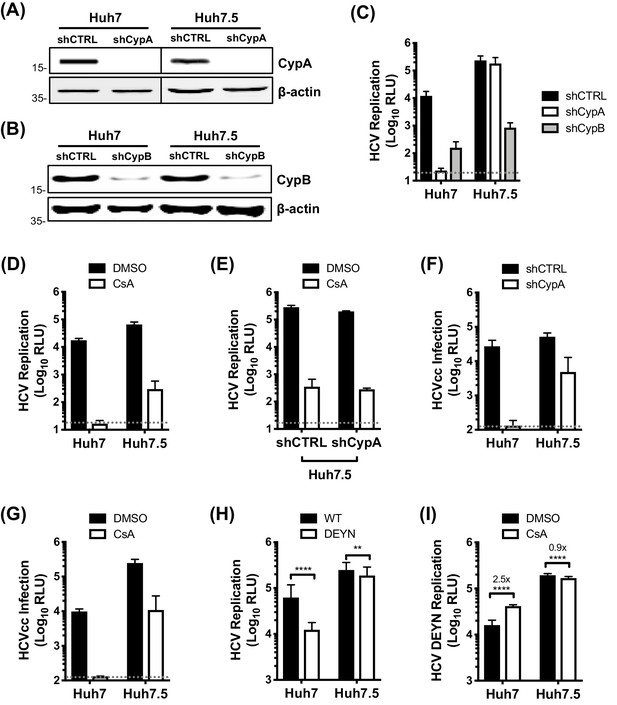
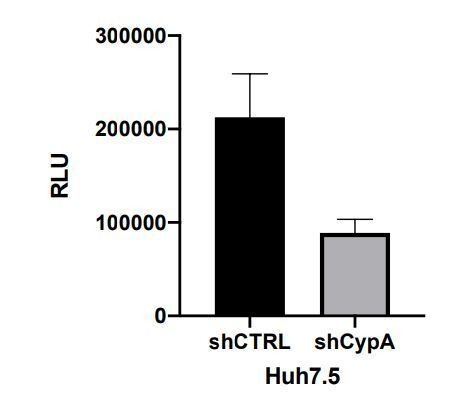
CypA is critical for HCV replication in Huh7 cells, but not in Huh7.5 cells.
(A) Western blot detecting CypA (A) or CypB (B) expression in Huh7 and Huh7.5 cells transduced with Cyp specific shRNA-expressing lentiviruses as shown. Actin was detected as a loading control. (C) Evaluation of HCV replicon replication in CypA- and CypB-silenced cells. Luciferase reporter activity was measured at 4 and 48 hr post-electroporation (hpe) and is expressed as relative luciferase units (RLU) at 48 hpe. (D-E) Huh7 or Huh7.5 cells (silenced or not for CypA expression) were electroporated with replicon RNA and treated with 1 μM CsA at 4 hr post electroporation (hpe). Luciferase reporter activity was measured at 48 hr post-electroporation. (F) HCVcc infection in CypA-silenced Huh7 and Huh7.5 cells. Cells were infected with HCVcc (J6/JFH1-RLuc) and infection was assessed after 72 hr by measuring luciferase activity. (G) Huh7 or Huh7.5 cells were infected with HCVcc and treated with 1 μM CsA at 4 hr post-infection (hpi). After 72 hr, infection was measured by luciferase activity. (H) Replication of HCV NS5A wild-type (WT) and HCV CsA resistance mutant (NS5A D316E/Y317N; DEYN) in Huh7 and Huh7.5 cells. Cells were electroporated with in vitro transcribed replicon RNA as described above, and replication was assessed by luciferase activity at 48 hpe. (I) Huh7 or Huh7.5 cells were electroporated with HCV NS5A DEYN replicon RNA. After 4 hr, cells were treated with 1 μM CsA and replication was assessed by luciferase activity at 48 hpe. All graphs show relative luciferase units (RLU) expressed as means ± standard deviation from at least three independent experiments each performed in triplicate. Statistical significance was evaluated by t-test using GraphPad Prism (**** p-value<0.0001; ** p-value<0.01). Detection limits of the assays are shown by the dotted grey line.
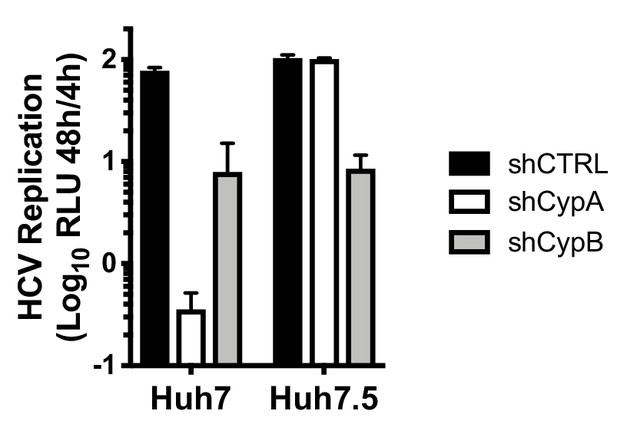
CypA depletion abrogates HCV replication in Huh7, but not Huh7.5 cells.
The 48 hr values from Figure 1C were normalised to the 4 hr RLU to compensate for potential differences in electroporation efficiency between cell lines, and are expressed as fold increase compared to 4 hr RLU.
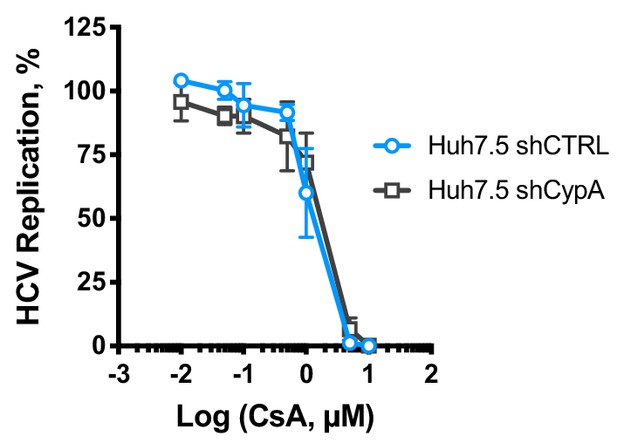
CsA is equally potent against HCV replication in CypA-depleted Huh7.5 cells.
Huh7.5 cells (silenced or not for CypA expression) were electroporated with replicon RNA and treated with serially diluted CsA at 4 hr post electroporation (hpe). Luciferase reporter activity was measured at 48 hr post-electroporation. HCV replication is expressed as a percentage relative to the DMSO control.
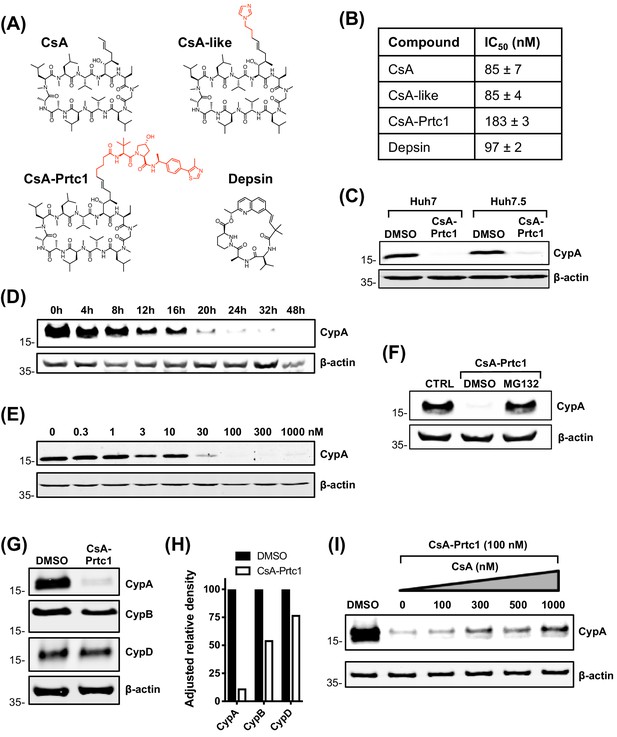
Structures and properties of distinct novel CypI.
(A) Structures of CypI used in this study; their effects on viral replication and cell viability are shown in Figure supplement 1. (B) CypI-CypA binding affinity measured by fluorescence polarisation using a fluorescein labelled CsA probe. (C) Western blot showing CsA-Prtc1-mediated degradation of CypA in Huh7 and Huh7.5 cells after 48 hr treatment with 1 μM CsA-Prtc1. (D) Analysis of CypA degradation at time points shown after 1 μM CsA-Prtc1 treatment in Huh7 cells detecting CypA expression by western blot. (E) Dose-response of CsA-Prtc1-mediated CypA degradation in Huh7 cells. Cells were treated with the indicated concentrations of CsA-Prtc1 for 48 hr, and CypA levels detected by western blot. (F) CsA-Prtc1-mediated degradation of CypA is proteasome-dependent. Cells were treated with CsA-Prtc1 (1 μM) with or without the proteasome inhibitor MG132 (10 μM) for 24 hr. CypA was detected by western blot. (G) CsA-Prtc1 specificity for CypA. Huh7 cells were treated with 1 μM CsA-Prtc1 for 24 hr and CypA, CypB or CypD detected by western blot. (H) Quantitation by densitometry of gel in (G) showing adjusted relative density normalised to the actin loading control. (I) CsA treatment rescues CypA from CsA-Prtc1-mediated degradation. Huh7 cells, treated for 24 hr with CsA-Prtc1 (100 nM), in the presence of increasing concentrations of CsA, were lysed and CypA expression detected by western blot. (C-I) One representative western blot is shown from at least two independent experiments; (D-E) quantitation by densitometry analysis (showing the combined data from the independent experiments) is shown in Figure supplement 2A-B.
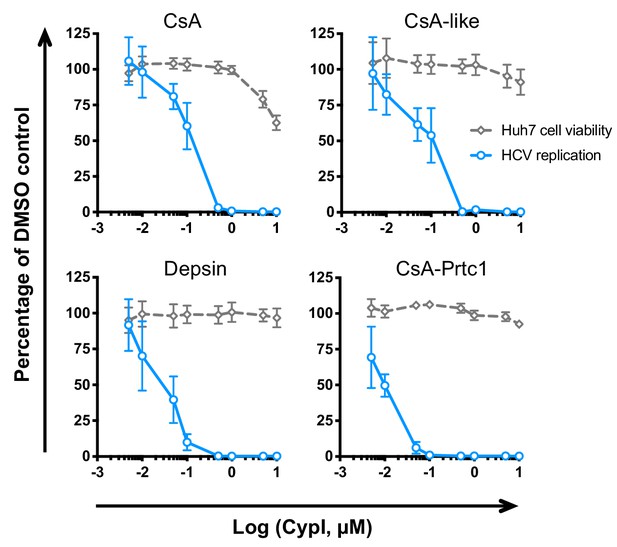
Novel CypI inhibit HCV replication and are not cytotoxic.
Approximately 2 × 106 Huh7 cells were electroporated with 5 μg HCV replicon RNA. CypI were added at four hpe. Replication (blue line) was measured by luciferase activity at 48 hpe and is expressed as percentage relative to DMSO-treated control. Cell viability (grey dashed line) was measured by Alamar Blue assay. Graphs show means ± standard deviation from three independent experiments each performed in triplicate.
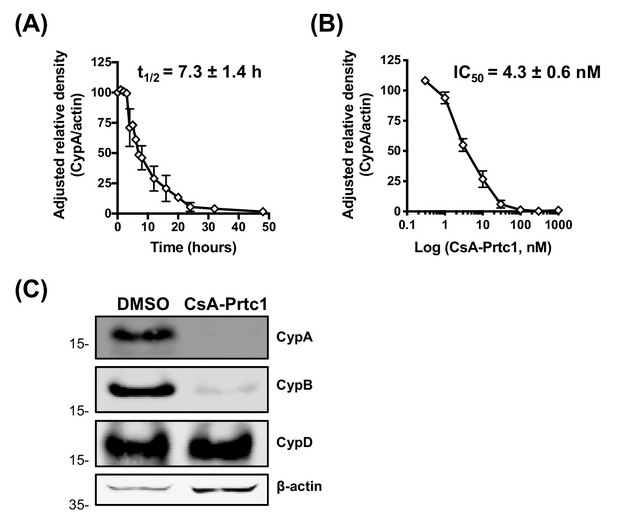
Characterisation of CsA-Prtc1 activity.
(A–B) Quantitation by densitometry analysis showing adjusted relative density normalised to the actin loading control for the time-course and dose-response shown in Figure 2D–E. Huh7 cells were treated with CsA-Prtc1 at 1 μM (A) or the indicated concentrations (B). Lysates were collected at the indicated time points (A) or after 48 hr of treatment (B) and CypA expression was evaluated by western blot. One representative blot for each is shown in Figure 2D–E. (C) Huh7 cells were treated with CsA-Prtc1 (1 μM). Lysates were collected 48 hr later and evaluated for CypA, CypB and CypD expression by western blot.
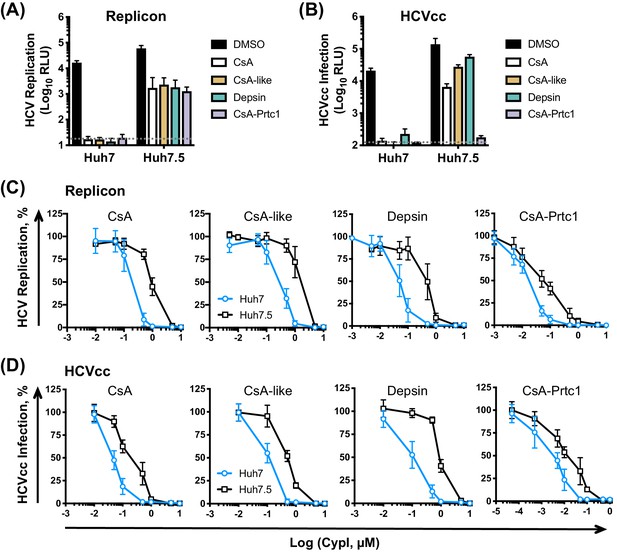
CypI are more potent against HCV replication and infection in Huh7 cells than in Huh7.5 cells.
(A) CypI more potently inhibit HCV replication in Huh7 cells than in Huh7.5 cells. Huh7 or Huh7.5 cells electroporated with HCV replicon RNA were treated with 1 μM CypI at four hpe and replication was measured by luciferase activity after 48 hr. (B) CypI more potently inhibit HCVcc infection in Huh7 cells than in Huh7.5 cells. Cells infected with HCVcc were treated with 1 μM CypI at four hpi and replication was measured by luciferase activity after 72 hr. (C-D) Dose-response analyses comparing antiviral activity of CypI in Huh7 and Huh7.5 cells. Cells were electroporated with HCV replicon RNA (C) or infected with HCVcc (D) and treated with increasing concentrations of CypI four later. Replication or infection was measured by luciferase activity after 48 hr (C) or 72 hr (D), and is expressed as a percentage relative to the DMSO vehicle-treated control. All graphs show means ± standard deviation from at least three independent experiments each performed in triplicate. Detection limits of the assays are shown by the dotted grey line.
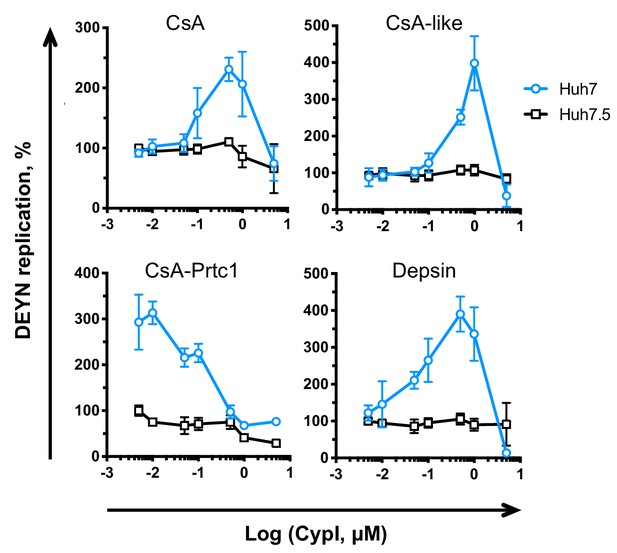
DEYN replication is enhanced by CypI treatment in Huh7 cells.
Approximately 2 × 106 Huh7 cells were electroporated with 5 μg HCV NS5A DEYN replicon RNA. CypI were added at four hpe. Replication was measured by luciferase activity at 48 hpe and is expressed as percentage relative to DMSO-treated control. Graphs show means ± standard deviation from two independent experiments each performed in triplicate.
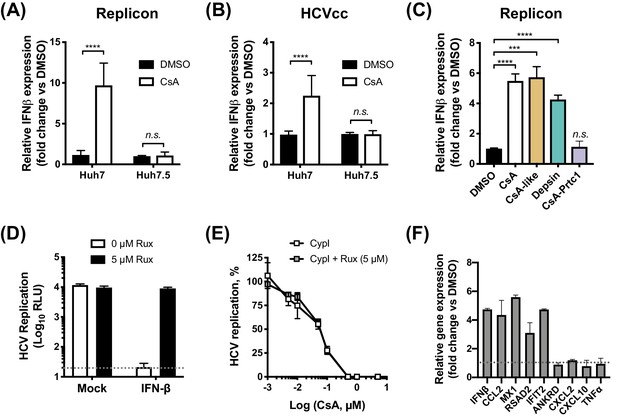
CypI induce expression of IFN-β and antiviral genes in Huh7, but not Huh7.5, cells.
(A–C) Cells electroporated with HCV replicon RNA (A, C) or infected with HCVcc (B) were treated with 5 μM CsA (A, B) or CypI (C) 4 hr later. After 48 hr, RNA was extracted and expression of IFN-β mRNA was evaluated by qRT-PCR. Data were normalised by GAPDH expression and are expressed as fold change compared to the DMSO vehicle-treated control. (D-E) CypI potency does not depend on IFN signalling. HCV replication in Huh7 cells, electroporated as described above, and treated with IFN-β (5 ng/mL) or CypI, in the presence or absence of the Jak/STAT inhibitor ruxolitinib (Rux). Rux treatment rescued HCV replication from IFN-β inhibition (D) but not from CypI (E). (F) CsA treatment induces expression of a subset of antiviral genes in HCV-replicating Huh7 cells. RNA expression of IFN-β, CCL2, MX1, RSAD2 IFIT2, ANKRD, CXCL2, CXCL10 and TNFα mRNA was evaluated by qRT-PCR at 48 hpe in Huh7 cells electroporated with HCV replicon RNA and treated with CsA (5 μM) at four hpe. Data were normalised by GAPDH expression and are expressed as fold change compared to the DMSO vehicle-treated control. All graphs show means ± standard deviation from at least three independent experiments each performed in triplicate. Statistical significance was evaluated by t-test using GraphPad Prism (**** p-value<0.0001; *** p-value<0.001; n.s. (not significant), p-value>0.05). Detection limits of the assay (D) or gene expression in DMSO-treated cells (set as 1) (F) are shown by the dotted grey line.
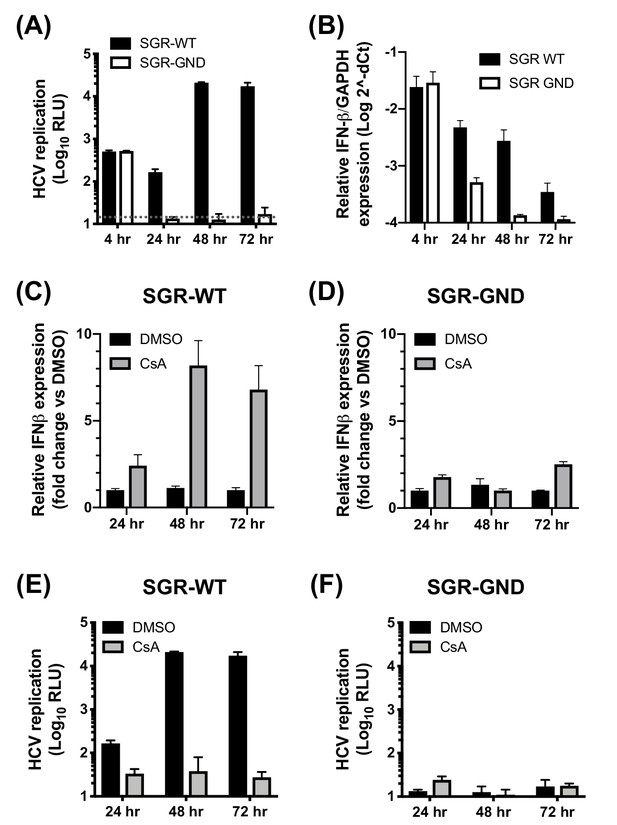
Induction of IFN-β expression by CsA depends on HCV replication.
Approximately 2 × 106 Huh7 cells were electroporated with 5 μg wild-type HCV replicon RNA (SGR-WT) or polymerase-defective HCV replicon RNA (SGR-GND). (A) Replication was measured by luciferase activity at 4, 24, 48 and 72 hpe and is expressed as RLU. (B) RNA was extracted at the indicated time points and expression of IFN-β mRNA was evaluated by qRT-PCR. Data were normalized by GAPDH expression and is expressed on a log scale as 2^-dCt. (C-F) Huh7 cells were electroporated with wild-type (C, E) or polymerase-defective (D, F) HCV replicon RNA as described above. At four hpe, 5 μM CsA or DMSO vehicle was added. HCV replication was measured by luciferase reporter activity at the indicated time points (E-F), while RNA was extracted from parallel samples and expression of IFN-β mRNA was evaluated by qRT-PCR (C-D). Data were normalised by GAPDH expression and is expressed as fold change compared to the DMSO vehicle-treated control. Graphs show means ± standard error from two independent experiments each done in quadruplicate.
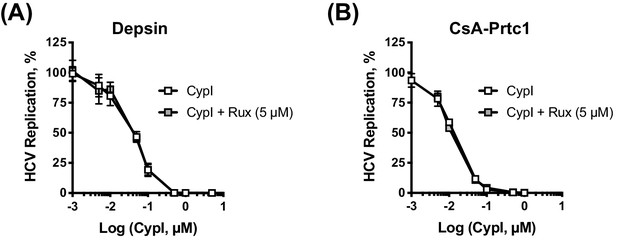
Inhibition of IFN-β signalling by ruxolitinib does not affect CypI potency.
Dose-response curves of CypI (A-B) in HCV-replicating cells treated with Rux (as in Figure 4D–E). Graphs show means ± standard deviation from at least three independent experiments each performed in triplicate.
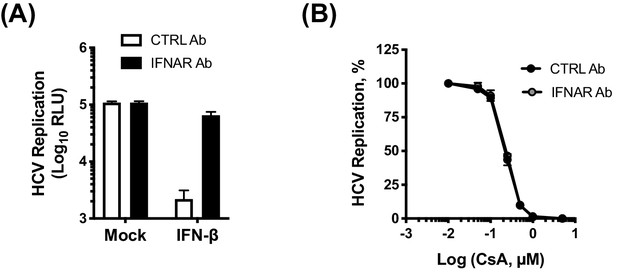
Inhibition of IFN-β signalling by IFNAR antibody does not affect CsA potency.
(A) Huh7 cells were treated with 0.025 ng/mL of IFN-beta, in the presence of CTRL antibody or IFNAR antibody (2 µg/mL). (B) CsA antiviral activity against HCV in cells treated with CTRL antibody or IFNAR antibody (2 µg/mL). HCV replication is expressed as a percentage relative to the DMSO control. Graphs show two independent experiments.
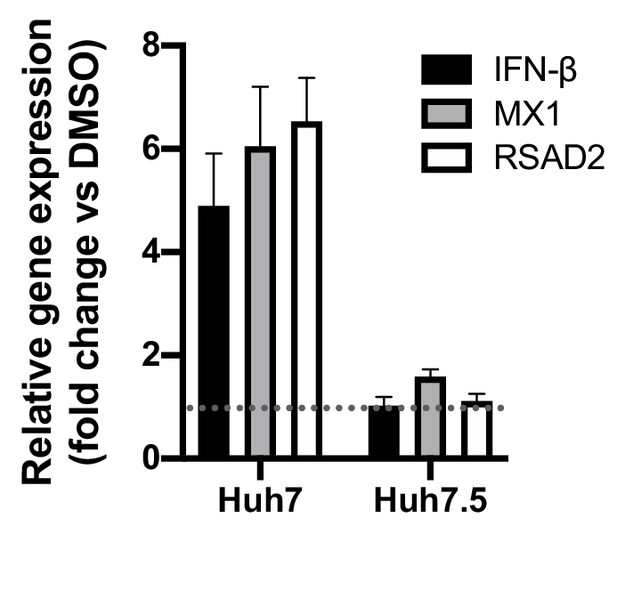
CsA induces expression of antiviral genes in Huh7, but not Huh7.5, cells.
Approximately 2 × 106 Huh7 or Huh7.5 cells were electroporated with 5 μg HCV replicon RNA. At four hpe, DMSO vehicle or CsA (5 μM) was added. RNA was extracted at 48 hpe and expression of IFN-β, MX1 or RSAD2 mRNA was evaluated by qRT-PCR. Data were normalised by GAPDH expression and is expressed as fold change compared to the DMSO vehicle-treated control. Graph shows mean ± standard error from two independent experiments each done in quadruplicate.
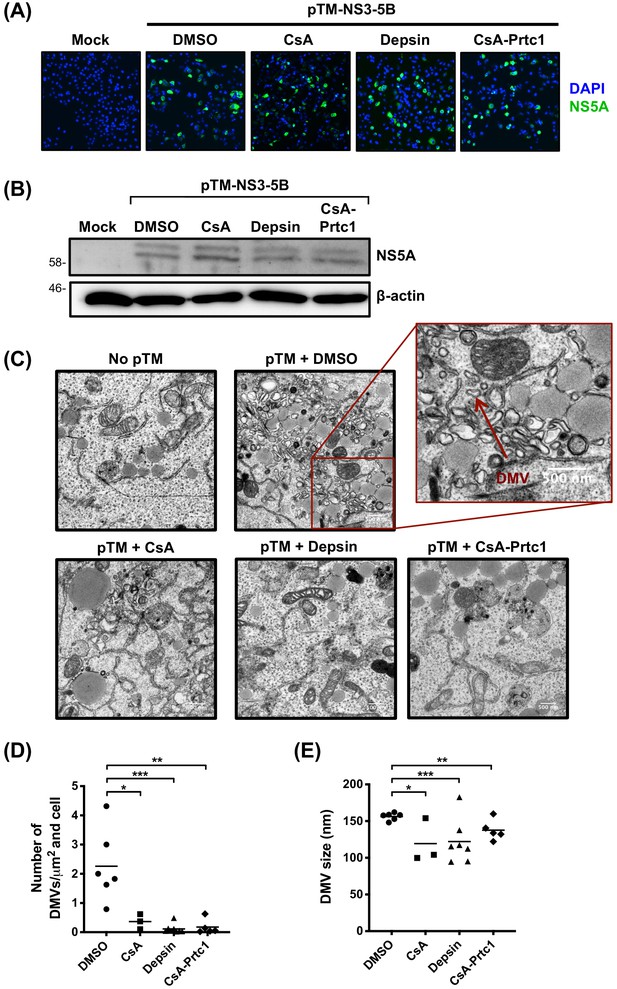
Antiviral CypI disrupt formation of the HCV replication organelle.
(A-E) Huh7-Lunet/T7 cells were transfected with pTM_NS3-5B and treated with CypI (5X EC90) at 4 hours post-transfection. Transfection efficiency and NS5A expression were evaluated 24 hours later by immunofluorescence (A) or Western blot for NS5A (B). (C) Representative electron micrographs showing the effect of CypI treatment on DMV formation. (D-E) The number and size of DMVs in 3-7 different cells per condition were quantitated using ImageJ. Statistical significance was evaluated by t-test using GraphPad Prism (* p-value < 0.05; ** p-value < 0.01; *** p-value < 0.001).
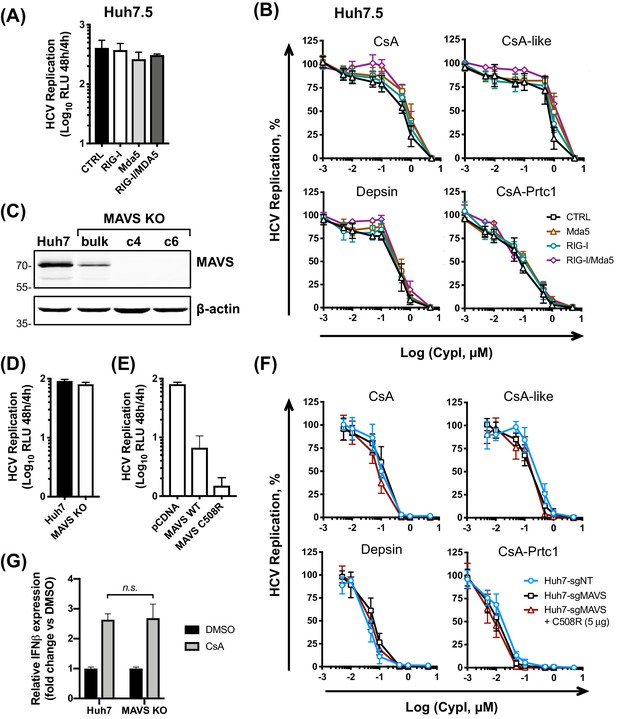
The RLR/MAVS pathway does not contribute to the antiviral potency of CypI.
Huh7.5 cells stably expressing RIG-I, Mda5 or both were electroporated with HCV replicon RNA and treated with increasing concentrations of CypI at four hpe. Replication was measured by luciferase activity at 48 hpe and is expressed as RLU (A) or percentage relative to the DMSO vehicle-treated control (B). Expression of RIG, Mda5 or both did not significantly affect HCV replication at 48 hr (A) or CypI dose-response curves in Huh7.5 cells (B). (C) Western blot detecting MAVS in single cell cloned Huh7 cells following MAVS knockout by CRISPR/Cas9. Huh7-sgMAVS cells were electroporated with 5 μg HCV replicon RNA in the presence or absence of plasmid encoding wild type MAVS (MAVS-WT) or mutant MAVS-C508R (conferring NS3/4A protease resistance). CypI were added at four hpe. Replication was measured by luciferase activity at 48 hpe and is expressed as RLU (D) or percentage relative to DMSO-treated control (E). HCV replication was not affected by knockout of MAVS (D) but was decreased by transfection of plasmid encoding MAVS-C508R (E). The presence or absence of MAVS did not affect the CypI dose response curves (F). (F) Huh7 or Huh7 MAVS KO cells were electroporated with HCV replicon RNA as described above, and treated with 5 μM CsA at four hpe. At 48 hpe, RNA was extracted and expression of IFN-β mRNA was evaluated by qRT-PCR. Data were normalised by GAPDH expression and is expressed as fold change compared to the DMSO vehicle-treated control. (A-F) All graphs show means ± standard deviation from at least three independent experiments each performed in triplicate. Statistical significance was evaluated by t-test using GraphPad Prism (n.s., not significant; p-value>0.05).
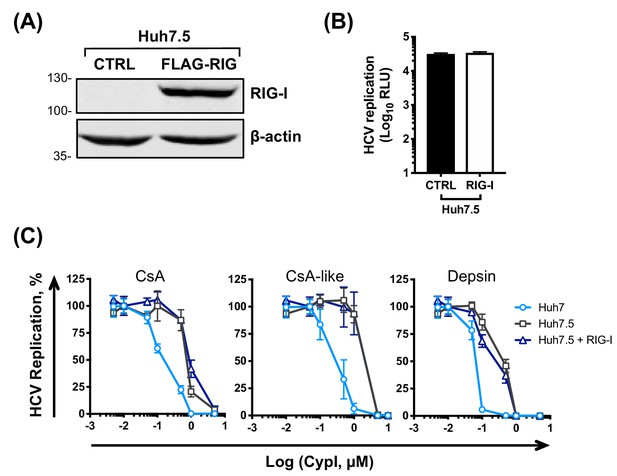
Expression of RIG-I in Huh7.5 cells does not affect CypI potency.
Approximately 2 × 106 Huh7.5 were electroporated with 5 μg HCV replicon RNA in the presence or absence of 5 μg FLAG-RIG-I plasmid. (A) Western blot showing RIG-I expression in the transfected cells. (B) CypI were added at four hpe. Replication was measured by luciferase activity at 48 hpe and is expressed as RLU (B) or percentage relative to DMSO-treated control (C). Graphs show means ± standard deviation from two independent experiments each performed in triplicate.
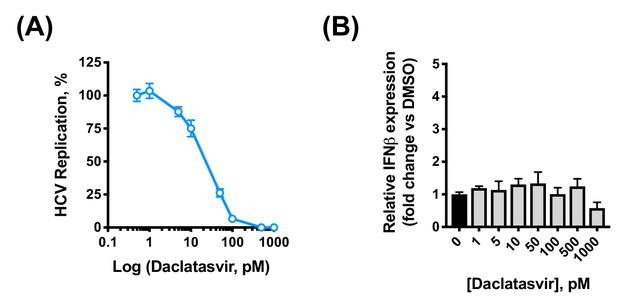
Daclatasvir treatment does not induce IFN expression in HCV-replicating Huh7 cells.
Approximately 2 × 106 Huh7.5 were electroporated with 5 μg HCV replicon RNA. At four hpe, DMSO vehicle or increasing concentrations of daclatasvir were added. (A) Replication was measured by luciferase activity at 48 hpe and is expressed as RLU. (B) RNA was extracted at 48 hpe and expression of IFN-β mRNA was evaluated by qRT-PCR. Data were normalised by GAPDH expression and is expressed as fold change compared to the DMSO vehicle-treated control. Graphs show means ± standard error from two independent experiments each done in triplicate (A) or quadruplicate (B).
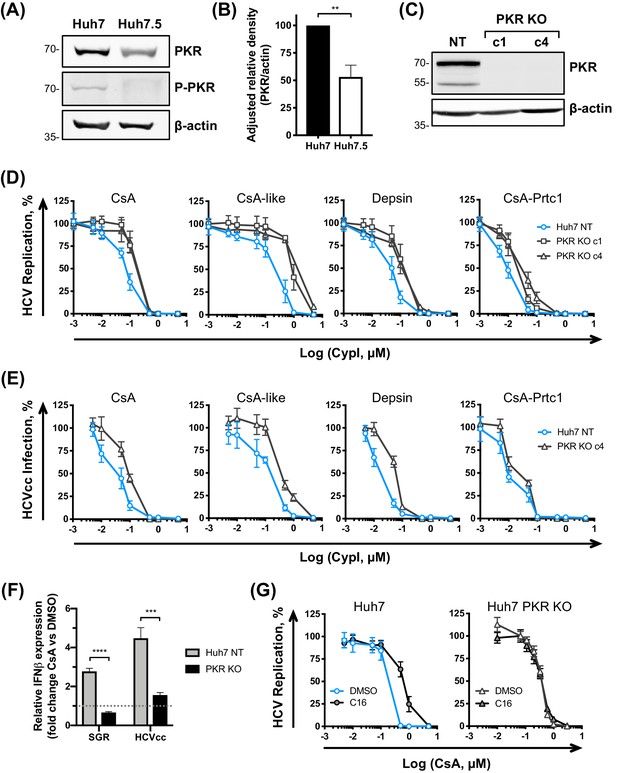
PKR modulates the antiviral potency of CypI against HCV.
(A) PKR expression and phosphorylation is reduced in Huh7.5 cells. Huh7 or Huh7.5 cells electroporated with in vitro transcribed HCV replicon RNA were lysed at 48 hpe, and PKR expression and phosphorylation assessed by western blot. One representative blot out of three independent experiments is shown. (B) Quantitation of band density from three independent experiments showing adjusted relative density normalised to the actin loading control. (C) Western blot detecting PKR expression in single cell cloned Huh7 cells following PKR knockout by CRISPR/Cas9. (D) CypI potency against HCV replication is decreased in the absence of PKR. Non targeted Huh7 (Huh7 (NT)) or Huh7 PKR KO clones 1 (c1) or 4 (c4) were electroporated with in vitro transcribed HCV replicon RNA and CypI added at four hpe. Replication was measured by luciferase activity at 48 hpe and is expressed as percentage relative to DMSO-treated control. (E) Huh7 NT or PKR KO cells clone 4 (c4) were infected with HCVcc and treated with increasing concentrations of CypI at four hpi. Replication was measured by luciferase activity after 72 hr and is expressed as percentage relative to DMSO-treated control. (F) Huh7 NT or PKR KO cells were electroporated with HCV replicon RNA or infected with HCVcc, and treated with 5 μM CsA at four hpe. At 48 hpe, RNA was extracted and expression of IFN-β mRNA was evaluated by qRT-PCR. Data were normalised by GAPDH expression and is expressed as fold change compared to the DMSO vehicle-treated control. (G) Huh7 or Huh7 PKR KO cells were electroporated as described above, and at four hpe were treated with increasing concentrations of CsA in the presence or absence of the PKR inhibitor C16 (1 μM). C16 decreased CypI potency in Huh7 cells, but not in Huh7 PKR KO cells. (A-F) All graphs show means ± standard deviation from at least three independent experiments each performed in triplicate. Statistical significance was evaluated by t-test using GraphPad Prism (**** p-value<0.0001; *** p-value<0.001; ** p-value<0.005). Gene expression in DMSO-treated cells (set as 1) (F) shown by the dotted grey line.
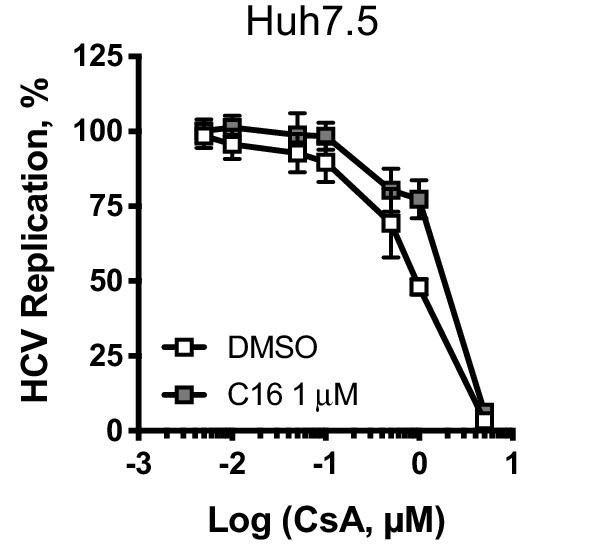
PKR inhibitor C16 only minimally affects CsA potency in Huh7.5 cells.
Huh7.5 cells were electroporated as described above, and at four hpe were treated with increasing concentrations of CsA in the presence or absence of the PKR inhibitor C16 (1 μM). C16 only minimally decreased CypI potency in Huh7.5 cells. Mean ± standard deviation from two independent experiments each performed in triplicate.
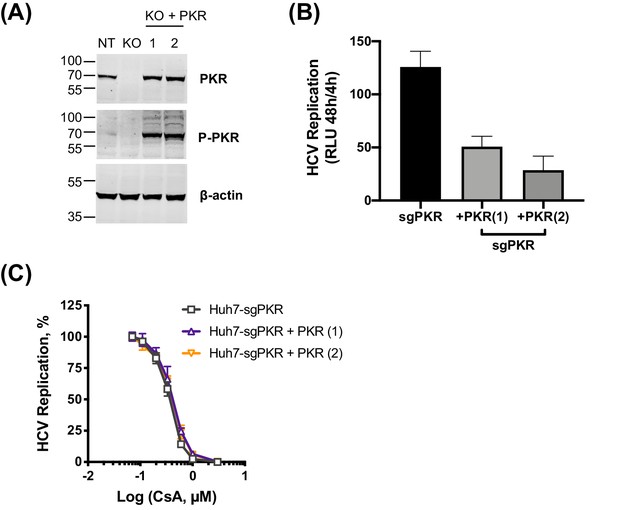
PKR overexpression does not affect HCV sensitivity to CsA.
(A) Western blot showing expression of PKR in Huh7 NT, Huh7 PKR KO (c4) or Huh7 PKR KO cells stably overexpressing PKR. (B) HCV replication in Huh7 PKR KO cells is inhibited by ectopic expression of PKR. (C) Expression of ectopic PKR does not affect HCV sensitivity to CsA.
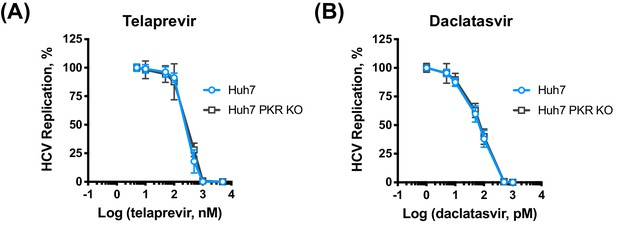
PKR does not affect HCV sensitivity to telaprevir or daclatasvir.
Huh7 NT or Huh7 PKR KO c4 were electroporated with in vitro transcribed HCV replicon RNA, and the indicated concentrations of telaprevir (A) or daclatasvir (B) were added at four hpe. Replication was measured by luciferase activity at 48 hpe and is expressed as percentage relative to DMSO-treated control. All graphs show means ± standard deviation from three independent experiments each performed in triplicate.
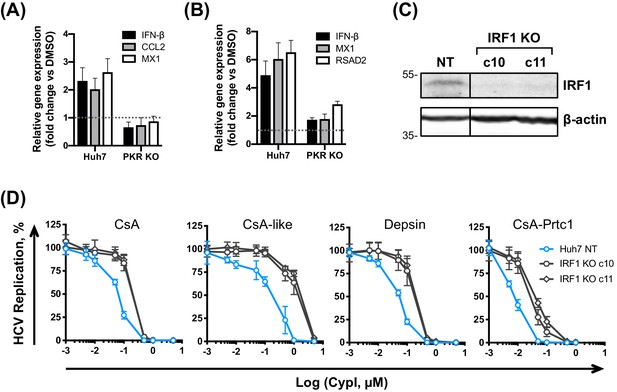
PKR induces IRF1-dependent intrinsic antiviral responses in HCV-replicating CypI-treated Huh7 cells.
(A–B) Induction of IRF1 target gene expression in HCV-replicating (A) or HCV-infected (B) cells depends on PKR. Expression of IFN-β, CCL2, MX1 or RSAD2 mRNA was evaluated by qRT-PCR at 48 hpe in Huh7 NT, or PKR KO cells, electroporated with HCV replicon RNA or infected with HCVcc and treated with 5 μM CsA at four hpe. Data were normalised by GAPDH expression and are expressed as fold change compared to the DMSO vehicle-treated control. (C) Western blot detecting IRF1 in Huh7 cells following IRF1 knockout by CRISPR/Cas9 and single cell cloning. (D) CypI potency against HCV replication was decreased in the absence of IRF1. HCV replication in Huh7 NT or IRF1 KO cells, measured by luciferase activity at 48 hpe, after CypI addition at four hpe, expressed as percentage relative to DMSO-treated control. (A-D) All graphs show means ± standard deviation from two or three independent experiments each performed in triplicate. Gene expression in DMSO-treated cells (set as 1) (A–B) is shown by the dotted grey line.
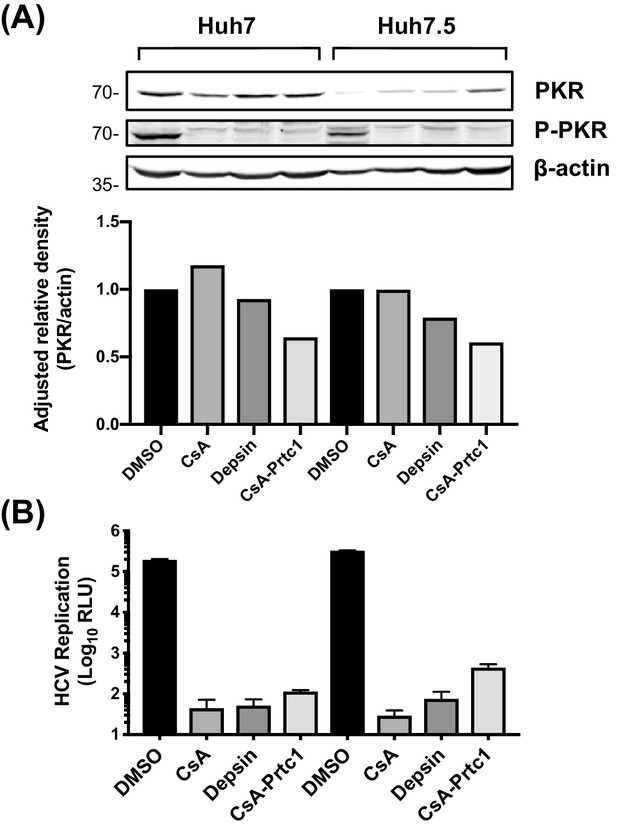
CypI treatment inhibits PKR autophosphorylation at T446.
Huh7 or Huh7.5 cells, electroporated with HCV replicon RNA and treated with fully inhibitory concentrations of CypI (5 μM) 4 hr later, were extracted and PKR expression and phosphorylation were evaluated by western blot (A) and HCV replication (B) at 48 hr. (A) Quantitation of PKR band density showing adjusted relative density normalised to the actin loading control.
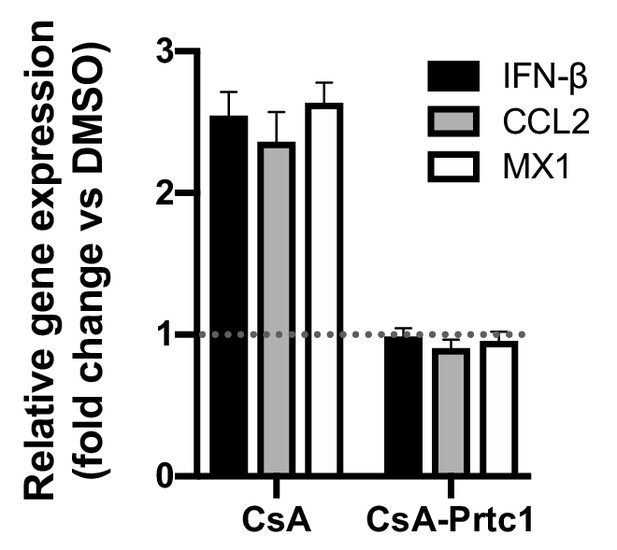
CsA, but not CsA-Prtc1, induces expression of antiviral genes.
Approximately 2 × 106 Huh7 cells were electroporated with 5 μg HCV replicon RNA. At four hpe, DMSO vehicle or CypI (5 μM) were added. RNA was extracted at 48 hpe and expression of IFN-β, CCL2 or MX1 mRNA was evaluated by qRT-PCR. Data were normalised by GAPDH expression and is expressed as fold change compared to the DMSO vehicle-treated control. Graph shows mean ± standard error from three independent experiments each done in quadruplicate.
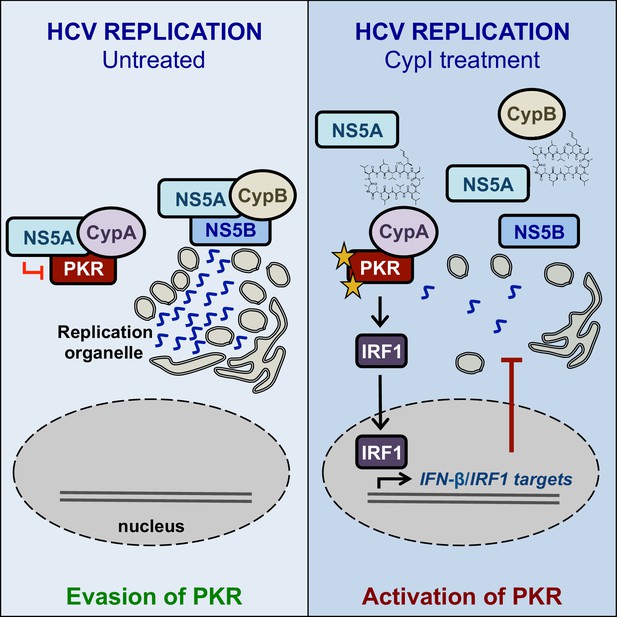
Working model for the proposed roles of Cyps in HCV replication, and the proposed antiviral mechanisms of CypI against HCV.
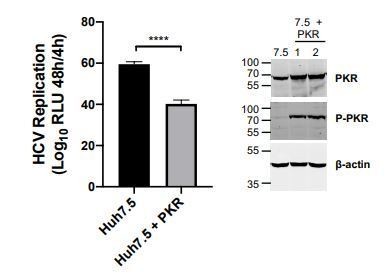
Huh7 Cells were transduced with lentiviral vector encoding PKR (pSCRPSY-EIF2AK2); transduced cells were selected by addition of puromycin.
Expression and activation of PKR was evaluated by Western blot. HCV replication was evaluated as described in the manuscript. HCV replication was slightly decreased in PKR overexpressing Huh7.5 cells. Statistical significance was evaluated by t-test using GraphPad Prism (**** p-value < 0.0001).
Tables
Comparison of CypI IC50 against HCV replication or infection in different cell lines.
| Replication (SGR) | IC50 (μM) | |||
|---|---|---|---|---|
| Cell line | CsA | CsA-like | Depsin | CsA-Prtc1 |
| Huh7 | 0.188 ± 0.032 | 0.266 ± 0.037 | 0.043 ± 0.005 | 0.016 ± 0.002 |
| Huh7.5 | 0.955 ± 0.123 | 1.374 ± 0.197 | 0.336 ± 0.053 | 0.055 ± 0.007 |
| Huh7.5-CTRL | 0.373 ± 0.085 | 0.527 ± 0.154 | 0.206 ± 0.043 | 0.070 ± 0.013 |
| Huh7.5-RIG-I | 0.560 ± 0.105 | 0.686 ± 0.181 | 0.243 ± 0.054 | 0.093 ± 0.019 |
| Huh7.5-Mda5 | 0.754 ± 0.160 | 0.950 ± 0.247 | 0.302 ± 0.060 | 0.100 ± 0.018 |
| Huh7.5-RIG-I/Mda5 | 0.820 ± 0.206 | 1.234 ± 0.298 | 0.351 ± 0.085 | 0.101 ± 0.021 |
| Huh7 NT c7 | 0.068 ± 0.008 | 0.184 ± 0.026 | 0.037 ± 0.004 | 0.009 ± 0.001 |
| Huh7 MAVS KO | 0.112 ± 0.019 | 0.160 ± 0.023 | 0.056 ± 0.008 | 0.010 ± 0.001 |
| Huh7 MAVS KO + C508R | 0.075 ± 0.014 | 0.141 ± 0.028 | 0.045 ± 0.006 | 0.008 ± 0.001 |
| Huh7 PKR KO c1 | 0.176 ± 0.034 | 0.971 ± 0.186 | 0.125 ± 0.021 | 0.021 ± 0.003 |
| Huh7 PKR KO c4 | 0.172 ± 0.040 | 1.249 ± 0.327 | 0.091 ± 0.015 | 0.028 ± 0.004 |
| Huh7 IRF1 KO c10 | 0.183 ± 0.042 | 1.219 ± 0.224 | 0.170 ± 0.039 | 0.027 ± 0.005 |
| Huh7 IRF1 KO c11 | 0.193 ± 0.047 | 1.593 ± 0.284 | 0.192 ± 0.045 | 0.040 ± 0.006 |
| Huh7 + DMSO | 0.216 ± 0.050 | |||
| Huh7 + C16 | 0.553 ± 0.098 | |||
| Huh7.5 + DMSO | 0.879 ± 0.131 | |||
| Huh7.5 + C16 | 1.750 ± 0.495 | |||
| HCVcc infection | IC50 (μM) | |||
| Cell line | CsA | CsA-like | Depsin | CsA-Prtc1 |
| Huh7 | 0.043 ± 0.012 | 0.091 ± 0.020 | 0.096 ± 0.016 | 0.003 ± 0.001 |
| Huh7.5 | 0.182 ± 0.037 | 0.316 ± 0.083 | 1.084 ± 0.373 | 0.014 ± 0.003 |
| Huh7 NT c7 | 0.033 ± 0.005 | 0.123 ± 0.023 | 0.018 ± 0.003 | 0.012 ± 0.002 |
| Huh7 PKR KO c4 | 0.095 ± 0.019 | 0.433 ± 0.101 | 0.051 ± 0.011 | 0.023 ± 0.005 |
Additional files
-
Supplementary file 1
Synthesis of novel CypI.
- https://cdn.elifesciences.org/articles/52237/elife-52237-supp1-v1.docx
-
Supplementary file 2
Oligo sequences.
- https://cdn.elifesciences.org/articles/52237/elife-52237-supp2-v1.doc
-
Supplementary file 3
Key resources table.
- https://cdn.elifesciences.org/articles/52237/elife-52237-supp3-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/52237/elife-52237-transrepform-v1.docx