Targeted induction of a silent fungal gene cluster encoding the bacteria-specific germination inhibitor fumigermin
Figures
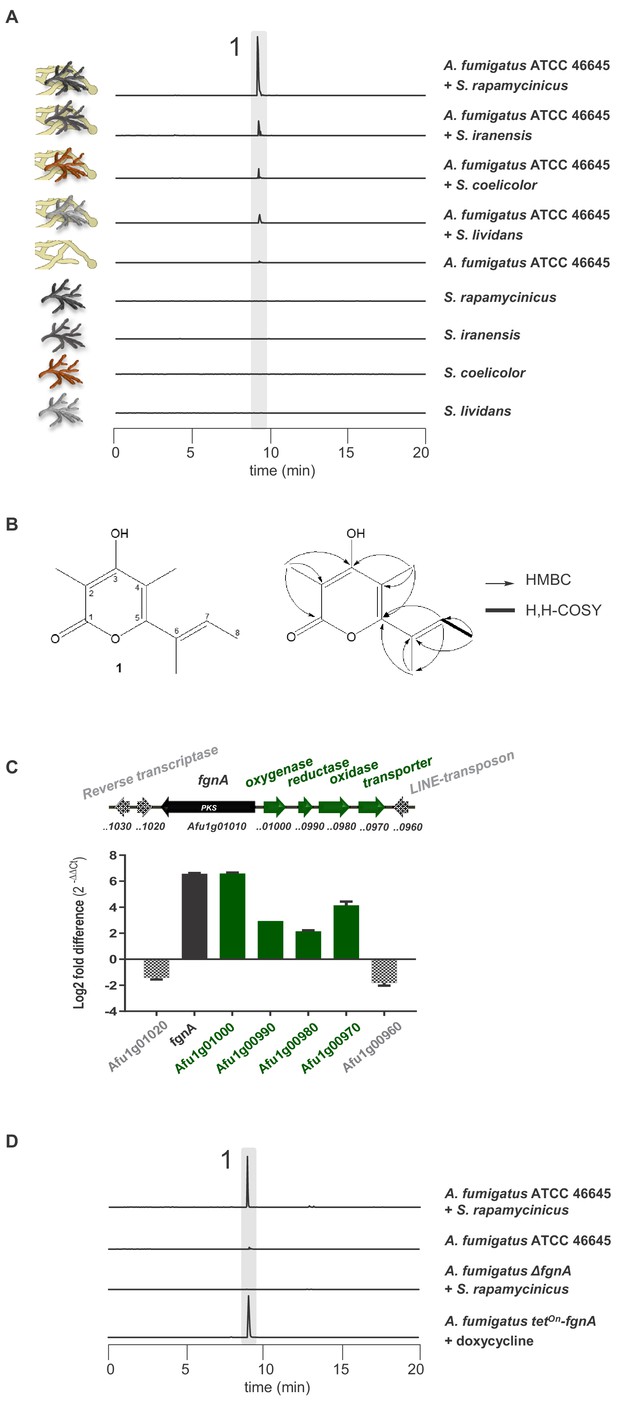
Induction of SM production in A. fumigatus by Streptomyces spp. and identification of the gene responsible for the SM production.
(A) LC-MS analysis of supernatants of A. fumigatus strains after 12 h in axenic or in co-culture with indicated Streptomyces species showing EIC traces for m/z 195 [M+H]+, corresponding to the newly formed compound 1, which is indicated by the highlighted strip. (B) Structure and 2D-NMR correlations of compound 1, fumigermin. (C) Transcription analysis of the A. fumigatus ATCC 46645 fgn cluster and adjacent genes determined by qRT-PCR after 5 h of co-cultivation with S. rapamycinicus. Relative mRNA levels were compared to the β-actin gene transcript levels. The relative expression values are visualized as relative log2 fold difference (2-ΔΔCt) of transcript levels for each gene in co-culture with S. rapamycinicus compared to transcript levels of the respective gene in A. fumigatus ATCC 46645 in axenic culture. Data are representative of three biological replicates with three technical replicates. Error bars indicate standard error of the mean. (D) EIC traces of supernatants of the different A. fumigatus strains in co-cultivation with S. rapamycinicus or under inducing conditions are shown for m/z 195 [M+H]+, which corresponds to fumigermin (1).
-
Figure 1—source data 1
Transcriptional analysis of the fgn cluster.
- https://cdn.elifesciences.org/articles/52541/elife-52541-fig1-data1-v1.xlsx
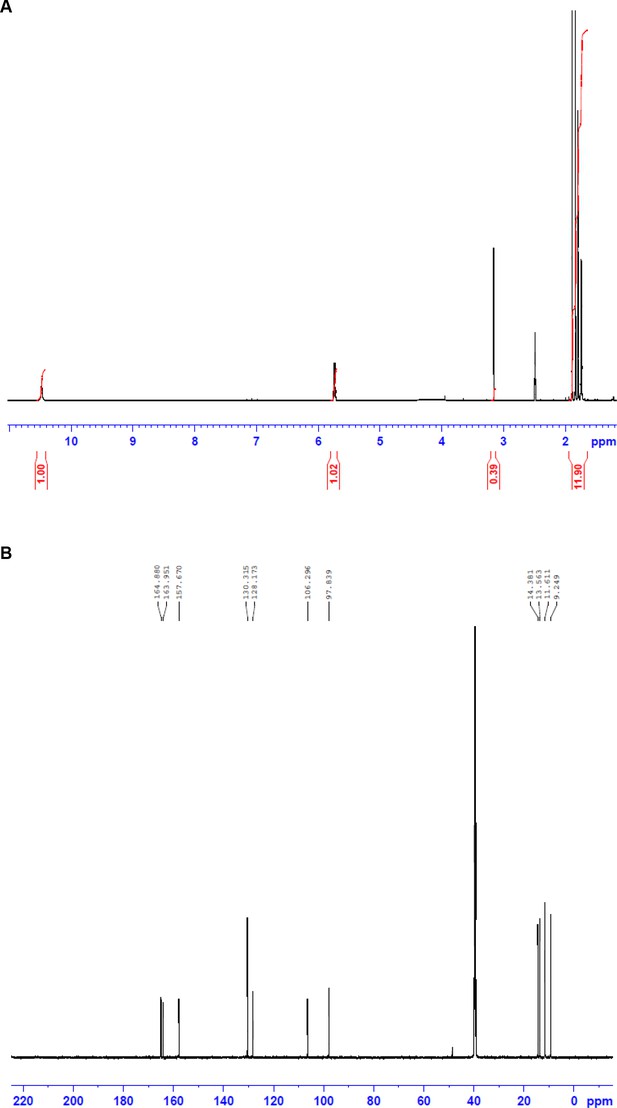
NMR spectra of fumigermin.
(A) 1H NMR spectrum of 1. (B) 13C NMR spectrum of 1.
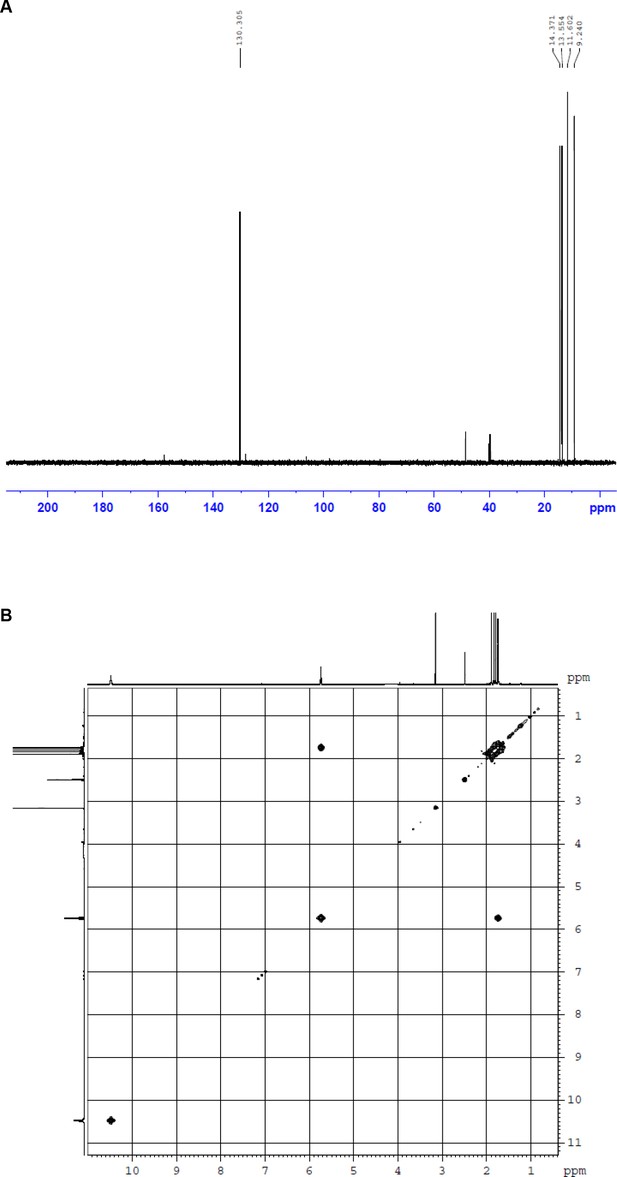
NMR spectra of fumigermin.
(A) DEPT135 NMR spectrum of 1. (B) H,H-COSY NMR spectrum of 1.
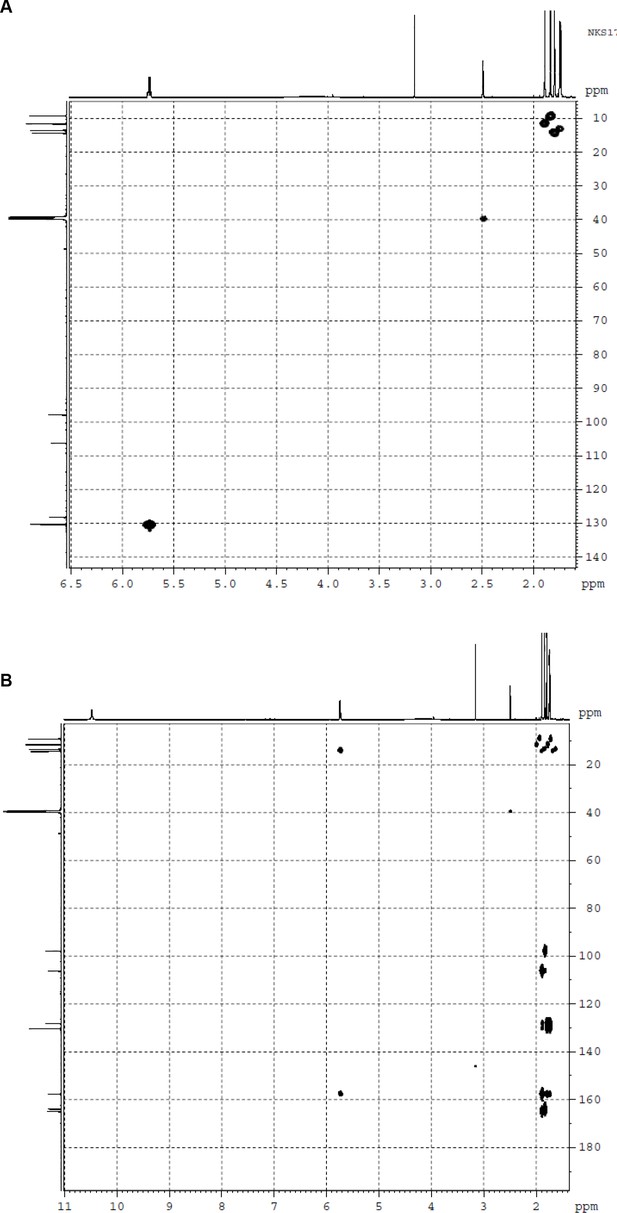
NMR spectra of fumigermin.
(A) HSQC NMR spectrum of 1. (B) HMBC NMR spectrum of 1.
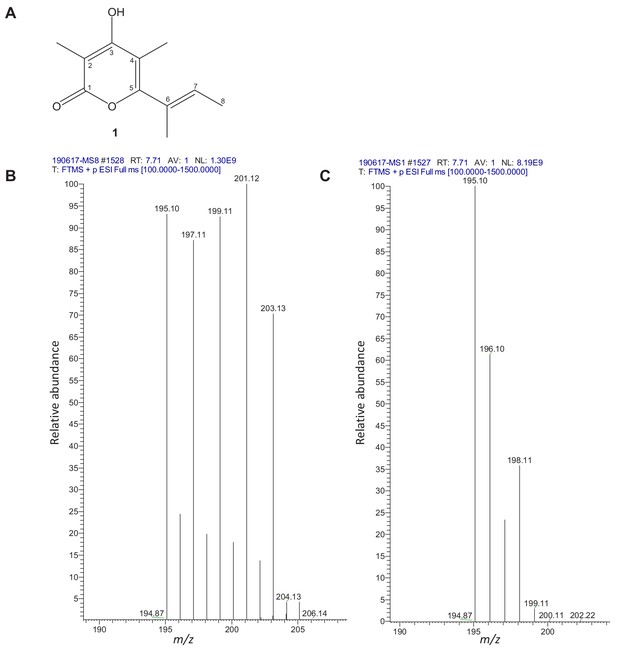
Structure of fumigermin (1).
(A) NMR data of 1. Chemical shifts of 1:1H-NMR (600 MHz, d6-DMSO): 10.50 (brs) 3-OH; 5.75 (m) H-7; 1.91 (s) 4-CH3; 1.85 (s) 2-CH3; 1.81 (t, 1.3) 6-CH3; 1.76 (dd, 6.9; 1.1) H-8. 13C-NMR (150 MHz, d6-DMSO): 164.9 C-3; 163.9 C-1; 157.7 C-5; 130.3 C-7; 128.2 C-6; 106.3 C-4; 97.8 C-2; 14.4 6-CH3; 13.6 C-8; 11.6 4-CH3; 9.2 2-CH3. HRESI-MS 195.1015 [M+H]+ (calcd. C11H15O3 195.1016). (B,C) Feeding experiments with labeled precursors to elucidate the biosynthesis of fumigermin. (B) HRESI-MS positive mode TIC spectrum of 13C-labeled fumigermin, after feeding of (2-13C)acetate, showing an increase in the observed fumigermin mass from the naturally occurring m/z 195 [M+H]+ to a mass of m/z 203 [M+H]+, indicating incorporation of eight 13C atoms. (C) HRESI-MS positive mode TIC spectrum of 13C-labeled fumigermin, after feeding of (methyl-13C)methionine, showing an increase in the observed fumigermin mass from m/z 195 [M+H]+ to m/z 198 [M+H]+, indicating incorporation of three 13C atoms.
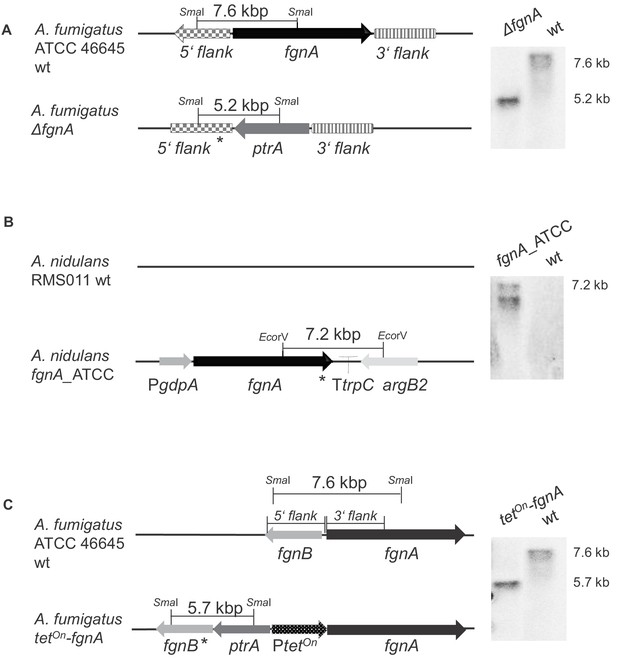
Verification of fungal transformant strains by Southern blot.
(A) Genomic situation in the A. fumigatus wt and the ΔfgnA mutant strain. DNA of wt and mutant strain were digested with SmaI overnight. The probe (*) directed against the 5’ flank was PCR-amplified using primers P45/P46. Genomic situation in the wt and mutant strain. Expected wt band at 7.6 kb, expected band at 5.2 kb characteristic of the deletion strain. (B) Genomic situation in the A. nidulans wt (does not encode the gene) and in the A. nidulans fgnA_ATCC transformant strain with an ectopic integration of the fgnA construct. Genomic DNA of A. nidulans wt and transformants was digested overnight with EcoRV. The probe (*) was directed against the 3’ end region of fgnA and amplified using primers P7/P10. Expected band in transformants at 7.2 kb. (C) Genomic situation in the wt and in the A. fumigatus tetOn-fgnA mutant strain. Genomic DNA of wt and transformants was digested overnight with SmaI. The probe (*) was directed against the 3’ flank and PCR-amplified using primers P45/P46. Expected wt band at 7.6 kb, expected band characteristic of mutant at 5.7 kb.
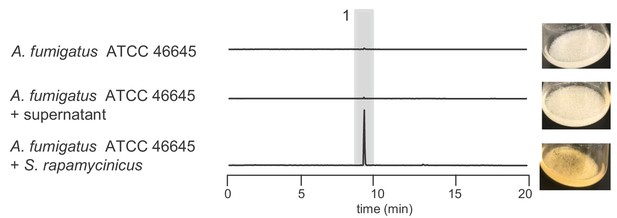
Verification of inducibility of fumigermin biosynthesis by S. rapamycinicus supernatant in A. fumigatus using LC-MS metabolic analysis.
LC-MS analysis showing EIC traces of supernatants of A. fumigatus ATCC 46645 after 12 h growth in axenic culture, with added S. rapamycinicus supernatant or in co-culture with S. rapamycinicus. Traces are shown for m/z 195 [M+H]+, which corresponds to fumigermin (1).
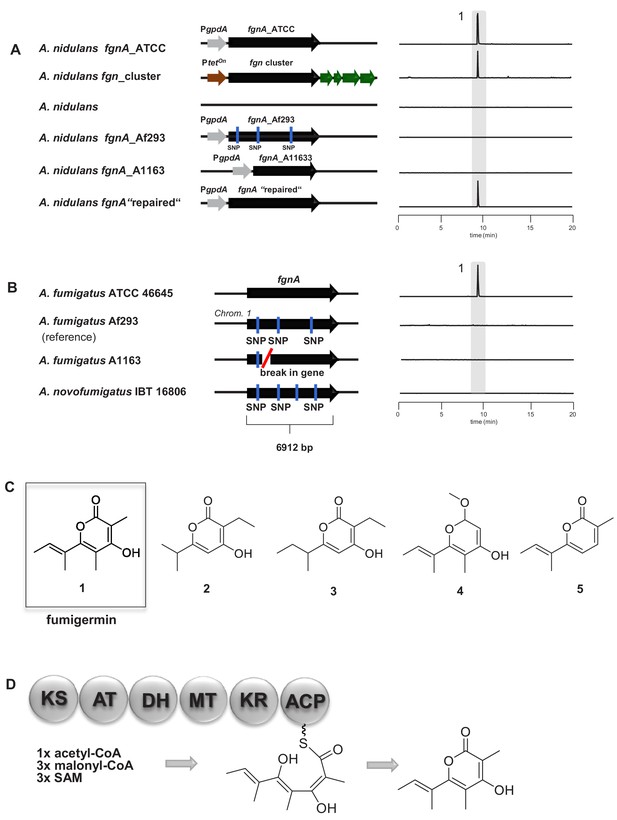
Molecular analysis of the fgnA PKS gene and structure of its biosynthesized product fumigermin.
(A) Heterologous expression of fgnA genes from the indicated A. fumigatus strains in the host A. nidulans. The entire fgn cluster was transferred and expressed in the host A. nidulans. A ‘repaired’ fgnA based on the gene of A1163 was expressed in A. nidulans. (B) PKS gene fgnA in different indicated A. fumigatus strains. SNPs and a gene break are marked. EIC traces of supernatants of the different A. fumigatus strains in co-cultivation with S. rapamycinicus are shown in (A) and (B) for m/z 195 [M+H]+, which corresponds to fumigermin (1). (C) Comparison of the discovered α-pyrone polyketide compound fumigermin (1) and the structurally related bacterial pyrones germicidin A (2), germicidin B (3), nectriapyrone (4) and gibepyrone (5). (D) FgnA enzyme domains and proposed biosynthesis of fumigermin.
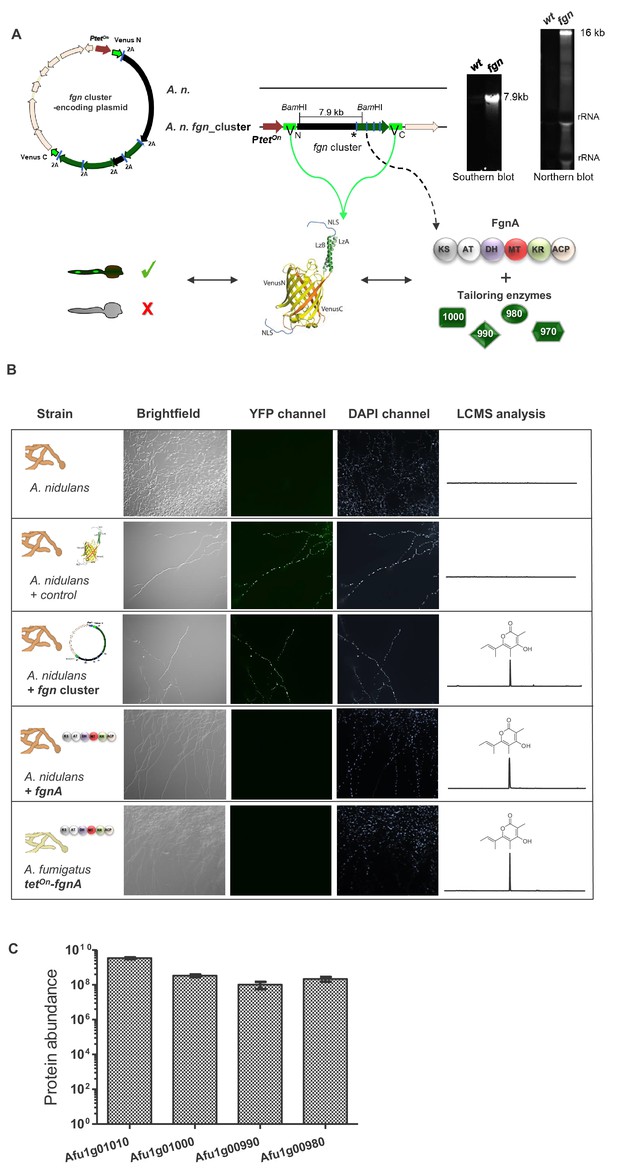
Expression of the A. fumigatus fgn cluster in the heterologous host A. nidulans.
(A) Strategy for the expression of all cluster genes and fluorescence-based screening of transformants. Verification of A. nidulans transformants by Southern and Northern blots. The previously described heterologous expression tool based on post-translational cleavage of proteins containing viral 2A peptides (Unkles et al., 2014; Hoefgen et al., 2018) was used to express all fgn cluster genes from a single polycistronic mRNA. To facilitate selection of transformants harboring the entire plasmids, we employed a gene encoding a split-Venus fluorescent protein that, in addition, contained a nuclear localization sequence. In the plasmid, the gene cluster was thus flanked by two genes, each encoding a half of the Venus gene. Consequently, when all genes of the plasmid are expressed, the Venus halves (N+C) are also produced and bind to each other to form a functional Venus fluorescent protein. As a result, fluorescence in the fungal nuclei indicates translation of all encoded proteins. The cluster construct was placed under the control of the tetOn promoter, allowing inducible expression of the genes from a polycistronic mRNA. Transformants showing nuclear fluorescence were further confirmed by Southern blot: genomic situation in the wt and in the A. nidulans transformant strain. Genomic DNA of wt and transformants was digested overnight with BamHI. The probe (*) was directed against the fgnA gene (primers P47/P48) and used for Southern and Northern blots. For Southern blot the expected band indicative of a transformant with the fgn cluster is at 7.9 kb, and for Northern blot at 16 kb for the entire fgn cluster polycistronic mRNA. In silico modeling of the split-Venus protein was carried out with PyMOL software (DeLano Scientific, LLC). (B) Fluorescence imaging of A. nidulans wt and trasformant strains and comparative metabolic analysis of fumigermin production in different Aspergillus strains. Mycelia were grown on AMM agar with supplements when required. The YFP channel has a light emission at λ = 527 nm. Samples were excited at λ = 514 nm. Fluorescence is shown in false color (green). Fungal nuclei were stained with Hoechst 34580. EIC traces of supernatants of the different strains are shown for m/z 195 [M+H]+, which corresponds to fumigermin (1). (C) Detection of produced A. fumigatus proteins in the heterologous host A. nidulans. The bar plot shows the abundance levels of the fgn cluster proteins based on the precursor ion (three biological replicates, three analytical replicates, shown with standard error of the mean) in strain A. nidulans fgn_cluster, which harbors a plasmid containing the entire fgn cluster genes, each separated from the others by 2A sequences and flanked by genes each encoding a half of the split-Venus protein.
-
Figure 2—figure supplement 1—source data 1
Global proteome of A. nidulans fgn_cluster.
- https://cdn.elifesciences.org/articles/52541/elife-52541-fig2-figsupp1-data1-v1.xlsx
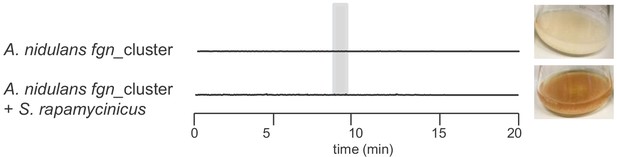
Verification of inducibility of fumigermin biosynthesis by S. rapamycinicus in a heterologous host context using LC-MS metabolic analysis.
LC-MS analysis showing EIC chromatograms of supernatants of strain A. nidulans fgn_cluster after growth for 12 h in axenic culture or in co-culture with S. rapamycinicus, without added doxycycline. Traces are shown for m/z 195 [M+H]+, which corresponds to fumigermin.
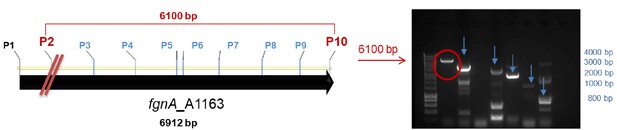
PCR strategy to locate the breakage of gene fgnA from A. fumigatus A1163.
Left, schematic of fgnA gene. Right, agarose gel of PCR products. P1/P10 did not lead to the amplification of a PCR product, suggesting a gene breakage. Primers annealing approximately every 800 bp of fgnA were designed. Increasing sizes of PCR products were obtained with the sequential primer pairs P10/P9, P10/P8, P10/P7 and so on, to P10/P3 (blue arrows). Primer pair P10/P2 gave the longest DNA fragment of 6100 bp (red circle). The red line marks the deduced approximate area of the fgnA gene breakage.
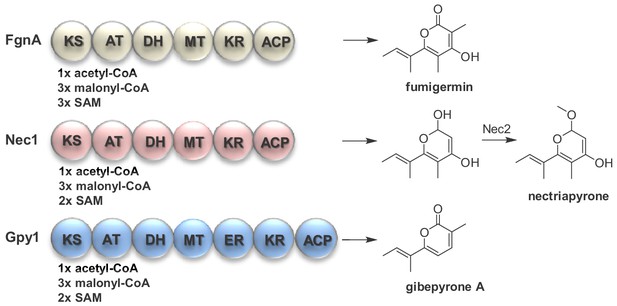
Comparison of the biosyntheses of fungal α-pyrones.
Fumigermin is biosynthesized by the type I PKS FgnA of A. fumigatus. Nectriapyrone is biosynthesized by the type I PKS Nec1 of P. oryzae. Gibepyrone is biosynthesized by the type I PKS Gpy1 of F. fujikuroi.
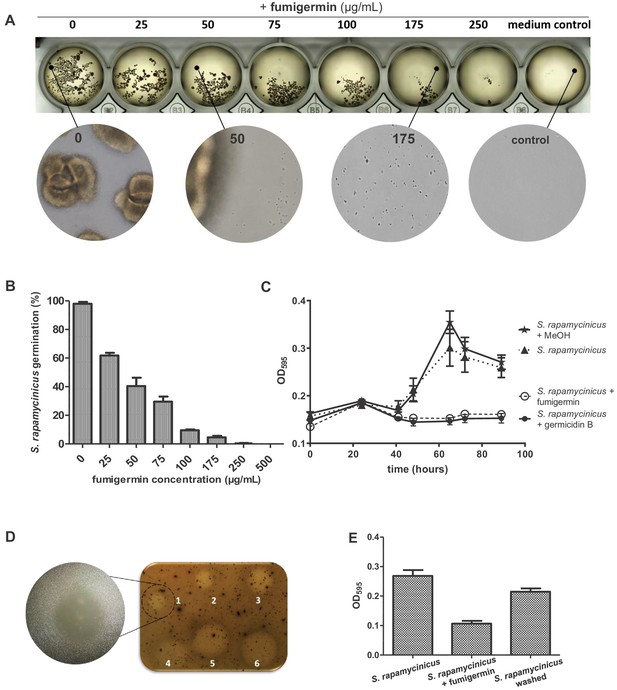
Activity of fumigermin against germination of S. rapamycinicus spores.
(A) 107 spores of S. rapamycinicus were inoculated in 500 µL M79 medium in a 48-well plate and treated with increasing fumigermin concentrations. Image taken after 5 days of growth at 28°C, 500 rpm. Magnified areas zoom in on an untreated sample, sample treated with 50 µg/mL or 175 µg/mL fumigermin and a control sample containing medium with no spores. (B) OD595 measurements were taken for each well using a Tecan device. The mean OD595 value of the untreated S. rapamycinicus samples was set as 100% germination, and used to calculate the relative germination percentage of the fumigermin-treated samples. (C) Graph of 90-h-long monitoring of S. rapamycinicus spore germination inhibition at 28°C using germicidin B and fumigermin (250 μg/mL). Controls were treated with MeOH or remained untreated. An increase in the OD595 value is indicative of S. rapamycinicus spore germination. Data in (B, C) correspond to three biological samples, each with three technical replicates. Error bars indicate standard error of the mean. (D) Qualitative inhibition of formation of S. rapamycinicus aerial mycelia by fumigermin. Oatmeal agar plates with embedded S. rapamycinicus spores were point-inoculated with different concentrations of fumigermin: (1) 12.5 μg, (2) 25 μg, (3) 50 μg, (4) 75 μg, (5) 100 μg, (6) 250 μg and incubated for 1 week at 28°C. Left, zoom of area (1). (E) Reversibility of germination inhibition. S. rapamycinicus spores were incubated with 250 μg/mL fumigermin. After 5 days, the spores did not germinate; when recovered, washed and placed in fresh medium, the spores grew and formed mycelia.
-
Figure 3—source data 1
OD595 measurement of S. rapamycinicus growth.
- https://cdn.elifesciences.org/articles/52541/elife-52541-fig3-data1-v1.xlsx
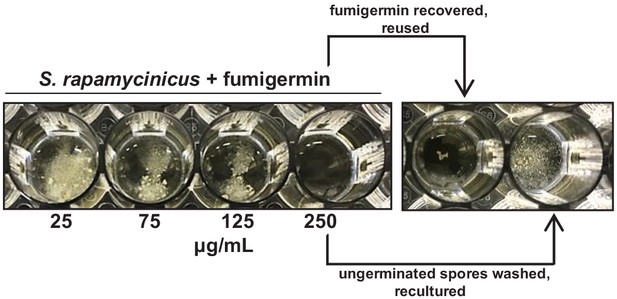
Stability of fumigermin and reversibility of its inhibitory action.
Top arrow, recovered fumigermin was further used to treat S. rapamycinicus spores. The re-used fumigermin inhibited the spore germination in a second round of usage, indicating the stability of the compound. Bottom arrow, ungerminated S. rapamycinicus spores previously treated with fumigermin were washed to remove fumigermin and given fresh medium. The spores were subsequently able to grow to form mycelia.

Germination assay of S. lividans and S. coelicolor treated with fumigermin.
107 spores of S. lividans or S. coelicolor were inoculated in 500 μL M79 medium in a 48-well plate and treated with increasing concentrations of fumigermin (0, 250, 500 or 1000 μg/mL). Shown are bacterial cultures after 3 days of growth with fumigermin at 28°C and orbital shaking at 500 rpm.
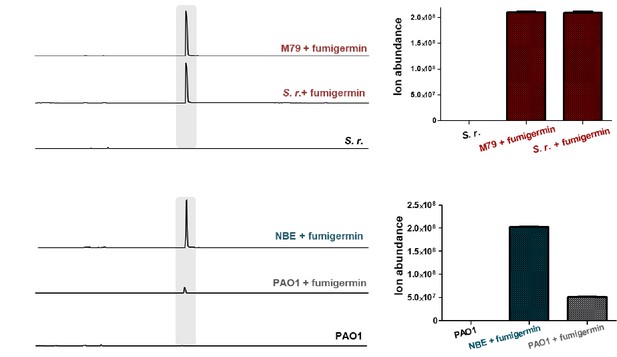
S. rapamycinicus cannot degrade fumigermin (top), but Pseudomonas aeruginosa can (bottom).
(A) LC-MS analysis of 1 mg of fumigermin added to bacterial cultures (3-day-old S. rapamycinicus pre-cultures; P. aeruginosa OD600 = 0.8). After 24 h, the cultures were analyzed using LC-MS. EIC traces shown for m/z 195 [M+H]+, corresponding to fumigermin, of control bacterial cultures without fumigermin (S. r. = S. rapamycinicus, PAO1 = P. aeruginosa), S. r. or PAO1 with 1 mg added fumigermin, or medium controls (M79 medium for S. r., NBE medium for PAO1) with 1 mg added fumigermin. Highlighted strip corresponds to fumigermin. (B) Fumigermin amount based on the intensity of the fumigermin ion of m/z 195 [M+H]+ in the EIC of the LC-MS analysis. Data are representative of three biological replicates with three technical replicates.
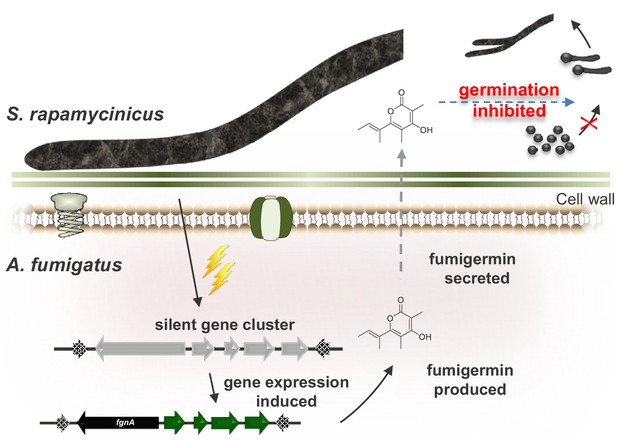
Model of the interaction between S. rapamycinicus and A. fumigatus ATCC 46645, which results in production of fumigermin.
During the co-cultivation of the two microorganisms, the bacterium S. rapamycinicus induces expression of the fgn gene cluster of the fungus A. fumigatus ATCC 46645, which leads to production of the SM fumigermin. Fumigermin is secreted and further inhibits germination of S. rapamycinicus spores.
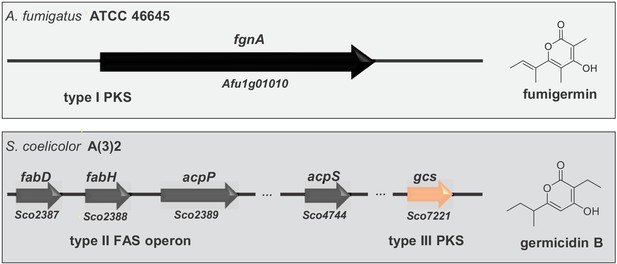
Alignment of genes involved in biosynthesis of the fungal compound fumigermin and the bacterial compound germicidin B.
Germicidin synthase is a type III bacterial PKS, while FgnA is a fungal iterative type I PKS. The synthesis of germicidins is coupled with fatty acid biosynthesis in S. coelicolor - the branched precursors are synthesized by FabD and FabH, after which they are further loaded onto and modified by Gcs (Chemler et al., 2012). In case of A. fumigatus, FgnA catalyzes the condensation of malonyl-CoA units to an acetyl-CoA starter unit independently of the fatty acid biosynthesis pathway. There is no striking similarity between Gcs and FgnA.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Aspergillus fumigatus) | Af293 | Nierman et al., 2005 | Wild-type strain, MAT1-2 | |
| Strain, strain background (Aspergillus fumigatus) | A1163 | da Silva Ferreira et al., 2006 | Wild-type strain, MAT1-1 | |
| Strain, strain background (Aspergillus fumigatus) | ATCC 46645 | Langfelder et al., 1998 | Wild-type strain, MAT1-1 | |
| Strain, strain background (A. novofumigatus) | IBT 16806 | Hong et al., 2005 | Wild-type strain | |
| Strain, strain background (Aspergillus fumigatus) | ΔfgnA | This paper | ATCC 46645 background, ptrA::fgnA, PTR | |
| Strain, strain background (Aspergillus fumigatus) | tetOn-fgnA | This paper | ATCC 46645 background, tetOn-fgnA, PTR | |
| Strain, strain background (Aspergillus nidulans) | RMS011 | Stringer et al., 1991 | pabaA1, yA2; ΔargB::trpCΔB, trpC801, veA1 | |
| Strain, strain background (Aspergillus nidulans) | fgnA_ATCC | This paper | RMS011 background, pUC18-gpdA- fgnA_ATCC, ArgB2+ | |
| Strain, strain background (Aspergillus nidulans) | fgnA_Af293 | This paper | RMS011 background, pUC18-gpdA- fgnA_Af293, ArgB2+ | |
| Strain, strain background (Aspergillus nidulans) | fgnA_A1163_repaired | This paper | RMS011 background, pUC18-gpdA-fgnA_A1163*, ArgB2+ | |
| Strain, strain background (Aspergillus nidulans) | fgn_cluster | This paper | RMS011 background, fgnA,Afu1g01000,Afu1g00990,Afu1g00980,Afu1g00970, ArgB2+ | |
| Strain, strain background (Streptomyces rapamycinicus) | ATCC 29253 | Kumar and Goodfellow, 2008 | Wild-type strain | |
| Strain, strain background (Streptomyces iranensis) | HM35 | Hamedi et al., 2010 | Wild-type strain | |
| Strain, strain background (Streptomyces lividans) | PM02 | van Dissel et al., 2015 | Wild-type strain | |
| Strain, strain background (Streptomyces coelicolor) | A(3)2 | Erikson, 1955 | Wild-type strain | |
| Commercial assay or kit | NEB HiFi | New England Biolabs, Frankfurt, Germany | E2621L | |
| Commercial assay or kit | Universal RNA Purification Kit | Roboklon, Berlin, Germany | E3598 | |
| Chemical compound, drug | germicidin B | Cayman Chemical, Ann Arbor, USA | CAS number 150973-78-7 | |
| Chemical compound, drug | digoxigenin-11- dUT | Jena BioScience, Jena, Germany | Catalog number NU-803-DIGXS | |
| Software, algorithm | GraphPad Prism 5 | GraphPad Software Inc,La Jolla, USA | RRID:SCR_002798 | |
| Software, algorithm | Thermo Xcalibur Qual Browser | Thermo Scientific, Dreieich, Germany | ||
| Software, algorithm | Proteome Discoverer (PD) 2.2 | Thermo Scientific, Dreieich, Germany | RRID:SCR_014477 | |
| Software, algorithm | Shimadzu Class-VP software (version 6.14 SP1) | Shimadzu, Duisburg, Germany | 6.14 SP1 | |
| Other | Hoechst 34580 | Thermo Fischer Scientific, Dreieich, Germany | H21486 |
Additional files
-
Supplementary file 1
Putative function of the genes in the predicted PKS biosynthetic gene cluster.
Domain analysis using the CD-search tool (NCBI, Marchler-Bauer et al., 2017) of each of the cluster genes (shown in black and green), as well as the 5’ and 3’ neighboring genes. Based on similarity to known genes, the potential function of the encoded proteins was predicted. Gene sequences were downloaded from the FungiDB database (https://fungidb.org/fungidb/).
- https://cdn.elifesciences.org/articles/52541/elife-52541-supp1-v1.pptx
-
Supplementary file 2
Primers used in this study.
- https://cdn.elifesciences.org/articles/52541/elife-52541-supp2-v1.pptx
-
Supplementary file 3
qRT-PCR primers used in this study.
- https://cdn.elifesciences.org/articles/52541/elife-52541-supp3-v1.pptx
-
Supplementary file 4
Primers used for amplification of Southern blot probes.
- https://cdn.elifesciences.org/articles/52541/elife-52541-supp4-v1.pptx
-
Supplementary file 5
Putative orthologs of Fgn cluster proteins identified using BLASTp with a 50% coverage and 50% identity cut-off.
- https://cdn.elifesciences.org/articles/52541/elife-52541-supp5-v1.pptx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/52541/elife-52541-transrepform-v1.docx





