Cooperative interactions facilitate stimulation of Rad51 by the Swi5-Sfr1 auxiliary factor complex
Figures
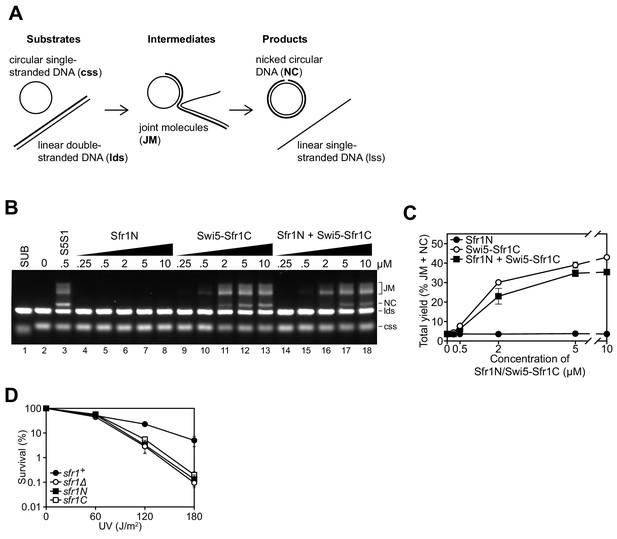
Sfr1N is essential for DNA repair mediated by Swi5-Sfr1.
(A) Schematic of the three-strand exchange assay. (B) Three-strand exchange reactions containing Rad51 with the indicated concentrations of Swi5-Sfr1, or Sfr1N and/or Swi5-Sfr1C, were incubated for 2 hr at 37 °C and DNA was separated by agarose gel electrophoresis to visualize substrates (css, lds), intermediates (JM) and products (NC). (C) The percentage of DNA signal per lane corresponding to JM and NC (total yield) was plotted against protein concentration. (D) Percentage of cell survival following acute UV irradiation for the indicated strains. For (C,D), mean values of three independent experiments ± s.d. are shown.
-
Figure 1—source data 1
Yield (%) of total DNA for data in Figure 1C.
Survival (%) following UV irradiation for data in Figure 1D.
- https://cdn.elifesciences.org/articles/52566/elife-52566-fig1-data1-v1.xlsx
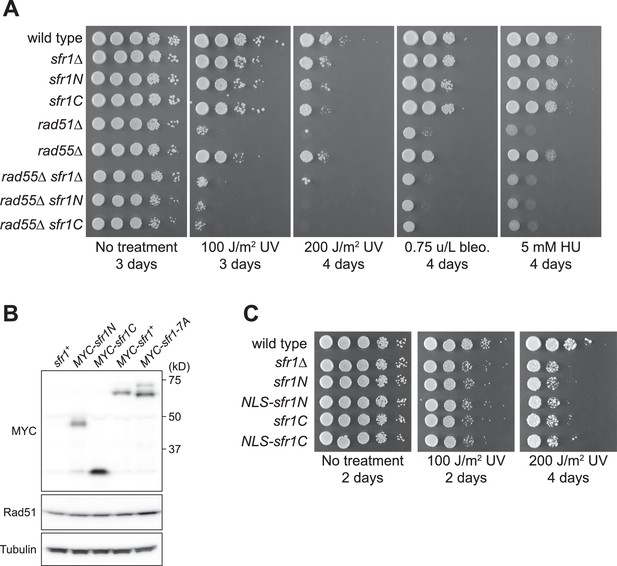
The DNA damage sensitivity of sfr1N/C is not due to protein instability and is not suppressed by fusion to an NLS.
(A,C) DNA damage sensitivity of the indicated strains was assessed. NLS, nuclear localization signal of the SV40 large T antigen. Bleo., bleomycin. HU, hydroxyurea. Note that sfr1Δ does not sensitize cells to the indicated doses of bleo. or HU unless Rad55/Rad57 is absent. (B) Proteins were extracted from log phase cultures, separated by SDS-PAGE and probed with the indicated antibodies. Tubulin serves as a loading control.
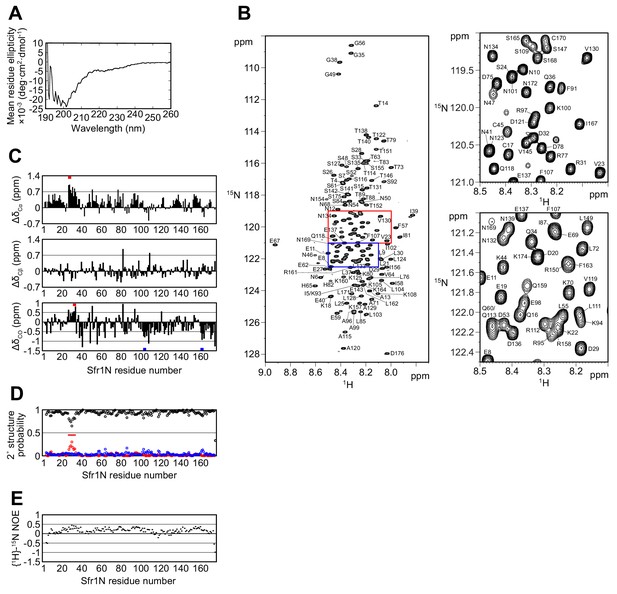
The N-terminal half of Sfr1 is an intrinsically disordered and flexible domain.
(A) Far-UV CD spectrum of Sfr1N. (B) 1H-15N HSQC spectrum of Sfr1N. Regions outlined in red and blue are enlarged in the top right and bottom right panels, respectively. (C) Secondary chemical shifts of 13Cα (top), 13Cβ (middle), and 13CO (bottom) were obtained and plotted against the corresponding position in Sfr1N. Values that suggest potential α-helix and β-strand formation are indicated with red and blue bars, respectively. (D) TALOS+ prediction of secondary structure probabilities. Black, red, and blue circles indicate the probabilities of random coils, α-helices, and β-strands, respectively. (E) {1H}-15N heteronuclear NOE was measured for Sfr1N. All analyzed main chain NHs showed NOE values of less than 0.44, with a protein-wide average of 0.15.
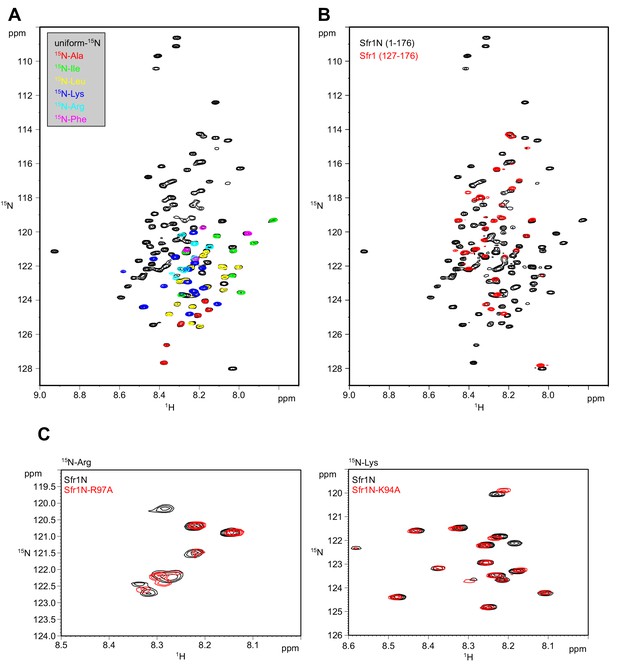
Validation of Sfr1N signals by 15N labeling.
(A) Superimposed 1H-15N HSQC spectra of Sfr1N with the indicated 15N-labeling. (B) Superimposed 1H-15N HSQC spectra of uniformly 15N-labeled Sfr1N and a truncated variant, Sfr1 (127-176). (C) Left, superimposed 1H-15N HSQC spectra of Arg-selectively 15N-labeled Sfr1N or Sfr1N-R97A. Right, superimposed 1H-15N HSQC spectra of Lys-selectively 15N-labeled Sfr1N or Sfr1N-K94A.
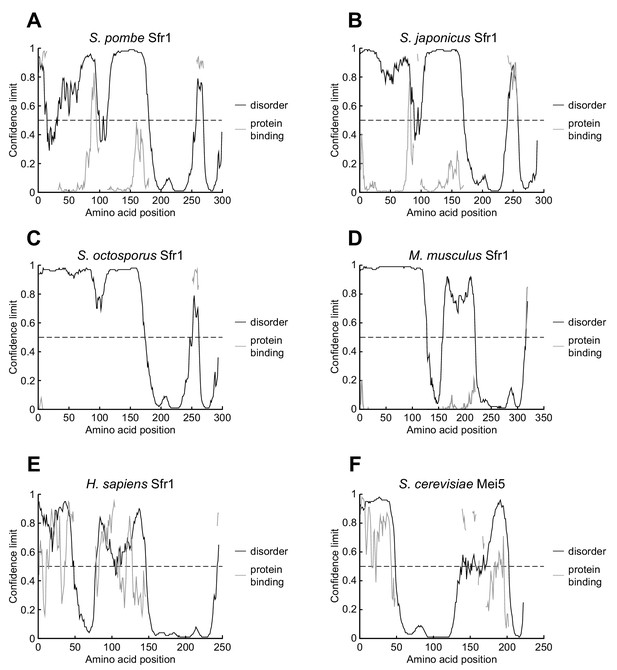
N-terminal intrinsically disordered domains may be an evolutionarily conserved feature of Sfr1.
(A–F) DISOPRED3 was used to predict the likelihood of disorder and protein binding in orthologs of Sfr1 from the indicated species. The likelihood of disorder is shown on the y-axis and residues of Sfr1 are displayed on the x-axis. The dashed line represents a confidence limit of 0.5, above which point DISOPRED3 predicts that a residue is more likely to be disordered than structured. Sequences were obtained from UniProt (S. pombe, Q9USV1; S. japonicus, B6JYP3; S. octosporus, S9R1E2; Mus musculus, Q8BP27; Homo sapiens, Q86XK3; S. cerevisiae, P32489).
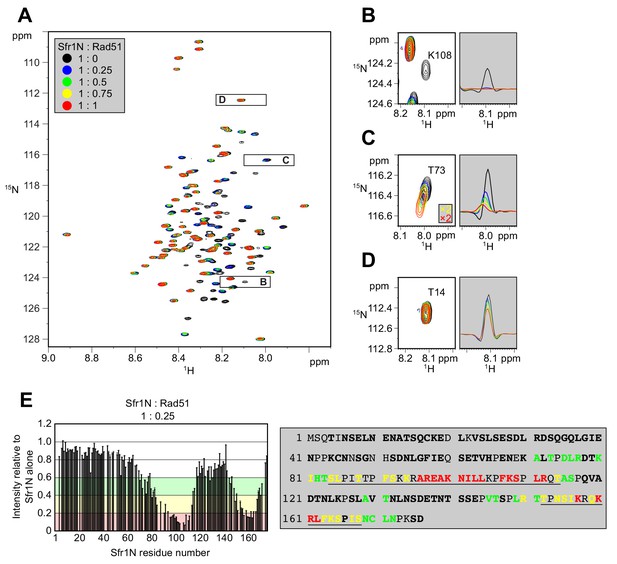
Two sites within Sfr1N interact with Rad51.
(A) Superimposed 1H-15N HSQC spectra of 15N-labeled Sfr1N in the absence of Rad51 or in the presence of Rad51 at the indicated molecular ratios. (B–D) Enlarged signals from K108, T73, and T14 (left), with slices along the 1H axis (right). For (C), the threshold of the yellow and red spectra was set 2x lower than the other spectra to show the signals clearly. (E) Signal intensity ratio of Sfr1N residues in the presence of Rad51 to those in the absence of Rad51 (left) and features of the Sfr1N amino acid sequence (right). Reductions in signal intensity of 40–60% (green), 60–80% (yellow), and >80% (red) are highlighted, along with the corresponding residues. Underlined residues correspond to Site 1 (S84 to T114) and Site 2 (T152 to S168), where the most significant signal attenuations were observed. Residues not in bold include prolines, unassigned residues, and residues with overlapped signals.
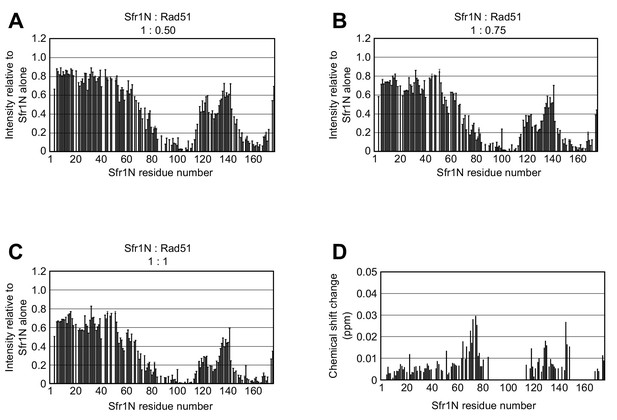
Changes in Sfr1N NMR signal intensity in response to Rad51 titration.
(A–C) NMR signal intensity of Sfr1N in the presence of the indicated ratio of Rad51 relative to the intensity observed in the absence of Rad51. (D) Chemical shift changes observed for the main chain amide NHs of Sfr1N induced by the addition of equimolar Rad51. Chemical shift changes were calculated using the formula , where ΔδH and ΔδN represent the 1H and 15N chemical shift differences, respectively. The residues that were severely attenuated by the addition of Rad51 were not analyzed and are shown as blanks.
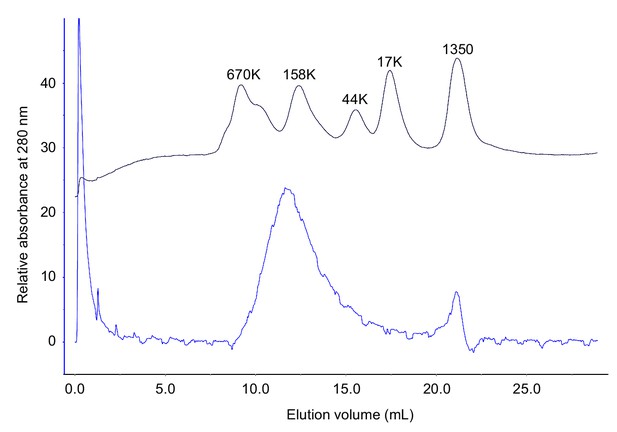
Molecular sizing of S. pombe Rad51.
Rad51 was analyzed by size-exclusion chromatography. The blue and navy lines indicate the elution profiles of Rad51 and molecular weight standards, respectively.
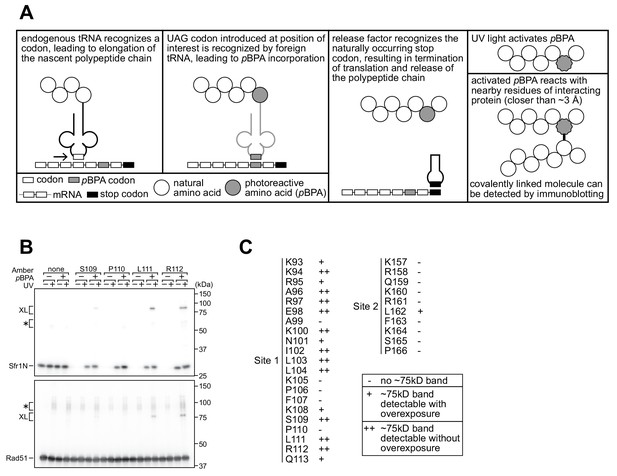
Residues within sites 1 and 2 can be site-specifically crosslinked to Rad51.
(A) Schematic of the crosslinking assay. (B) Example of Sfr1N-Rad51 crosslinking. The indicated residues were replaced with an amber codon, or the amber codon was omitted as a negative control (‘none’). XL, specific crosslinks (defined in the text). *non-specific crosslinks. (C) Summary of crosslinking results for all residues examined.
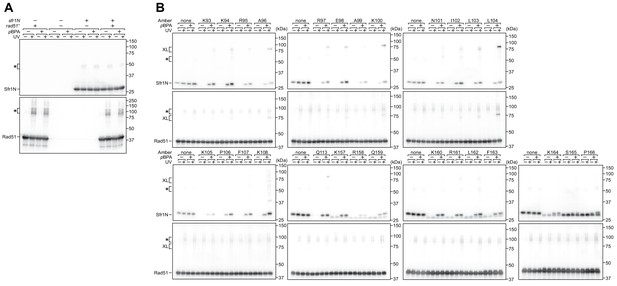
Nonspecific and specific crosslinking of Sfr1N to Rad51.
(A) The extent of non-specific crosslinking was assessed by individual and co-expression of Sfr1N and Rad51 in E. coli. (B) Specific crosslinking was assessed by expressing Rad51 with a version of Sfr1N in which a residue of interested was mutated to TAG, which encodes the photoreactive amino acid pBPA in this system.
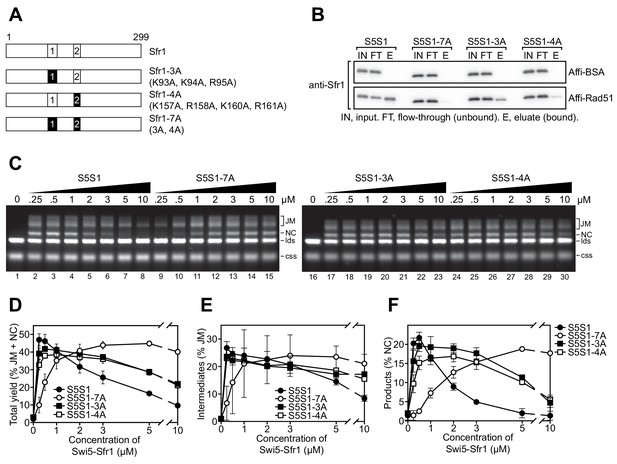
Interaction of sites 1 and 2 with Rad51 is important for the stimulation of Rad51-driven strand exchange.
(A) Schematic of Sfr1-Rad51 interaction site mutants. (B) The interaction of Swi5-Sfr1 (S5S1, wild type or mutants) with Rad51 was investigated by a pull-down assay. Affi-BSA is a control for nonspecific binding. (C) Three-strand exchange reactions were performed with the indicated concentrations of Swi5-Sfr1 (wild type or mutants). The percentage of DNA signal per lane corresponding to total yield (D), JM (E) or NC (F) was plotted against Swi5-Sfr1 concentration. For (D–F), mean values of three independent experiments ± s.d. are shown.
-
Figure 5—source data 1
Yield (%) of total DNA for data in Figure 5D–F.
- https://cdn.elifesciences.org/articles/52566/elife-52566-fig5-data1-v1.xlsx
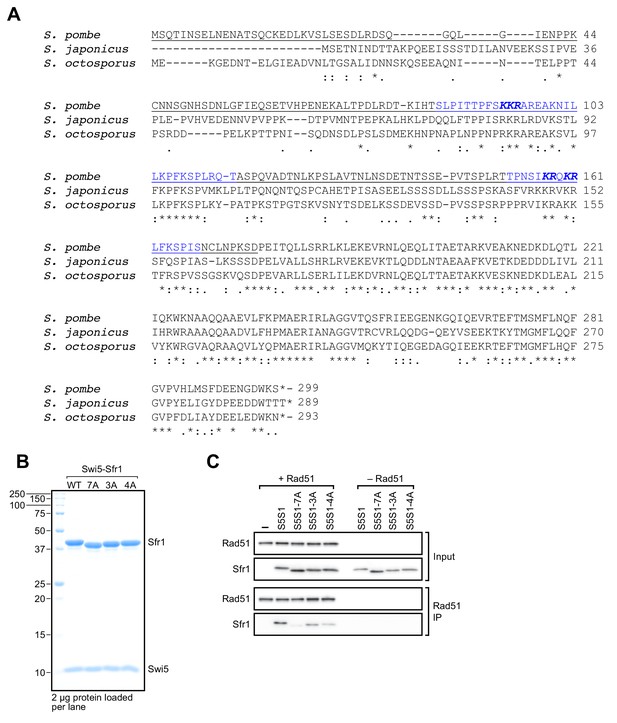
Identification of residues within sites 1 and 2 that are important for the functional interaction with Rad51.
(A) Sequence alignment of Sfr1 homologs from the genus Schizosaccharomyces were generated with Clustal Omega. S. pombe Sfr1N (residues 1–176) is underlined, with Sites 1 and 2 shown in blue. Bold italic typeface indicates consecutive positively charged residues that were targeted for mutation. (B) CBB-stained gel of purified Swi5-Sfr1 (wild type and mutants). 7A has mutations in all bold residues from (A), whereas 3A and 4A only contain mutations in bold residues from Sites 1 and 2, respectively. Size markers are annotated on the left in kDa. Note that Sfr1 migrates slower than expected from its calculated molecular weight (~33.6 kD), as previously reported (Haruta et al., 2006). This property is attributable to its N-terminal half (Kuwabara et al., 2012), hence why mutation of residues in Sites 1 and 2 affects its electrophoretic mobility. (C) Rad51 was mixed with Swi5-Sfr1 (S5S1, wild type or mutants; input), complexes were IP'd with anti-Rad51 antibodies and the contents of these IPs were assessed by immunoblotting. ‘–" indicates the omission of Swi5-Sfr1.
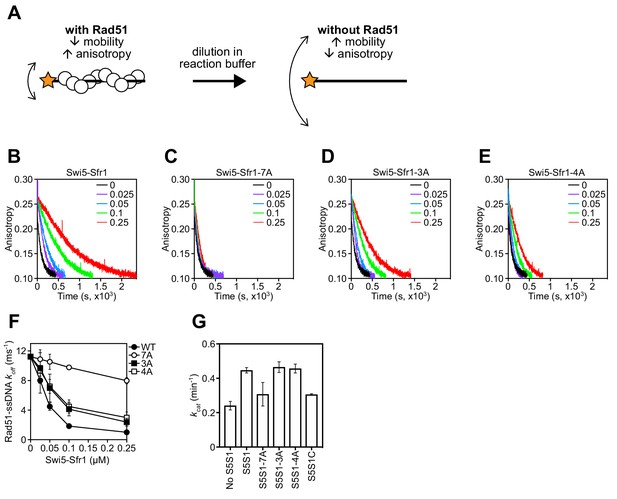
Rad51 filament stabilization and ATPase stimulation requires interactions with sites 1 and 2.
(A) Schematic of the fluorescence anisotropy assay. Rad51 monomers (white circles) are depicted forming a filament on an oligonucleotide (black line) labeled with the TAMRA fluorophore (orange star). (B–E) The anisotropy of fluorescently labeled ssDNA in complex with Rad51 was monitored following induction of filament collapse in the presence of Swi5-Sfr1 (wild type or mutants) at the indicated concentrations (µM). (F) koff values were determined for Rad51-ssDNA complexes at the indicated concentrations of Swi5-Sfr1. (G) Rad51-dependent ATP turnover was determined in the presence of the indicated Swi5-Sfr1 variant. For (F,G), mean values of three independent experiments ± s.d. are shown.
-
Figure 6—source data 1
Anisotropy values for data in Figure 6B–E.
- https://cdn.elifesciences.org/articles/52566/elife-52566-fig6-data1-v1.xlsx
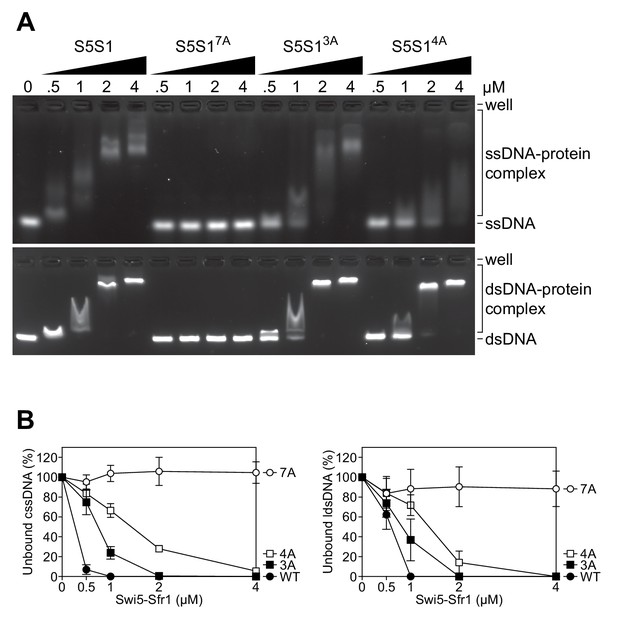
DNA binding is differentially defective in Site 1 and Site 2 mutants.
(A) Swi5-Sfr1 (S5S1, wild type or mutants) was incubated with ssDNA (top) or dsDNA (bottom) and protein-DNA complexes were resolved by agarose gel electrophoresis. (B) The amount of unbound ssDNA (left) or dsDNA (right) was quantified and expressed relative to the no protein lane, which was set to 100%. For (B), mean values of three independent experiments ± s.d. are shown.
-
Figure 6—figure supplement 1—source data 1
Unbound DNA (%) for data in Figure 6—figure supplement 1B.
- https://cdn.elifesciences.org/articles/52566/elife-52566-fig6-figsupp1-data1-v1.xlsx
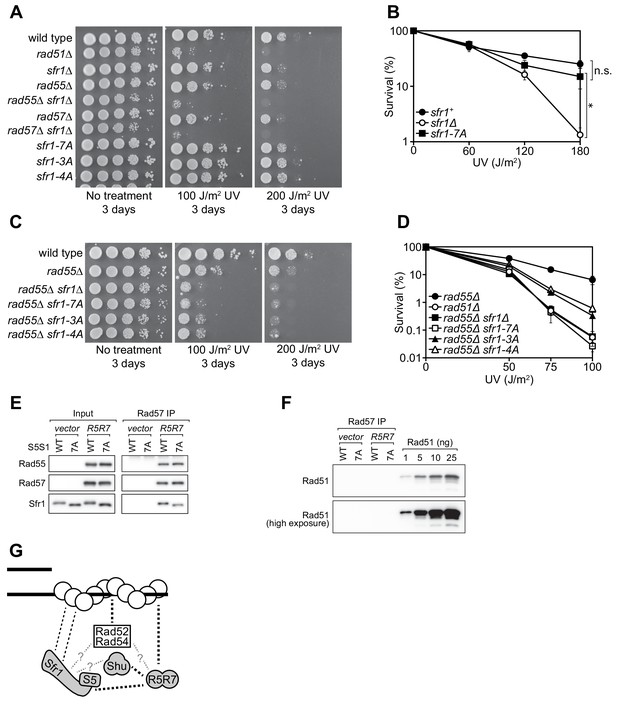
Rad55-Rad57 facilitates Swi5-Sfr1-dependent DNA repair.
(A,C) 10-fold serial dilutions of the indicated strains were spotted onto rich media with or without acute UV treatment and grown at 30 °C. (B,D) Percentage of cell survival was determined after acute exposure to UV. (E) The soluble cell lysate from S. pombe strains carrying the REP1 and REP2 expression vectors without inserts (vector) or encoding Rad55-Protein A and Rad57 (R5R7) was incubated with IgG-agarose. Proteins were eluted with 3C protease, for which a cleavage site exists between Rad55 and Protein A. This eluate was mixed with purified Swi5-Sfr1 (S5S1, wild type or 7A; input) and subjected to IP with anti-Rad57 antibody. These IPs were probed with the indicated antibodies. (F) Rad57 IPs from (E) and the indicated amounts of purified Rad51 were probed with an anti-Rad51 antibody. (G) Molecular bridging model. Rad51 monomers (white circles) are shown as a filament on ssDNA (solid black lines). Dashed black lines represent known physical interactions. Dashed gray lines with question marks represent potential physical interactions. Swi5-Sfr1 is recruited to Rad51 via two redundant mechanisms, one requiring its direct interaction with Rad51 and the other requiring Rad55-Rad57. See Discussion for details. For (B,D), mean values of three independent experiments ± s.d. are shown. Statistical analysis in (B) was by one-way ANOVA followed by Tukey’s multiple comparisons test. *, p<0.05. n.s., not significant (p>0.05).
-
Figure 7—source data 1
Survival (%) following UV irradiation for data in Figure 7B and Figure 7D.
Survival (%) following UV irradiation for data in Figure 7D.
- https://cdn.elifesciences.org/articles/52566/elife-52566-fig7-data1-v1.xlsx
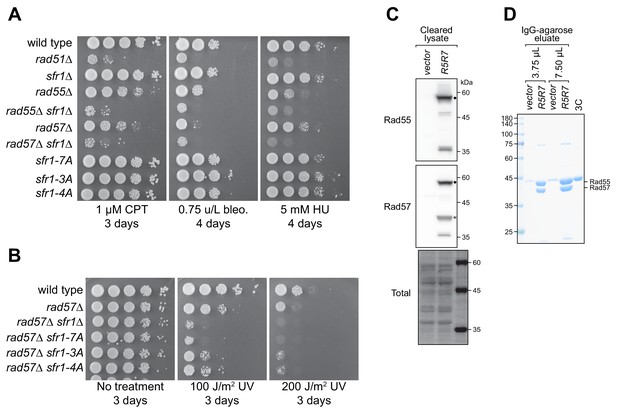
DNA repair in Sfr1-Rad51 interaction mutants is dependent on Rad57.
(A,B) DNA damage sensitivity of the indicated strains was assessed. ‘No treatment’ plate for (A) is shown in Figure 7A. CPT, camptothecin. Bleo., bleomycin. HU, hydroxyurea. Note that sfr1Δ cells are not sensitive to the indicated doses of CPT, bleo. or HU unless Rad55/Rad57 is absent. (C) S. pombe was transformed with the REP1 and REP2 expression vectors (vector) or the same vectors encoding Rad55-Protein A and Rad57 (R5R7). Soluble cellular extracts were probed with the indicated antibodies. Black dot indicates the Rad55-Protein A band, which is also reactive to the anti-Rad57 antibody due to the Protein A tag. Asterisk indicates the Rad57 band. Total, Ponceau S-stained membrane. (D) Soluble extracts from (C) were incubated with IgG-agarose and bound proteins were eluted via addition of 3C protease, which cleaves between Rad55 and Protein A. The indicated amount of this eluate was analyzed by SDS-PAGE and CBB staining. 2 µg of 3C protease was loaded on the rightmost lane as a control. Size markers are indicated on the left in kDa.
Additional files
-
Supplementary file 1
Key Resources Table.
- https://cdn.elifesciences.org/articles/52566/elife-52566-supp1-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/52566/elife-52566-transrepform-v1.docx





