Indirect sexual selection drives rapid sperm protein evolution in abalone
Figures
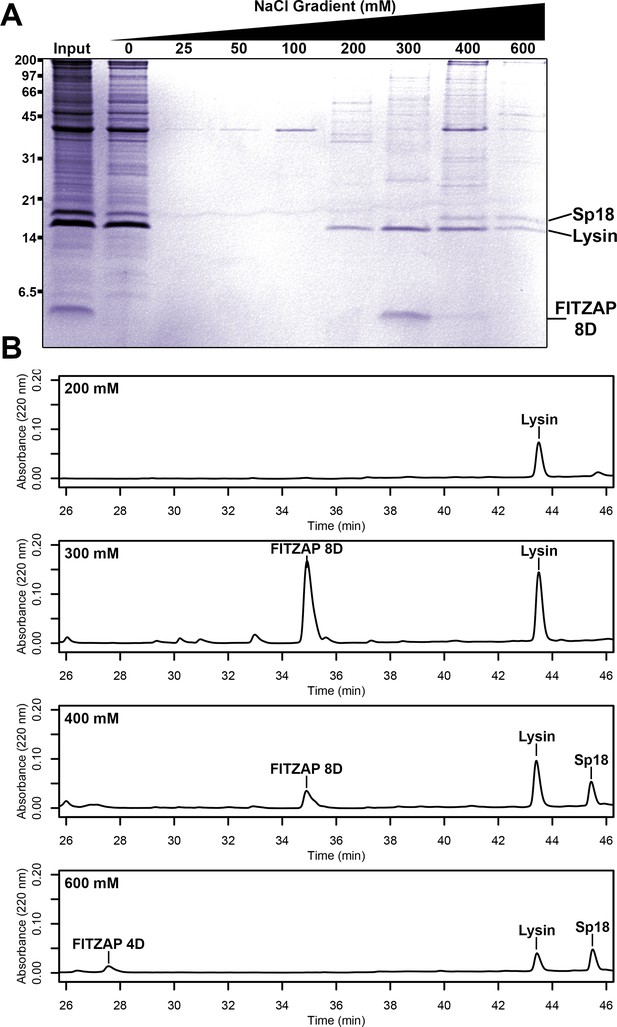
Co-purification of FITZAP and cationic acrosomal proteins by anion exchange.
(A) SDS-PAGE of proteins from red abalone sperm lysate separated by anion exchange. Despite their high net positive charges, lysin and sp18 co-eluted with FITZAP isoforms in the highest salt fractions. (B) RP-HPLC analysis of select anion exchange fractions which contained lysin. The peak of lysin elution at 300 mM coincides with the peak elution of FITZAP-8D, with both sp18 and FITZAP-4D eluting under higher salt conditions.
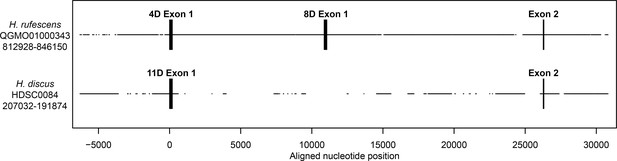
Genomic structure of FITZAP locus in red and disk abalone.
Comparison of FITZAP genomic loci between the red and disk abalone draft genomes. Each FITZAP coding sequencing is contained in two exons: exon 1 contains the signal peptide, poly-aspartate array, lysin binding region, and the furin cleavage site, while exon 2 contains the majority of the FITZAP C-terminal peptide. The multiple FITZAP isoforms are produced through alternative splicing of optional exon 1 sequences, with the red FITZAP 4D exon 1 being orthologous to disk FITZAP-11D.
-
Figure 1—figure supplement 1—source data 1
Sequence alignment of FITZAP loci from red and disk abalone draft genomes.
- https://cdn.elifesciences.org/articles/52628/elife-52628-fig1-figsupp1-data1-v2.txt
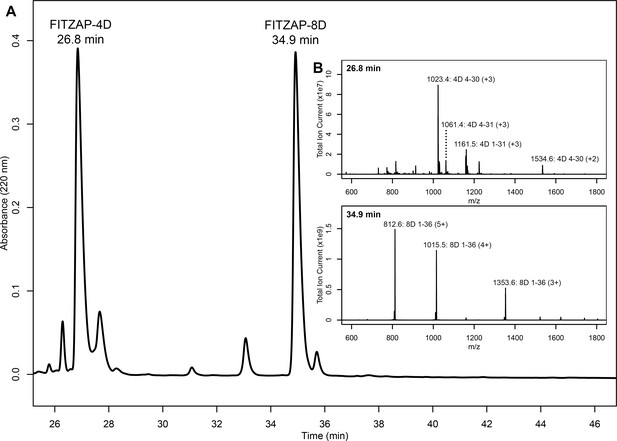
Purification and mass spectral analysis of natural red FITZAP-4D and 8D.
(A) Separation of anion exchange-enriched red abalone sperm lysate, with major peaks at 26.8 and 34.9 min being FITZAP-4D and 8D, respectively. (B) Mass spectral analysis of the FITZAP fractions show masses consistent with processing of FITZAP precursors by furin and carboxypeptidase B to liberate the C-terminal peptide from the longer N-terminal peptide containing the poly-aspartate region.
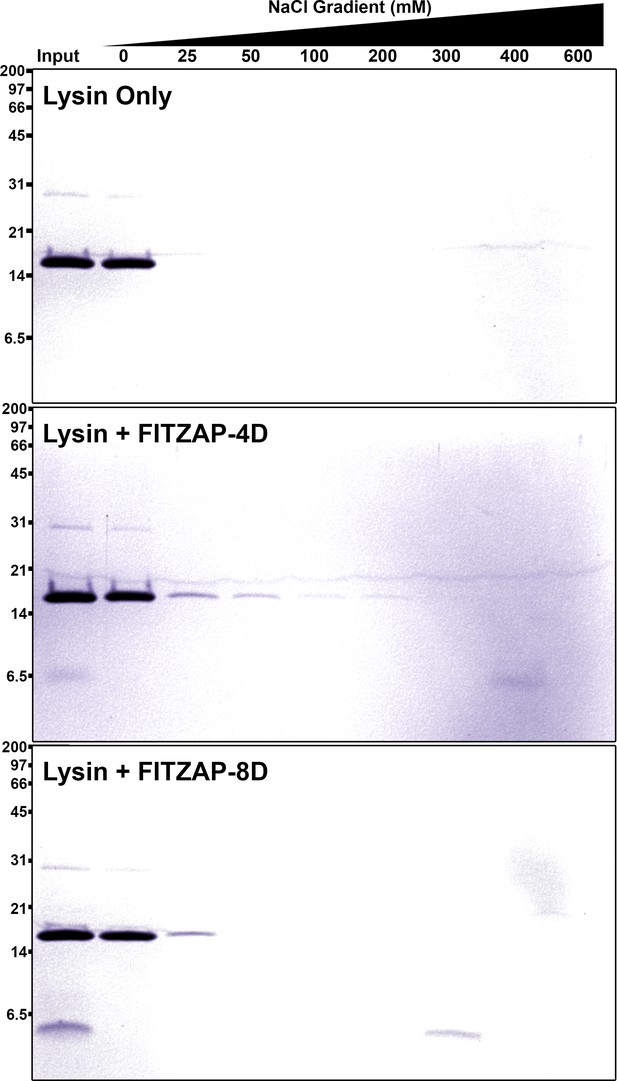
SAX chromatography of lysin and FITZAP in vitro.
Purified lysin has no innate affinity for the anion exchange resin, yet its elution is retarded when mixed with either FITZAP isoform.
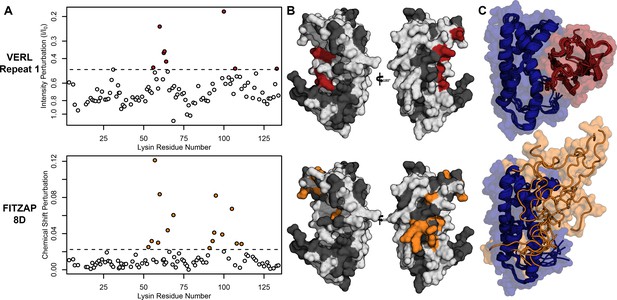
Lysin recognizes VERL and FITZAP through a common binding interface.
(A) NMR perturbation of red lysin upon conspecific binding of either VERL repeat 1 (data from Wilburn et al., 2018) or FITZAP-8D (1.5 molar equivalents). Two regions near residue 60 and residue 100 of lysin are perturbed in both cases (dashed line: median + 1.5 * interquartile range). (B) Mapping of perturbation onto a lysin solution structure (PDB 5utg) shows spatial clustering of these two regions to a single binding surface of lysin. (C) Docking of VERL repeat 1 (based on PDB 5mr3 by Raj et al., 2017) and FITZAP-8D (based on restraints from paramagnetic relaxation enhancement) supports that FITZAP is an intrinsically disordered protein that shields the VERL binding interface of lysin.
-
Figure 2—source data 1
Lysin NMR perturbation values for VERLr1 and FITZAP-8D.
- https://cdn.elifesciences.org/articles/52628/elife-52628-fig2-data1-v2.csv
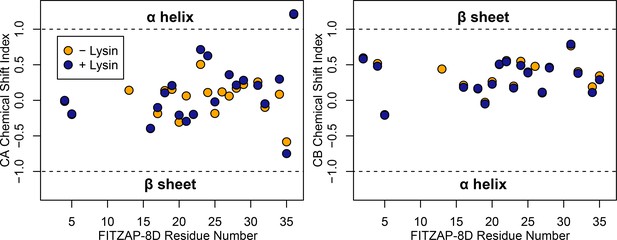
FITZAP is intrinsically disordered when free and bound to lysin.
CA and CB chemical shift indices for FITZAP-8D support that it is an intrinsically disordered protein, in both the presence and absence of equimolar lysin.
-
Figure 2—figure supplement 1—source data 1
FITZAP chemical shift indices.
- https://cdn.elifesciences.org/articles/52628/elife-52628-fig2-figsupp1-data1-v2.csv
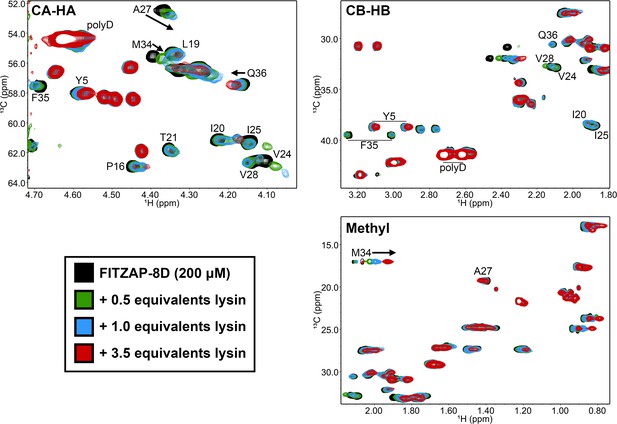
Lysin binding causes few changes in FITZAP 13C chemical shifts.
Comparison of 13C-HSQC spectra of FITZAP (200 μM) in the presence of different concentrations of F104A lysin approaching saturation conditions (3.5 equivalents ≈ 92% saturated, based on Kd estimates by fluorescence polarization). The small changes in both the CA-HA and CB-HB regions support that FITZAP remains intrinsically disordered when bound to lysin. Furthermore, no 13C and only 1H chemical shift perturbations were observed in the methyl region.
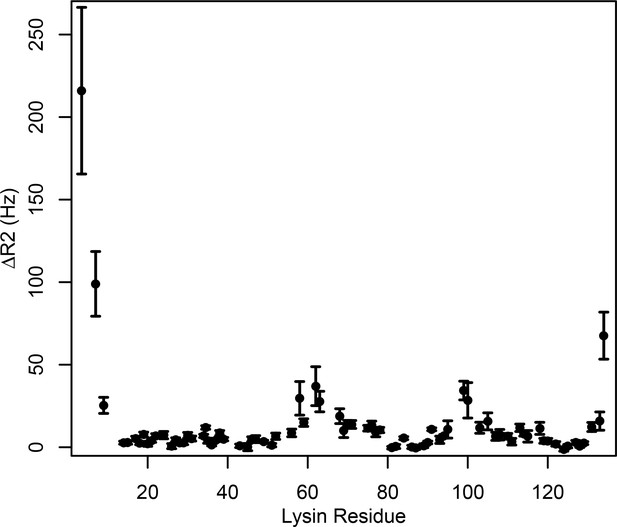
Intermolecular PRE measurements between 15N-lysin and FITZAP-8D.
Changes in 15N transverse relaxation rates were calculated between 15N-lysin and FITZAP-8D with and without Cu2+ bound to the N -terminal ATCUN motif of FITZAP-8D (reported as mean ± standard error).
-
Figure 2—figure supplement 3—source data 1
PRE measurements of lysin-FITZAP interactions.
- https://cdn.elifesciences.org/articles/52628/elife-52628-fig2-figsupp3-data1-v2.csv
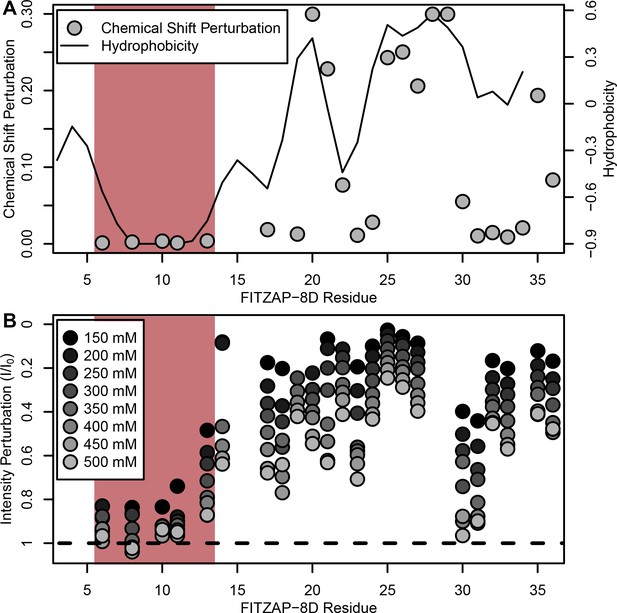
NMR perturbation of FITZAP-8D upon lysin binding.
(A) Backbone amide chemical shift perturbation of FITZAP-8D upon addition of lysin under seawater conditions (500 mM NaCl). The solid line denotes the relative hydrophobicity of the sequence which correlates well with the chemical shift perturbation, supporting that FITZAP-lysin interactions under high-salt conditions are driven by hydrophobic packing. The poly-aspartate region is highlighted in red and shows no chemical shift perturbation under seawater conditions. (B) Intensity perturbation of FITZAP-8D by binding of lysin under different ionic strengths ranging from approximately intracellular levels (150 mM) to seawater conditions (500 mM). Differences in intensity perturbation as a function of salt are in part driven by the salt dependence on lysin-FITZAP Kd (Figure 5), but the weak perturbation observed in the poly-aspartate region only under low salt conditions support that these residues are involved in some form of molecular interaction with lysin (likely salt bridges as part of FITZ complexes) only under intracellular conditions.
-
Figure 3—source data 1
FITZAP chemical shift perturbations by lysin binding.
- https://cdn.elifesciences.org/articles/52628/elife-52628-fig3-data1-v2.csv
-
Figure 3—source data 2
FITZAP intensity perturbation by lysin at differing salt concentrations.
- https://cdn.elifesciences.org/articles/52628/elife-52628-fig3-data2-v2.csv
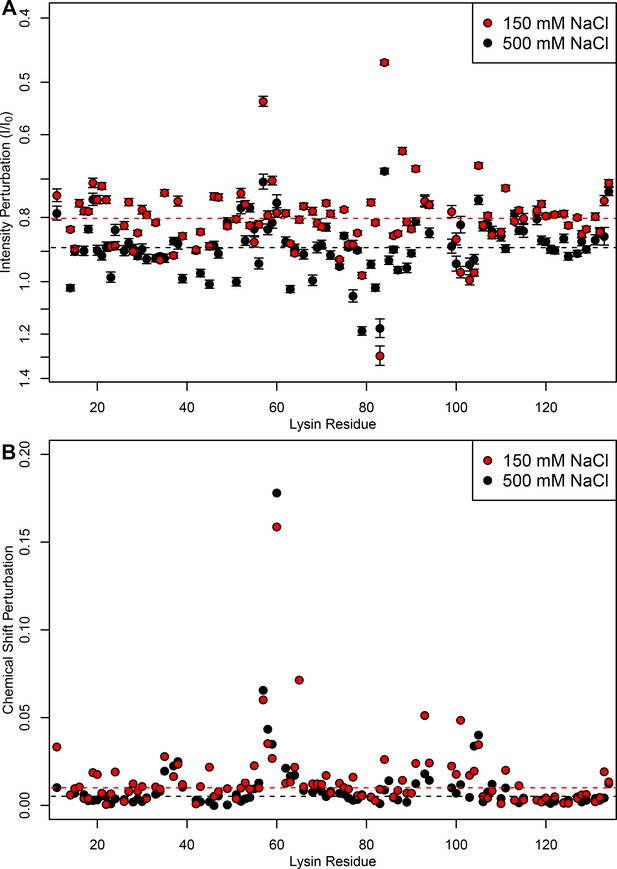
FITZ complexes experience different exchange rates in intracellular vs seawater conditions.
(A) Intensity perturbation and (B) chemical shift perturbation of 15N-lysin (100 μM) in the presence of FITZAP-8D (100 μM) under both intracellular (150 mM) and seawater (500 mM) salt conditions, with dotted lines representing the median of each condition. Under intracellular salt concentrations, there is greater intensity perturbation, reflective of greater peak broadening and faster transverse relaxation from increased molecular weight of FITZAP binding lysin. However, despite lower affinity under higher salt concentrations, similar if not greater chemical shift perturbation is observed under seawater ionic strengths. Chemical shift perturbations are observed under fast exchange dynamics (ns-μs scales), while intensity perturbations imply slower dynamics (ms scales), such that this perturbation suggests that lysin-FITZAP are in slower exchange under intracellular salt conditions. Intensity perturbation is reported as mean ± standard error.
-
Figure 3—figure supplement 1—source data 1
Lysin intensity and chemical shift perturbation by FITZAP binding at differing salt concentrations.
- https://cdn.elifesciences.org/articles/52628/elife-52628-fig3-figsupp1-data1-v2.csv
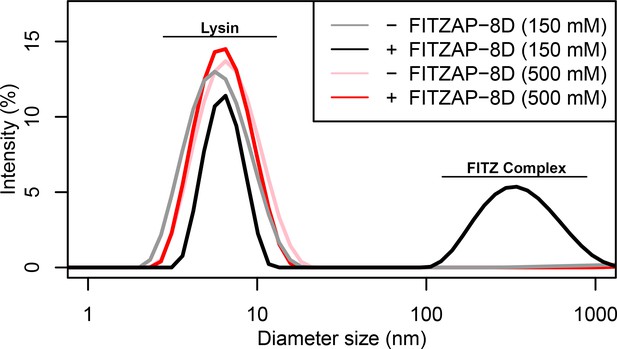
FITZ complex formation is dependent on both FITZAP and the ionic environment.
Dynamic light scattering measurements of lysin with and without equimolar FITZAP-8D at intracellular (150 mM) and seawater (500 mM) salt conditions. Lysin and FITZAP associate into FITZ complexes with mean diameter of ~400 nm only under intracellular salt conditions.
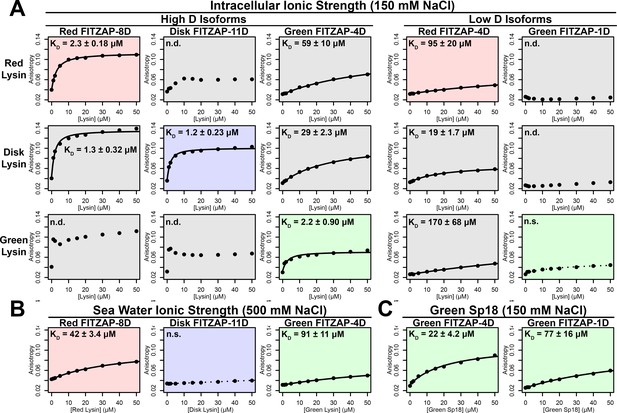
FITZAP interactions with cationic acrosomal proteins within and between species.
(A) Lysin-FITZAP interactions were measured by fluorescence polarization under approximately intracellular salt conditions for all combinations within and between species of red, disk, and green abalone (conspecific interactions are shaded as red, blue, and green, respectively). Low micromolar binding affinities were observed for conspecific interactions of lysin with high-D FITZAP isoforms. (B) Binding affinities between conspecific lysin and FITZAP high-D isoforms are substantially reduced under extracellular seawater conditions. (C) Green sp18 shows tighter binding to conspecific FITZAP-4D compared to FITZAP-1D; however, the relative affinity of sp18 for 4D over 1D (77/22 = 3.5 X) is greater than that of lysin (>100 X), suggesting that sp18 has higher preference for low D isoforms compared to lysin. (n.s. = not significant at p<0.05; n.d. = not determined if anisotropy was not monotonically positive and consistent with single-state binding; Kd reported as mean ± standard error).
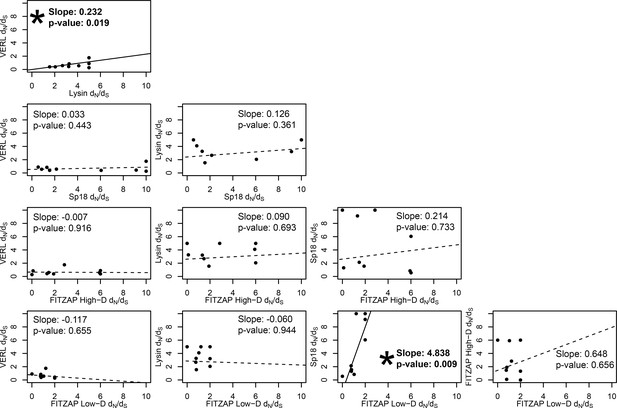
Test of sequence coevolution between abalone reproductive genes by correlated branch dN/dS.
Comparison of branch dN/dS estimates for all combinations of lysin, VERL, sp18, FITZAP low-D, and FITZAP high-D across the Pacific abalone clade. Based on methods by Clark et al. (2009), correlation between these values was assessed by weighted linear regression, with a significant positive slope suggesting coevolution. Significant correlations were found between lysin-VERL (as in Clark et al., 2009) and in sp18-FITZAP low-D.
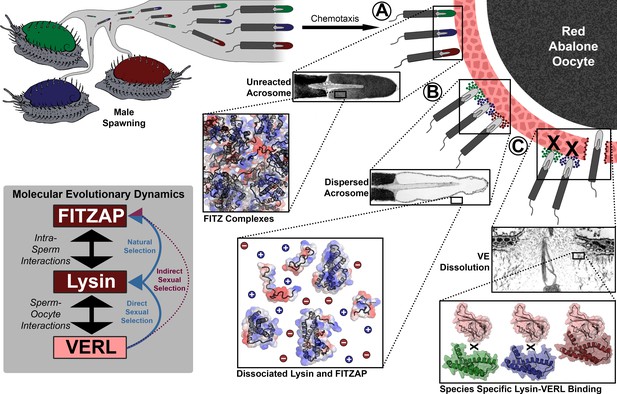
Rapid evolution of FITZAP is due to indirect sexual selection from VERL.
Male abalone have overlapping habitat ranges and spawning periods such that there is opportunity for hybridization to occur. Sperm are attracted to eggs via chemotaxis and bind to the vitelline envelope (A). At this stage, the acrosome is still intact (Lewis et al., 1982) with lysin and FITZAP tightly packaged as FITZ complexes. Binding to the VE causes the sperm to acrosome react (B), releasing its contents into highly ionic seawater which disperses lysin and FITZAP. Liberated lysin can now dissolve the VE by binding VERL domains (C); these interactions are species-specific and provide one barrier to hybridization (Swanson and Vacquier, 1997). Because the rapid evolution of lysin is driven by direct sexual selection to maintain sperm-egg interactions, and FITZAP is co-evolving to bind the same interface, VERL imposes indirect sexual selection onto FITZAP. Microscopy images adapted from Lewis et al. (1982) and Sakai et al., 1982.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | 5-alpha | New England Biolabs | C2987 | Chemically competent cells for cloning and sequencing |
| Strain, strain background (Escherichia coli) | Rosetta2 | Novagen | 71400 | Chemically competent cells for protein expression |
| Biological sample (Haliotis rufescens) | red abalone sperm | The Abalone Farm | Freshly isolated from male abalone testis | |
| Biological sample (Haliotis fulgens) | green abalone lysin | Collected from 32°51’00’ N, 117°16’34’ W | Purified from sperm by cation exchange chromatography, and provided by Vic Vacquier | |
| Recombinant DNA reagent | pCR4-TOPO | Thermo-Fisher | K457540 | Cloning of DNA for sequencing |
| Recombinant DNA reagent | pET11d | Novagen | 69439 | Cloning of DNA for protein expression |
| Recombinant DNA reagent | MBP-FITZAP | This paper | See Supplementary file 1 | |
| Sequence-based reagent | FITZAP F’ | This paper | 5’-ATGAGGGTTRTTCTAATT-3’ | |
| Sequence-based reagent | Sp18 F’ | This paper | 5’-GGAAACAGTATGAGGTYTTTGSTGCTT-3’ | |
| Peptide, recombinant protein | MBP-TEV protease | Sigma-Aldrich | T4455 | |
| Commercial assay or kit | BCA Protein Assay | Pierce | 23225 | |
| Software, algorithm | RStudio v.1.0.136 | https://rstudio.com/ | RRID:SCR_000432 | |
| Software, algorithm | RAxML v8.2.12 | https://cme.h-its.org/exelixis/software.html | RRID:SCR_006086 | |
| Software, algorithm | Fast Statistical Alignment (FSA) v.1.15.9 | http://fsa.sourceforge.net/ | RRID:SCR_016114 | |
| Software, algorithm | PAML v4 | http://abacus.gene.ucl.ac.uk/software/paml.html | RRID:SCR_014932 | |
| Software, algorithm | NMRFAM-Sparky | https://nmrfam.wisc.edu/nmrfam-sparky-distribution/ | ||
| Software, algorithm | Xplor-NIH v.2.48 | https://nmr.cit.nih.gov/xplor-nih/ | ||
| Software, algorithm | TALOS-N | https://spin.niddk.nih.gov/bax/software/TALOS-N/ | ||
| Software, algorithm | PyMOL v.1.8 | https://github.com/schrodinger/pymol-open-source | RRID:SCR_000305 |
Additional files
-
Supplementary file 1
FASTA sequences of FITZAP expression constructs.
DNA sequences amplified by fusion PCR and cloned into pET11d for recombinant bacterial expression.
- https://cdn.elifesciences.org/articles/52628/elife-52628-supp1-v2.txt
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/52628/elife-52628-transrepform-v2.docx





