Sex-determining genes distinctly regulate courtship capability and target preference via sexually dimorphic neurons
Figures
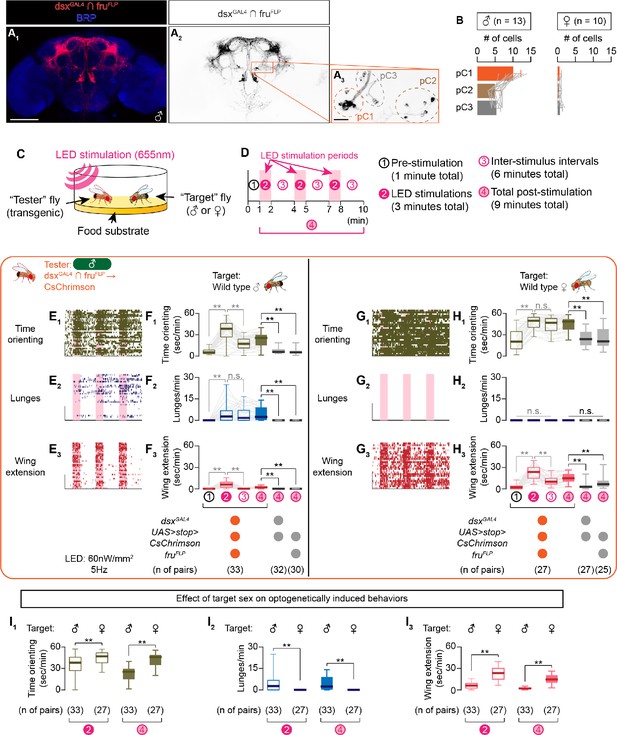
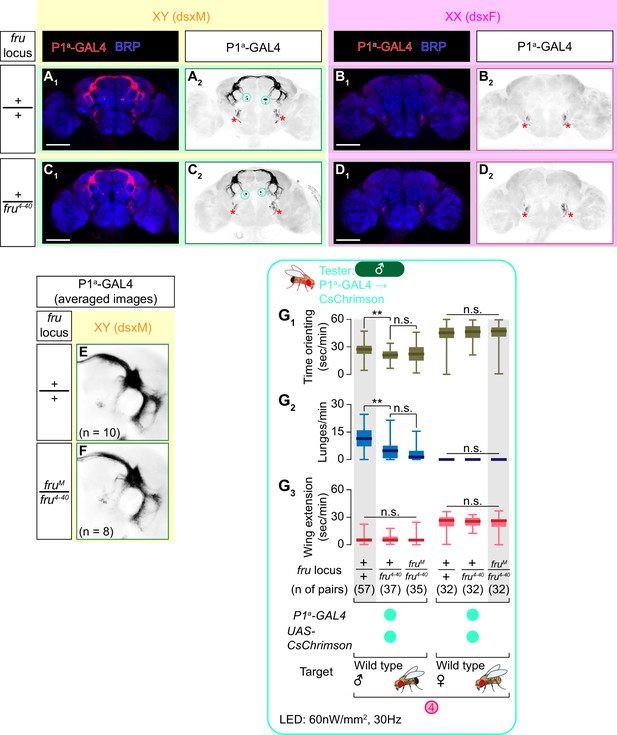
Sex of the target fly influences behaviors triggered by the optogenetic activation of social behavior-promoting neurons.
(A) Expression of CsChrimson:tdTomato under the control of dsxGAL4 and fruFLP (red in A1, black in A2,3) in a male brain is visualized together with a neuropil marker BRP (blue in A1) by immunohistochemistry. Labeled cell body clusters are enlarged in A3. Scale bar: 100 μm (A1), 10 μm (A3). (B) Mean number of cell bodies per hemibrain visualized by anti-DsRed antibody in male (left) and female (right) brains. (C) Schematics of the design of behavioral assays. (D) Schematics of the optogenetic stimulation paradigm. Time windows 1–4 represent periods in which behavioral parameters are pooled and calculated in subsequent panels. (E, G) Rasters of behaviors (indicated in left) performed by male tester flies that express CsChrimson:tdTomato under the control of dsxGAL4 and fruFLP. Pink bar: LED-on periods, horizontal bar: 10 min (also see D), vertical bar: 10 flies. LED stimulation condition is indicated at the bottom. (F, H): Boxplots of time orienting (F1, H1), lunges (F2, H2), and wing extension (F3, H3) by the tester flies during the time windows 1–4 (see D). Testers’ genotypes and pair numbers are indicated below the plots. Gray lines represent single testers. In gray: **p<0.01, n.s. p>0.05 (Kruskal-Wallis one-way ANOVA and post-hoc Wilcoxon signed rank test). In black: **p<0.01, n.s., p>0.05 (Kruskal-Wallis one-way ANOVA and post-hoc Mann-Whitney U-test). Target flies are either wild-type group-housed males (E, F) or mated females (G, H). (I) Comparison of time orienting (I1), lunges (I2), and wing extension (I3) performed by tester flies that express CsChrimson:tdTomato under the control of dsxGAL4 and fruFLP in males (data replotted from F, H) toward male or female target flies (indicated above). Number of pairs tested and time windows compared are indicated below the panels. **p<0.01 (Mann-Whitney U-test). For detailed methods to quantify behaviors, see Materials and methods.
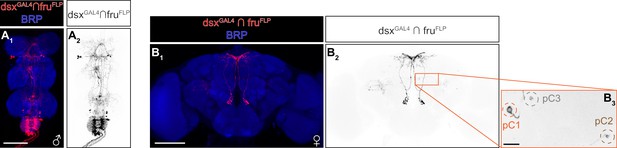
Additional characterization of the dsxGAL4 ∩ fruFLP neurons.
(A, B) Expression of CsChrimson:tdTomato under the control of dsxGAL4 and fruFLP (red in A1, B1, black in A2, B2,3) in a male ventral nerve cord (A) and a female brain (B) is visualized together with a neuropil marker BRP (blue in A1, B1) by immunohistochemistry. Labeled cell body clusters in B2 are enlarged in B3). Scale bar: 100 μm (A1, B1), 10 μm (B3).
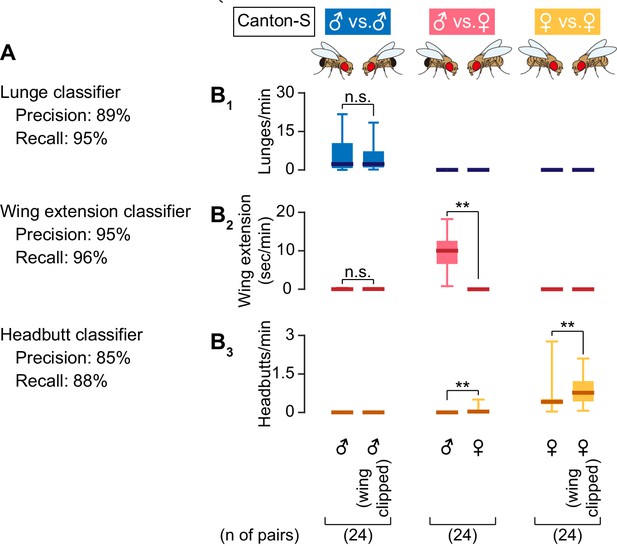
Performance of automated behavior classifiers.
(A) Precision and recall values of the three behavioral classifiers used in this study. See Materials and methods for the definition of precision and recall. (B) Boxplots of lunges (B1), wing extensions (B2), and headbutts (B3), performed by pairs of wild-type male flies (singly reared for 6 days) (left), pairs of a wild-type male and a wild-type mated female (reared in same-sex groups for 6 days) (middle), and pairs of wild-type virgin female flies (singly reared for 6 days) (right) during 30 min assays. For male pairs and virgin female pairs, data from marked flies (whose wing tips were clipped) are shown on the right side. Pair numbers are indicated below the panel.
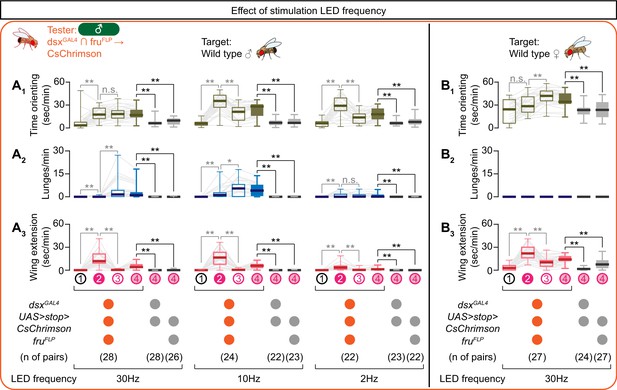
Behaviors induced by optogenetic stimulation of dsxGAL4 ∩ fruFLP neurons across various LED frequencies.
(A, B) Boxplots of time orienting (A1, B1), lunges (A2, B2) and wing extension (A3, B3) by the tester flies during the time windows 1–4 (see Figure 1D). Tester’s genotypes and pair numbers are indicated below the plots. Gray lines represent single testers. LED frequencies were varied as indicated below the plot. Target flies are wild-type group-housed males (A) or mated females (B). In gray: **p<0.01, n.s. p>0.05 (Kruskal-Wallis one-way ANOVA and post-hoc Wilcoxon signed rank test). In black: **p<0.01, n.s. p>0.05 (Kruskal-Wallis one-way ANOVA and post-hoc Mann-Whitney U-test).
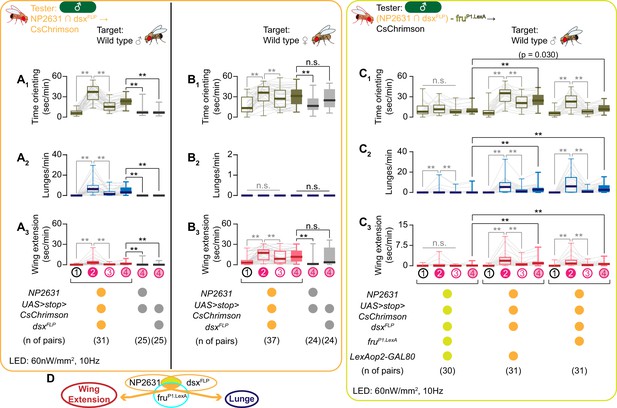
NP2631 ∩ dsxFLP neurons that express both dsx and fru can promote courtship and aggressive behaviors.
(A–C): Boxplots of time orienting (A1–C1), lunges (A2–C2), and wing extension (A3–C3) by the tester flies during the time windows 1–4 (see Figure 1D). Testers’ genotypes and pair numbers are indicated below the plots. Gray lines represent single testers. Target flies are either group-housed wild-type males (A, C) or mated females (B). Gray lines represent single testers. In gray: **p<0.01, n.s. p>0.05 (Kruskal-Wallis one-way ANOVA and post-hoc Wilcoxon signed rank test). In black: **p<0.01, n.s. p>0.05 (Kruskal-Wallis one-way ANOVA and post-hoc Mann-Whitney U-test). (D). A schematic summary. A subset of NP2631 ∩ dsxFLP neurons that express fru (labeled by the fruP1.LexA allele) (orange) can promote both lunges and wing extensions.
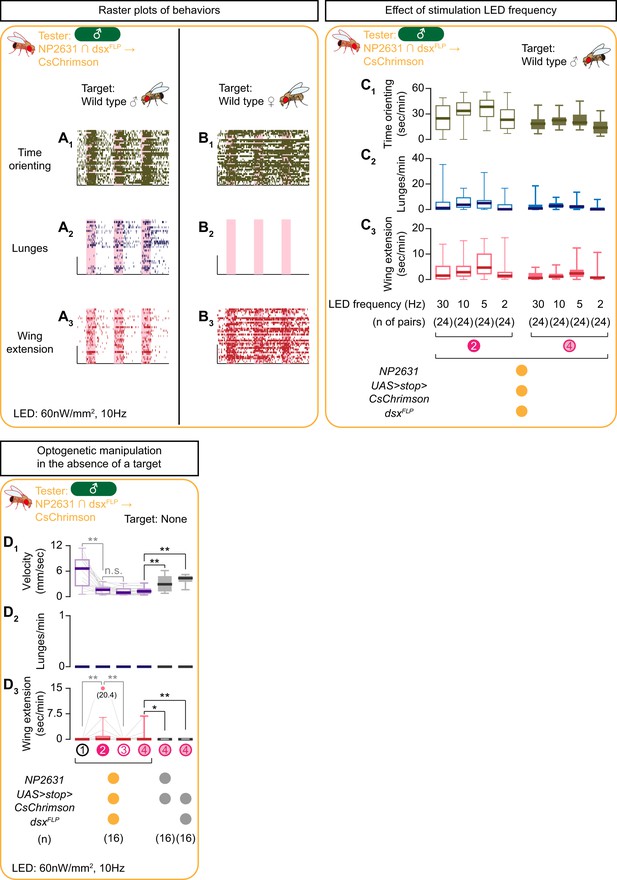
Detailed description of behaviors induced by the optogenetic activation of NP2631 ∩ dsxFLP neurons.
(A, B) Rasters of time orienting (A1, B1), lunges (A2, B2), and wing extensions (A3, B3) performed by male tester flies that express CsChrimson:tdTomato under the control of NP2631 and dsxFLP (the same datasets as used in Figure 2A, (B). Pink bar: LED-on periods, horizontal bar: 10 min, vertical bar: 10 flies. LED stimulation condition is indicated at the bottom. (C): Boxplots of time orienting (C1), lunges (C2), and wing extension (C3) by the tester flies during the time windows 2 (left) and 4 (right) (see Figure 1D). Tester’s genotype (indicated below) is the same as testers in A) and B). LED frequencies were varied as indicated below the plot. Target flies are wild-type group-housed males. (D) Boxplots of velocity (D1), lunges (D2), and wing extensions (D3) performed by the tester flies that express CsChrimson:tdTomato under the control of NP2631 and dsxFLP in the absence of a target fly. Testers’ genotypes and pair numbers are indicated below the plots. .
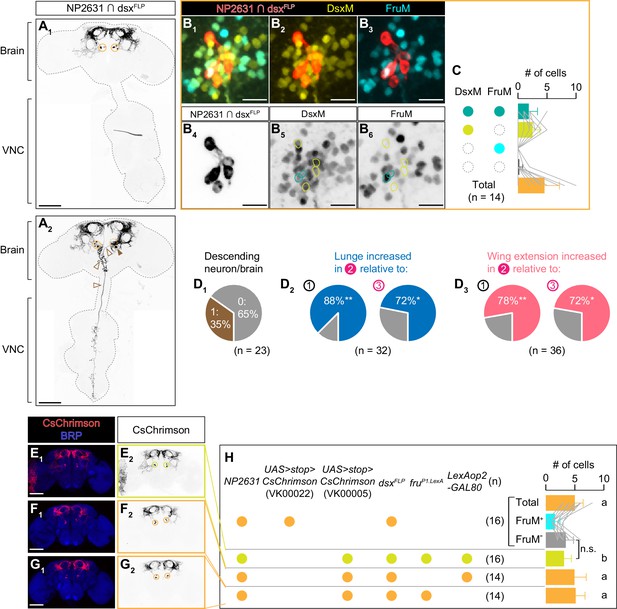
A detailed characterization of NP2631 ∩ dsxFLP neurons.
(A): Expression of CsChrimson:tdTomato in a representative male central nervous system (brain and ventral nerve cord (VNC)) under the control of NP2631 and dsxFLP is visualized by immunohistochemistry (scale bar: 100 µm). The P1/pC1 clusters at the posterior part of the brains are indicated in orange circles. The majority of brains had no other labeled neurons (A1), but 35% (see D1) of the examined brains (A2) also had a single neuron with a large cell body (brown arrowhead) that extend contralateral and descending projections (open brown arrowheads). (B) Representative image of NP2631 ∩ dsxFLP neurons (red in B1–B3), black in B4) visualized by immunohistochemistry against tdTomato along with immunoreactivity against DsxM (yellow in B1, 2, black in B5) and FruM (cyan in B1, 3, black in B6) (scale bar: 10 µm). Two CsChrimson:tdTomato-expressing neurons also express both DsxM and FruM (green circles in B5, 6) whereas four other neurons express DsxM only (cyan circles in B5, 6). (C) Mean number of P1/pC1 NP2631 ∩ dsxFLP neuronal cell bodies per hemibrain, categorized by immunoreactivity against DsxM and FruM. Gray lines represent single hemibrain. Error bars: S.D. (D) Pie charts that shows ratios of flies with a NP2631 ∩ dsxFLP descending neuron (brown in D1), flies that increased lunges toward male targets during LED stimulation (window two in Figure 1D) relative to during pre-stimulation (window one in Figure 1D) (blue in D2) left), or during inter-stimulus intervals (window three in Figure 1D) (blue in D2) right), flies that increased wing extensions toward female targets in window two relative to in window 1 (pink in D3) left), or in window 3 (pink in D3) right), among male flies that express CsChrimson:tdTomato under the control of NP2631 and dsxFLP. Behavioral data are from the same data set used in Figure 3—figure supplement 1I2 (D2) and I3 (D3), **p<0.01, Fisher’s exact test for the frequency of a descending neuron. (E–G) Expression of CsChrimson:tdTomato (red in E1–G1), black in E2–G2) in a representative male brain in genotypes shown in H) is visualized together with the neuropil marker BRP (blue in E1–G1) by immunohistochemistry (scale bar: 100 µm). (H): Mean number of cell bodies per hemibrain visualized by anti-DsRed antibody in each genotype shown in left. Brains from the top genotype were also co-immunostained by an anti-FruM antibody, and the breakdown of FruM-expressing cells and FruM-negative cells is also shown (gray lines represent single hemibrains). Error bars: S.D. Number of hemibrains examined are indicated to the left. Lowercase letters denote significance group (p<0.05, one-way ANOVA with post-hoc Tukey’s honestly significant difference test). n.s. p>0.05, Student’s t-test.
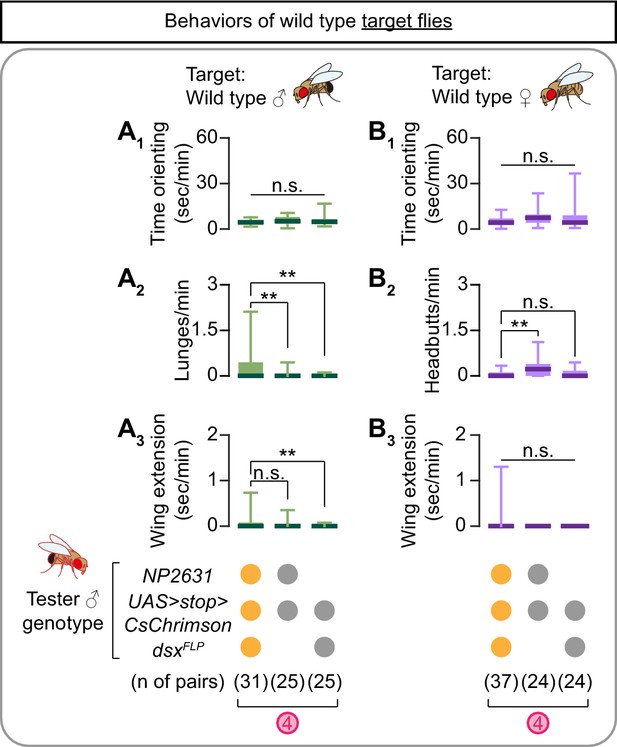
Behaviors of target flies remain largely unaffected by the optogenetic stimulation of NP2631 ∩ dsxFLP neurons in tester flies.
(A, B) Boxplots of time orienting (A1, B1), lunges (A2), headbutts (B2), and wing extension (A3, B3) by the target male (A) or female (B) flies during the time windows 4 (see Figure 1D) of the experiments shown in Figure 2A (A) and 2B (B). Testers’ genotypes and pair numbers are indicated below the plots. **p<0.01, n.s. p>0.05 (Kruskal-Wallis one-way ANOVA and post-hoc Mann-Whitney U-test).
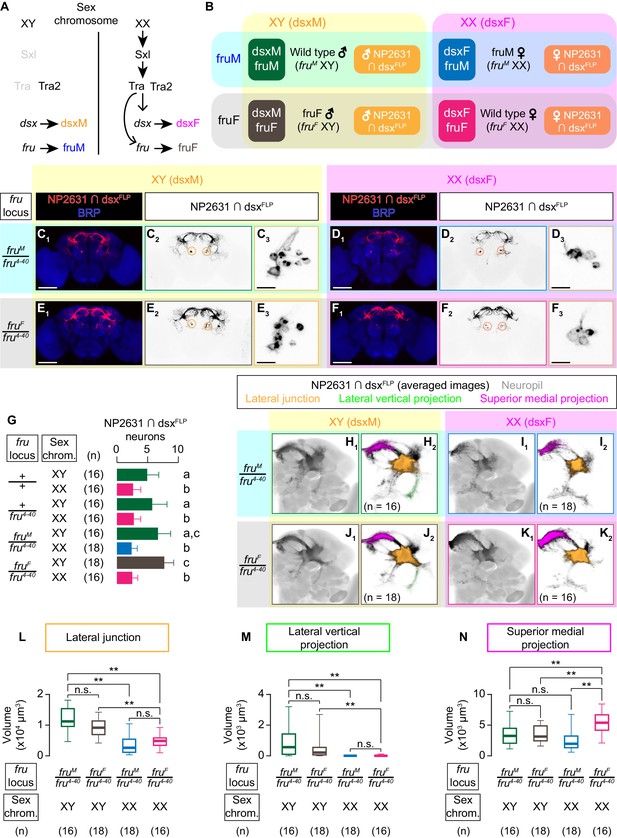
dsx specifies the sexual dimorphism of NP2631 ∩ dsxFLP neurons.
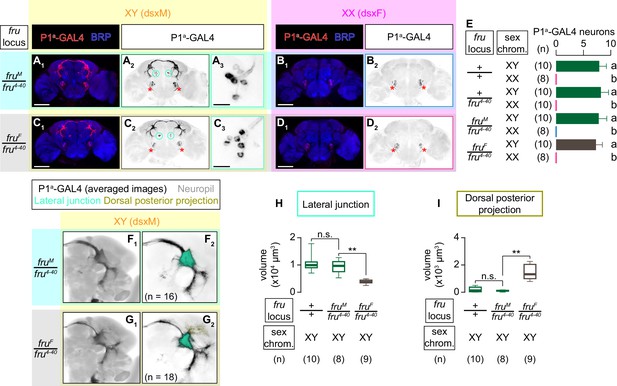
(A) Schematics of the sex-determination pathway in Drosophila. (B) Schematic of the four sex genotypes defined by dsx and fru splicing, and how NP2631 ∩ dsxFLP neurons are specified in each genotype (see following panels for details). (C–F) Expression of CsChrimson:tdTomato under the control of NP2631 and dsxFLP (red in C1–F1, black in C2,3-F2,3) in brains of a male (C), fruM female (D), fruF male (E), and female (F) is visualized together with a neuropil marker BRP (blue) by immunohistochemistry. Circle: soma (right cluster is enlarged in C3–F3). Scale bar: 100 μm (C1–F1), 10 μm (C3–F3). (G) Mean number of cell bodies per hemibrain visualized by anti-DsRed antibody in each genotype represented in C–F) and Figure 3—figure supplement 1A–D. (H–K) Z-projection of the registered and averaged images of CsChrimson:tdTomato expression under the control of NP2631 and dsxFLP (black in H1–K1) in male (H), fruM female (I), fruF male (J), and female (K). A part of the standard Drosophila brain is shown in gray in H2–K2). Number of used hemibrains are indicated in H2–K2). Lateral junction (orange), lateral vertical projection (green), and superior medial projection (magenta) are segmented and overlaid in H2-K2. L-M: Boxplot of volumes of lateral junction (L), lateral vertical projection (M), and superior medial projections (N). Genotypes and number of hemibrains are indicated below the plot. **p<0.01, n.s. p>0.05 (Kruskal-Wallis one-way ANOVA and post-hoc Wilcoxon signed rank test).
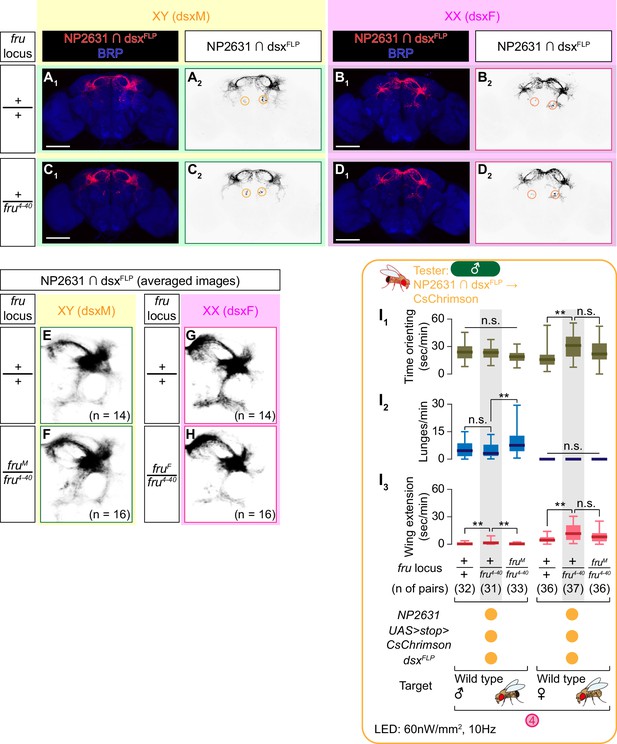
Heterozygosity of the fru4-40 allele leaves the neuroanatomy and function of NP2631 ∩ dsxFLP neurons largely unaltered.
(A–D) Expression of CsChrimson:tdTomato under the control of NP2631 and dsxFLP (red in A1–D1), black in A2,3-D2,3) in representative brains of a fru wild-type male (A), fru wild-type female (B), fru4-40 heterozygous male (C), and fru4-40 heterozygous female (D), is visualized together with a neuropil marker BRP (blue in A1–D1) by immunohistochemistry. Scale bar: 100 μm. Circle: soma. (E–H) Z-projection of the registered and averaged images of CsChrimson:tdTomato expression under the control of NP2631 and dsxFLP in fru wild-type male (E), fruM/fru4-40 male (F), duplication from Figure 3H), fru wild-type female (G), and fruF/fru4-40 female (H), duplication from Figure 3K). Number of used hemibrains are indicated. (I) Boxplots of time orienting (I1), lunges (I2), and wing extensions (I3) performed by male tester flies that express CsChrimson:tdTomato under the control of NP2631 and dsxFLP. Only values during the time window four are shown. Testers’ fru locus genotypes and pair numbers are indicated below the plots. Target flies are either group-housed wild-type males or mated females, as indicated. Plots with gray shades are replots of the data sets shown in Figure 2A and B. **p<0.01, n.s. p>0.05 (Kruskal-Wallis one-way ANOVA and post-hoc Mann-Whitney U-test).
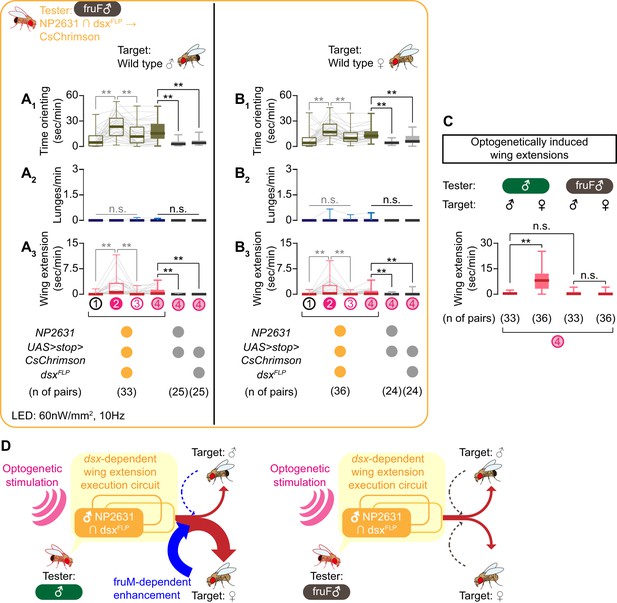
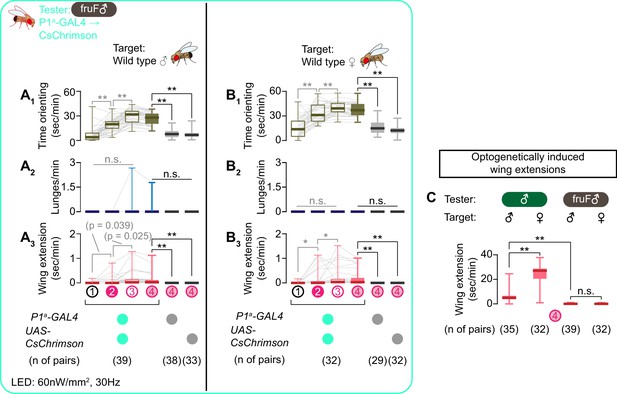
NP2631 ∩ dsxFLP neurons in fruF males promote courtship, but not aggressive, behavior.
(A, B) Boxplots of time orienting (A1, B1), lunges (A2, B2), and wing extension (A3, B3) by the tester flies during the time windows 1–4 (see Figure 1D). Testers’ genotypes and pair numbers are indicated below the plots. Gray lines represent single testers. Target flies are either group-housed wild-type males (A) or mated females (B). In gray: **p<0.01, n.s. p>0.05 (Kruskal-Wallis one-way ANOVA and post-hoc Wilcoxon signed rank test). In black: **p<0.01, n.s. p>0.05 (Kruskal-Wallis one-way ANOVA and post-hoc Mann-Whitney U-test). (C) Comparison of wing extension performed by flies that express CsChrimson:tdTomato under the control of NP2631 and dsxFLP in males (data from Figure 3—figure supplement 1I) or in fruF males (data from A, (B), during the time window 4. Sex of tester and target flies is indicated above the panels. Number of pairs tested is indicated below the panels. **p<0.01, n.s. p>0.05 (Mann-Whitney U-test). (D) Models of the function of NP2631 ∩ dsxFLP neurons in the context of dsx and fru.
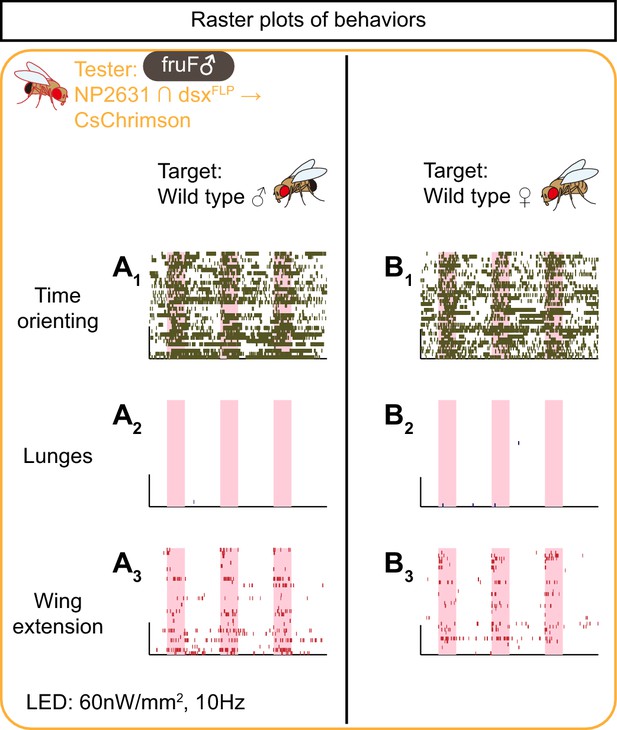
Rasters of behaviors induced by the optogenetic activation of NP2631 ∩ dsxFLP neurons in fruF males.
(A, B) Rasters of time orienting (A1, B1), lunges (A2, B2), and wing extensions (A3, B3) performed by fruF male tester flies that express CsChrimson:tdTomato under the control of NP2631 and dsxFLP (the same datasets as used in Figure 4A, (B). Pink bar: LED-on periods, horizontal bar: 10 min, vertical bar: 10 flies. LED stimulation condition is indicated at the bottom.
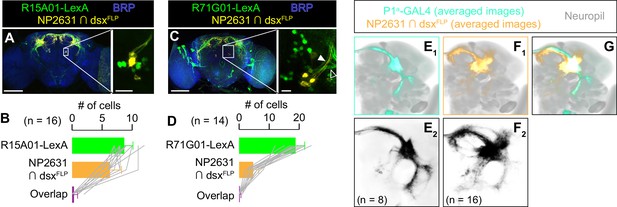
P1a neurons are distinct from NP2631 ∩ dsxFLP neurons.
(A, C) Expression of CsChrimson:tdTomato under the control of NP2631 and dsxFLP (yellow), GCaMP6f under the control of R15A01-LexA (green in A) or R71G01-LexA (green in C), and a neuropil marker BRP (blue) in a representative male brain are visualized by immunohistochemistry. Scale bar: 100 μm. An inset represents a magnified view of the posterior cell body cluster in the white rectangle (scale bar: 10 μm). An open arrowhead in C) indicates a neural tract from LexA-expressing neurons that appear distinct from the tract from neurons labeled by NP2631 and dsxFLP (white arrowhead). (B, D) Mean number of cell bodies per hemibrain with immunohistochemical signal by anti-DsRed antibody (orange), anti-GFP antibody (green), and both antibodies (purple) in brains of the genotype represented in A (B) or in C (D). In D, all LexA-expressing neurons located near NP2631 and dsxFLP neurons are included, although some may belong to different neuronal clusters. Error bars: S.D. Gray lines represent single hemibrains. (E–G) Z-projection of the registered and averaged images of CsChrimson:tdTomato expression under the control of P1a-GAL4 (cyan in E1, G, black in E2), and NP2631 and dsxFLP (orange in F1, G, black in F2; duplication from Figure 3H) neurons. A part of the standard Drosophila brain is shown in gray. Number of hemibrains used are indicated in E2 and F2.
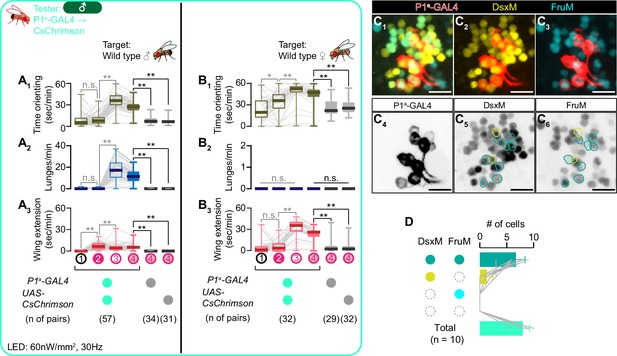
Detailed characterization of P1a neurons.
(A, B) Boxplots of time orienting (A1, B1), lunges (A2, B2), and wing extension (A3, B3) by the tester flies during the time windows 1–4 (see Figure 1D). Testers’ genotypes and pair numbers are indicated below the plots. Gray lines represent single testers. Target flies are either group-housed wild-type males (A) or mated females (B). Gray lines represent single testers. In gray: **p<0.01, *p<0.05, n.s. p>0.05 (Kruskal-Wallis one-way ANOVA and post-hoc Wilcoxon signed rank test). In black: **p<0.01, n.s. p>0.05 (Kruskal-Wallis one-way ANOVA and post-hoc Mann-Whitney U-test). (C) Representative image of P1a neurons (red in C1–C3, black in C4) visualized by immunohistochemistry against tdTomato along with immunoreactivity against DsxM (yellow in C1, 2, black in C5) and FruM (cyan in C1, 3, black in C6) (scale bar: 10 µm). Eight CsChrimson:tdTomato-expressing neurons also express both DsxM and FruM (green circles in C5, 6) whereas two other neurons express DsxM only (cyan circles in C5, 6). (D) Mean number of P1a neuronal cell bodies per hemibrain, categorized by immunoreactivity against DsxM and FruM. Gray lines represent single hemibrains. Error bars: S.D.
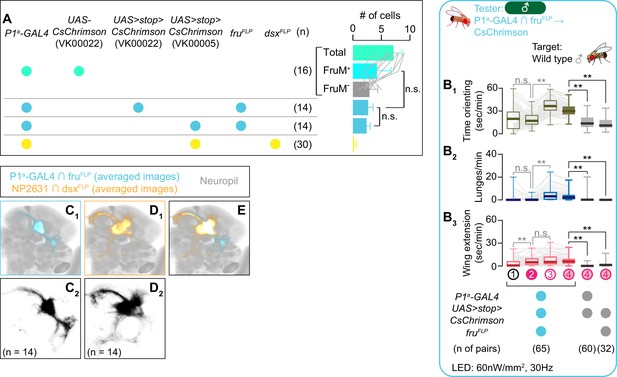
Detailed characterization of P1a ∩ fruFLP neurons.
(A) Mean number of cell bodies per hemibrain visualized by anti-DsRed antibody in each genotype shown in left. Brains from the top genotype were also co-immunostained by an anti-FruM antibody, and the breakdown of FruM-expressing cells and FruM-negative cells is also shown (gray lines represent single hemibrains). Error bars: S.D. Number of hemibrains examined are indicated to the left. Lowercase letters denote significance group (p<0.05, one-way ANOVA with post-hoc Tukey’s honestly significant difference test). n.s. p>0.05, Student’s t-test. (B) Boxplots of time orienting (B1), lunges (B2), and wing extension (B3) by the tester flies during the time windows 1–4 (see Figure 1D). Testers’ genotypes and pair numbers are indicated below the plots. Gray lines represent single testers. Target flies are group-housed wild-type males. Gray lines represent single testers. In gray: **p<0.01, *p<0.05, n.s. p>0.05 (Kruskal-Wallis one-way ANOVA and post-hoc Wilcoxon signed rank test). In black: **p<0.01, n.s. p>0.05 (Kruskal-Wallis one-way ANOVA and post-hoc Mann-Whitney U-test). (C–E) Z-projection of the registered and averaged images of CsChrimson:tdTomato expression under the control of P1a-GAL4 and fruFLP (cyan in C, E) and NP2631 and dsxFLP (orange in D, E; duplication from Figure 3—figure supplement 1E) neurons. A part of the standard Drosophila brain is shown in gray. Number of used hemibrains are indicated in C2 and D2. Note that the genotype of animals in B (cyan) and C is identical to the 3rd genotype from the top in A.
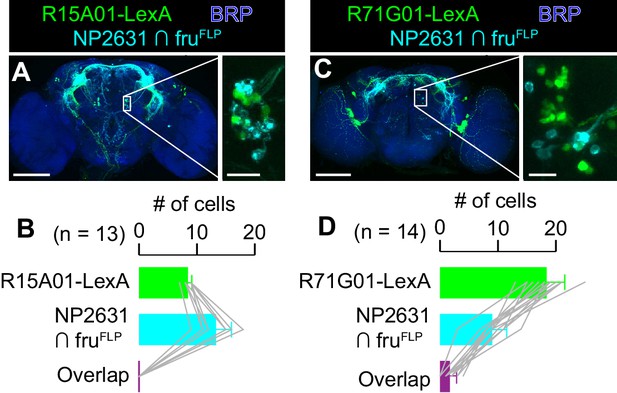
P1a and NP2631 ∩ fruFLP neurons do not overlap.
(A, C) Expression of CsChrimson:tdTomato under the control of NP2631 and fruFLP (cyan), GCaMP6f under the control of R15A01-LexA (green in A) or R71G01-LexA (green in C), and a neuropil marker BRP (blue) in a representative male brain are visualized by immunohistochemistry. Scale bar: 100 μm. An inset represents magnified view of the posterior cell body cluster in the white rectangle (scale bar: 10 μm). (B, D) Mean number of cell bodies per hemibrain with immunohistochemical signal by anti-DsRed antibody (cyan), anti-GFP antibody (green), and both antibodies (purple) in brains of the genotype represented in A (B) or in C (D). Error bars: S.D. Gray lines represent single hemibrains.
Both dsx and fru specify the sexual dimorphism of P1a neurons.
(A–D) Expression of CsChrimson:tdTomato under the control of P1a-GAL4 (red in A1–D1, black in A2,3-D2,3) in brains of a male (A), fruM female (B), fruF male (C), and female (D) is visualized together with a neuropil marker BRP (blue) by immunohistochemistry. Circle: soma (right cluster is enlarged in A3 and C3), red asterisk: sex-invariant background labeling (see Materials and methods for details). Scale bar: 100 μm (A1–D1), 10 μm (A3, C3). (E) Mean number of cell bodies per hemibrain visualized by anti-DsRed antibody in each genotype represented in A–D) and Figure 6—figure supplement 1A–D. (F, G) Z-projection of the registered and averaged images of CsChrimson:tdTomato expression under the control of P1a-GAL4 in male (F) and fruF male (G). A part of the standard Drosophila brain is shown in gray in F2, G2. Number of hemibrains used are indicated in F2) and G2). Lateral junction (cyan) and dorsal posterior projection (yellow) are segmented and overlaid in F2) and G2. H, I. Boxplot of volumes of lateral junction (H) and dorsal posterior projection (I). Genotypes and number of hemibrains are indicated below the plot. **p<0.01, n.s. p>0.05 (Kruskal-Wallis one-way ANOVA and post-hoc Mann-Whitney U-test).
Heterozygosity of the fru4-40 allele leaves the neuroanatomy and function of P1a neurons largely unaltered.
(A–D) Expression of CsChrimson:tdTomato under the control of P1a (red in A1–D1), black in A2,3-D2,3) in representative brains of a fru wild-type male (A), fru wild-type female (B), fru4-40 heterozygous male (C), and fru4-40 heterozygous female (D), is visualized together with a neuropil marker BRP (blue in A1–D1) by immunohistochemistry. Scale bar: 100 μm. Circle: soma, red asterisk: sex-invariant background labeling.( E, F) Z-projection of the registered and averaged images of CsChrimson:tdTomato expression under the control of P1a in fru wild-type male (E) duplication from Figure 5E) and fruM/fru4-40 male (F); duplication from Figure 6F). Number of used hemibrains are indicated. (G) Boxplots of time orienting (G1), lunges (G2), and wing extensions (G3) performed by male tester flies that express CsChrimson:tdTomato under the control of P1a. Only values during the time window four are shown (see Figure 1D). Testers’ fru locus genotypes and pair numbers are indicated below the plots. Target flies are either group-housed wild-type males or mated females, as indicated. Plots with gray shades are replots of the data sets shown in Figure 5—figure supplement 1A,B. **p<0.01, n.s. p>0.05 (Kruskal-Wallis one-way ANOVA and post-hoc Mann-Whitney U-test).
P1a neurons in fruF males are defective in promoting both courtship and aggressive behavior.
(A, B) Boxplots of time orienting (A1, B1), lunges (A2, B2), and wing extension (A3, B3) by the tester flies during the time windows 1–4 (see Figure 1D). Their genotypes and numbers are indicated below the plots. Gray lines represent single testers. Target flies are either group-housed wild-type males (A) or mated females (B). In gray: **p<0.01, *p<0.05, n.s. p>0.05 (Kruskal-Wallis one-way ANOVA and post-hoc Wilcoxon signed rank test). In black: **p<0.01, n.s. p>0.05 (Kruskal-Wallis one-way ANOVA and post-hoc Mann-Whitney U-test). (C) Comparison of wing extension performed by flies that express CsChrimson:tdTomato under the control of P1a in males (data from Figure 6—figure supplement 1G) or in fruF males (data from A, (B), during the time window 4. Sex of tester and target flies is indicated above the panels. Number of pairs tested is indicated below the panels. **p<0.01, n.s. p>0.05 (Mann-Whitney U-test).
Videos
Representative behavior of a male tester fly that expresses.
CsChrimson:tdTomato under the control of dsxGAL4 and fruFLP toward a wild-type male (Part 1) or a wild-type female (Part 2) target fly, at the onset and offset of LED stimulation.
Representative behavior of a male tester fly that expresses.
CsChrimson:tdTomato under the control of NP2631 and dsxFLP toward a wild-type male (Part 1) or a wild-type female (Part 2) target fly, at the onset and offset of LED stimulation.
3D-rendered average image of NP2631 ∩ dsxFLP neurons in male (green) and in female (magenta).
Gray represents a standard unisex Drosophila brain (Bogovic et al., 2018).
3D-rendered average image of NP2631 ∩ dsxFLP neurons in male (green; duplication from Video 3) and in fruF male (yellow).
Gray represents a standard unisex Drosophila brain (Bogovic et al., 2018).
3D-rendered average image of NP2631 ∩ dsxFLP neurons in female (magenta, duplication from Video 3) and in fruM female (blue).
Gray represents a standard unisex Drosophila brain (Bogovic et al., 2018).
Representative behavior of a fruF male tester fly that expresses.
CsChrimson:tdTomato under the control of NP2631 and dsxFLP toward a wild-type male (Part 1) or a wild-type female (Part 2) target fly, at the onset and offset of LED stimulation.
3D-rendered average image of P1a neurons (cyan) and NP2631 ∩ dsxFLP neurons (orange) in male.
Gray represents a standard unisex Drosophila brain (Bogovic et al., 2018).
3D-rendered average image of P1a neurons in male (green) and in fruF male (yellow).
Gray represents a standard unisex Drosophila brain (Bogovic et al., 2018).
Tables
Complete genotypes of Drosophila strains used in this study.
| Figure | Panel | Abbreviated genotype | COMPLETE GENOTYPE (‘Y’ represents the Y chromosome) |
|---|---|---|---|
| Figure 1 | A, B | dsxGAL4 ∩ fruFLP ♂ | w/Y; 20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00022/+; dsxGAL4/fruFLP |
| Figure 1—figure supplement 1 | A | ||
| Video 1 | |||
| Figure 1 | B | dsxGAL4 ∩ fruFLP ♀ | w/w; 20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00022/+; dsxGAL4/fruFLP |
| Figure 1—figure supplement 1 | B | ||
| Figure 1 | E-I | dsxGAL4, UAS > stop > CsChrimson, fruFLP | w/Y; 20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00022/+; dsxGAL4/fruFLP |
| Figure 1—figure supplement 1 | C, D | ||
| Figure 1 | E-H | dsxGAL4, UAS > stop > CsChrimson | w/Y; 20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00022/+; dsxGAL4/+ |
| Figure 1—figure supplement 1 | C, D | ||
| Figure 1 | E-H | UAS > stop > CsChrimson, fruFLP | w/Y; 20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00022/+; fruFLP/+ |
| Figure 1—figure supplement 1 | C, D | ||
| Figure 1—figure supplement 2 | B | Canton-S ♂ | +/Y; +/+; +/+ (Canton-S) |
| Figure 1—figure supplement 2 | B | Canton-S ♀ | +/+; +/+; +/+ (Canton-S) |
| Figure 2 | A, B | NP2631, UAS > stop > CsChrimson, dsxFLP | w/Y; NP2631/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00022; dsxFLP, fru4-40/+ |
| Figure 2—figure supplement 1 | A-C | ||
| Figure 2—figure supplement 3 | A, B | ||
| Video 2 | |||
| Figure 2 | A, B | NP2631, UAS > stop > CsChrimson | w/Y; NP2631/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00022; fru4-40/+ |
| Figure 2—figure supplement 3 | A, B | ||
| Figure 2 | A, B | UAS > stop > CsChrimson, dsxFLP | w/Y; +/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00022; dsxFLP, fru4-40/+ |
| Figure 2—figure supplement 3 | A, B | ||
| Figure 2 | C | NP2631, UAS > stop > CsChrimson, dsxFLP, fruP1. LexA, LexAop2-GAL80 | w/Y; NP2631/8XLexAop2-GAL80 in attP40; 20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00005, dsxFLP/fruP1. LexA |
| Figure 2 | C | NP2631, UAS > stop > CsChrimson, dsxFLP, LexAop2-GAL80 | w/Y; NP2631/8XLexAop2-GAL80 in attP40; 20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00005, dsxFLP/+ |
| Figure 2 | C | NP2631, UAS > stop > CsChrimson, dsxFLP, fruP1. LexA | w/Y; NP2631/+; 20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00005, dsxFLP/fruP1. LexA |
| Figure 2—figure supplement 1 | A-C | NP2631, UAS > stop > CsChrimson, dsxFLP | w/Y; NP2631/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00022; dsxFLP, fru4-40/+ |
| Figure 2—figure supplement 2 | A-D | NP2631 ∩ dsxFLP | w/Y; NP2631/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00022; dsxFLP/+ |
| Figure 2—figure supplement 2 | E, H | NP2631, UAS > stop > CsChrimson (VK00005), dsxFLP, fruP1. LexA, LexAop2-GAL80 | w/Y; NP2631/8XLexAop2-GAL80 in attP40; 20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00005, dsxFLP/fruP1. LexA |
| Figure 2—figure supplement 2 | F, H | NP2631, UAS > stop > CsChrimson (VK00005), dsxFLP, LexAop2-GAL80 | w/Y; NP2631/8XLexAop2-GAL80 in attP40; 20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00005, dsxFLP/+ |
| Figure 2—figure supplement 2 | G, H | NP2631, UAS > stop > CsChrimson (VK00005), dsxFLP, fruP1. LexA | w/Y; NP2631/+; 20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00005, dsxFLP/fruP1. LexA |
| Figure 3 | C, G, H, L-N | NP2631 ∩ dsxFLP, XY, fru locus: fruM/fru4-40 | w/Y; NP2631/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00022; dsxFLP, fru4-40/fruM |
| Figure 3—figure supplement 1 | F | ||
| Video 3, 4 | |||
| Figure 3 | D, G, I, L-N | NP2631 ∩ dsxFLP, XX, fru locus: fruM/fru4-40 | w/w; NP2631/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00022; dsxFLP, fru4-40/fruM |
| Video 5 | |||
| Figure 3 | E, G, J, L-N | NP2631 ∩ dsxFLP, XY, fru locus: fruF/fru4-40 | w/Y; NP2631/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00022; dsxFLP, fru4-40/fruF |
| Video 4 | |||
| Figure 3 | F, G, K-N | NP2631 ∩ dsxFLP, XX, fru locus: fruF/fru4-40 | w/w; NP2631/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00022; dsxFLP, fru4-40/fruF |
| Figure 3—figure supplement 1 | H | ||
| Video 3, 5 | |||
| Figure 3 | G | NP2631 ∩ dsxFLP, XY, fru locus: +/+ | w/Y; NP2631/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00022; dsxFLP/+ |
| Figure 3—figure supplement 1 | A, E | ||
| Figure 3 | G | NP2631 ∩ dsxFLP, XX, fru locus: +/+ | w/w; NP2631/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00022; dsxFLP/+ |
| Figure 3—figure supplement 1 | B, G | ||
| Figure 3 | G | NP2631 ∩ dsxFLP, XY, fru locus: +/fru4-40 | w/Y; NP2631/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00022; dsxFLP, fru4-40/+ |
| Figure 3—figure supplement 1 | C | ||
| Figure 3 | G | NP2631 ∩ dsxFLP, XX, fru locus: +/fru4-40 | w/w; NP2631/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00022; dsxFLP, fru4-40/+ |
| Figure 3—figure supplement 1 | D | ||
| Figure 3—figure supplement 1 | I | NP2631, UAS > stop > CsChrimson, dsxFLP, fru locus: +/+ | w/Y; NP2631/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00022; dsxFLP/+ |
| Figure 3—figure supplement 1 | I | NP2631, UAS > stop > CsChrimson, dsxFLP, fru locus: +/fru4-40 | w/Y; NP2631/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00022; dsxFLP, fru4-40/+ |
| Figure 3—figure supplement 1 | I | NP2631, UAS > stop > CsChrimson, dsxFLP, fru locus: fruM/fru4-40 | w/Y; NP2631/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00022; dsxFLP, fru4-40/fruM |
| Figure 4 | A-C | fruF ♂, NP2631, UAS > stop > CsChrimson, dsxFLP | w/Y; NP2631/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00022; dsxFLP, fru4-40/fruF |
| Figure 4—figure supplement 1 | A, B | ||
| Video 6 | |||
| Figure 4 | A, B | fruF ♂, NP2631, UAS > stop > CsChrimson | w/Y; NP2631/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00022; fru4-40/fruF |
| Figure 4 | A, B | fruF ♂, UAS > stop > CsChrimson, dsxFLP | w/Y; +/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00022; dsxFLP, fru4-40/fruF |
| Figure 4 | C | ♂, NP2631, UAS > stop > CsChrimson,dsxFLP | w/Y; NP2631/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00022; dsxFLP, fru4-40/fruM |
| Figure 5 | A, B | NP2631 ∩ dsxFLP, R15A01-LexA | w/Y; NP2631/13XLexAop2-IVS-GCaMP6f-p10 in su(Hw)attP5; R15A01-LexA in attP2/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00005, dsxFLP |
| Figure 5 | C, D | NP2631 ∩ dsxFLP, R71G01-LexA | w/Y; NP2631/13XLexAop2-IVS-GCaMP6f-p10 in su(Hw)attP5; 20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00005, dsxFLP/R71G01-LexA in attP2 |
| Figure 5 | E, G | P1a-GAL4 | w/Y; R15A01-p65AD:Zp in attP40/20XUAS-IVS-CsChrimson:tdTomato in VK00022; R71G01-Zp:GAL4DBD in attP2/+ |
| Video 7 | |||
| Figure 5 | F, G | NP2631 ∩ dsxFLP | w/Y; NP2631/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00022; dsxFLP, fru4-40/fruM |
| Video 7 | |||
| Figure 5—figure supplement 1 | A | P1a-GAL4, UAS-CsChrimson | w/Y; R15A01-p65AD:Zp in attP40/20XUAS-IVS-CsChrimson:tdTomato in VK00022; R71G01-Zp:GAL4DBD in attP2/+ |
| Figure 5—figure supplement 1 | A | P1a-GAL4 | w/Y; R15A01-p65AD:Zp in attP40/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00022; R71G01-Zp:GAL4DBD in attP2/+ |
| Figure 5—figure supplement 1 | A | UAS-CsChrimson | w/Y; +/20XUAS-IVS-CsChrimson:tdTomato in VK00022; +/+ |
| Figure 5—figure supplement 1 | B | P1a-GAL4, UAS-CsChrimson | w/Y; R15A01-p65AD:Zp in attP40/20XUAS-IVS-CsChrimson:tdTomato in VK00022; R71G01-Zp:GAL4DBD in attP2, fru4-40/fruM |
| Figure 5—figure supplement 1 | B | P1a-GAL4 | w/Y; R15A01-p65AD:Zp in attP40/10XUAS-IVS-GFP-p10 in VK00022; R71G01-Zp:GAL4DBD in attP2, fru4-40/fruM |
| Figure 5—figure supplement 1 | B | UAS-CsChrimson | w/Y; +/20XUAS-IVS-CsChrimson:tdTomato in VK00022; fru4-40/fruM |
| Figure 5—figure supplement 1 | C, D | P1a-GAL4 | w/Y; R15A01-p65AD:Zp in attP40/20XUAS-IVS-CsChrimson:tdTomato in VK00022; R71G01-Zp:GAL4DBD in attP2/+ |
| Figure 5—figure supplement 2 | A | P1a-GAL4, UAS-CsChrimson (VK00022) | w/Y; R15A01-p65AD:Zp in attP40/20XUAS-IVS-CsChrimson:tdTomato in VK00022; R71G01-Zp;GAL4DBD in attP2/+ |
| Figure 5—figure supplement 2 | A | P1a-GAL4, UAS > stop > CsChrimson (VK00022), fruFLP | w/Y; R15A01-p65AD:Zp in attP40/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00022; R71G01-Zp;GAL4DBD in attP2/fruFLP |
| Figure 5—figure supplement 2 | A | P1a-GAL4, UAS > stop > CsChrimson (VK00005), fruFLP | w/Y; R15A01-p65AD:Zp in attP40/+; 71G01-Zp;GAL4DBD in attP2/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00005, fruFLP |
| Figure 5—figure supplement 2 | A | P1a-GAL4, UAS > stop > CsChrimson (VK00005), dsxFLP | w/Y; R15A01-p65AD:Zp in attP40/+; 71G01-Zp;GAL4DBD in attP2/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00005, dsxFLP |
| Figure 5—figure supplement 2 | C, E | P1a-GAL4 ∩ fruFLP | w/Y; R15A01-p65AD:Zp in attP40/+; 71G01-Zp;GAL4DBD in attP2/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00005, fruFLP |
| Figure 5—figure supplement 2 | D, E | NP2631 ∩ dsxFLP | w/Y; NP2631/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00022; dsxFLP/+ |
| Figure 5—figure supplement 2 | B | P1a-GAL4,UAS > stop > CsChrimson, fruFLP | w/Y; R15A01-p65AD:Zp in attP40/+;R71G01-Zp:GAL4DBD in attP2/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00005, fruFLP |
| Figure 5—figure supplement 2 | B | P1a-GAL4, UAS > stop > CsChrimson | w/Y; R15A01-p65AD:Zp in attP40/+; R71G01-Zp;GaL4DBD in attP2/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00005 |
| Figure 5—figure supplement 2 | B | UAS > stop > CsChrimson,fruFLP | w/Y; R15A01-p65AD:Zp in attP40/+; +/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00005, fruFLP |
| Figure 5—figure supplement 3 | A, B | NP2631 ∩ fruFLP, R15A01-LexA | w/Y; NP2631/13XLexAop2-IVS-GCaMP6f-p10 in su(Hw)attP5; R15A01-LexA in attP2/20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00005, fruFLP |
| Figure 5—figure supplement 3 | C, D | NP2631 ∩ fruFLP, R71G01-LexA | w/Y; NP2631/13XLexAop2-IVS-GCaMP6f-p10 in su(Hw)attP5; 20XUAS > myr:TopHAT2 > CsChrimson:tdTomato in VK00005, fruFLP/R71G01-LexA in attP2 |
| Figure 6 | A, E, F, H, I | P1a-GAL4, XY, fru locus: fruM/fru4-40 | w/Y; R15A01-p65AD:Zp in attP40/20XUAS-IVS-CsChrimson:tdTomato in VK00022; R71G01-Zp:GAL4DBD in attP2, fru4-40/fruM |
| Figure 6—figure supplement 1 | F | ||
| Figure 6 | B, E | P1a-GAL4, XX, fru locus:fruM/fru4-40 | w/w; R15A01-p65AD:Zp in attP40/20XUAS-IVS-CsChrimson:tdTomato in VK00022; R71G01-Zp:GAL4DBD in attP2, fru4-40/fruM |
| Figure 6 | C, E, G-I | P1a-GAL4, XY, fru locus: fruF/fru4-40 | w/Y; R15A01-p65AD:Zp in attP40/20XUAS-IVS-CsChrimson:tdTomato in VK00022; R71G01-Zp:GAL4DBD in attP2, fru4-40/fruF |
| Figure 6 | D, E | P1a-GAL4, XX, fru locus:fruF/fru4-40 | w/w; R15A01-p65AD:Zp in attP40/20XUAS-IVS-CsChrimson:tdTomato in VK00022; R71G01-Zp:GAL4DBD in attP2, fru4-40/fruF |
| Figure 6 | E, H, I | P1a-GAL4, XY, fru locus: +/+ | w/Y; R15A01-p65AD:Zp in attP40/20XUAS-IVS-CsChrimson:tdTomato in VK00022; R71G01-Zp:GAL4DBD in attP2/+ |
| Figure 6—figure supplement 1 | A, E | ||
| Figure 6 | E | P1a-GAL4, XX, fru locus: +/+ | w/w; R15A01-p65AD:Zp in attP40/20XUAS-IVS-CsChrimson:tdTomato in VK00022; R71G01-Zp:GAL4DBD in attP2/+ |
| Figure 6—figure supplement 1 | B | ||
| Figure 6 | E | P1a-GAL4, XY, fru locus: +/fru4-40 | w/Y; R15A01-p65AD:Zp in attP40/20XUAS-IVS-CsChrimson:tdTomato in VK00022; R71G01-Zp:GAL4DBD in attP2, fru4-40/+ |
| Figure 6—figure supplement 1 | C | ||
| Figure 6 | E | P1a-GAL4, XX, fru locus: +/fru4-40 | w/w; R15A01-p65AD:Zp in attP40/20XUAS-IVS-CsChrimson:tdTomato in VK00022; R71G01-Zp:GAL4DBD in attP2, fru4-40/+ |
| Figure 6—figure supplement 1 | D | ||
| Figure 6—figure supplement 1 | G | P1a-GAL4, UAS-CsChrimson, fru locus: +/+ | w/Y; R15A01-p65AD:Zp in attP40/20XUAS-IVS-CsChrimson:tdTomato in VK00022; R71G01-Zp:GAL4DBD in attP2, +/+ |
| Figure 6—figure supplement 1 | G | P1a-GAL4, UAS-CsChrimson, fru locus: +/fru4-40 | w/Y; R15A01-p65AD:Zp in attP40/20XUAS-IVS-CsChrimson:tdTomato in VK00022; R71G01-Zp:GAL4DBD in attP2, fru4-40/+ |
| Figure 6—figure supplement 1 | G | P1a-GAL4, UAS-CsChrimson, fru locus: fruM/fru4-40 | w/Y; R15A01-p65AD:Zp in attP40/20XUAS-IVS-CsChrimson:tdTomato in VK00022; R71G01-Zp:GAL4DBD in attP2, fru4-40/fruF |
| Figure 6—figure supplement 2 | A-C | fruF ♂, P1a-GAL4, UAS-CsChrimson | w/Y; R15A01-p65AD:Zp in attP40/20XUAS-IVS-CsChrimson:tdTomato in VK00022; R71G01-Zp:GAL4DBD in attP2, fru4-40/fruF |
| Figure 6—figure supplement 2 | A | fruF ♂, P1a-GAL4 | w/Y; R15A01-p65AD:Zp in attP40/10XUAS-IVS-GFP-p10 in VK00022; R71G01-Zp:GAL4DBD in attP2, fru4-40/fruF |
| Figure 6—figure supplement 2 | A | fruF ♂, UAS-CsChrimson | w/Y; R15A01-p65AD:Zp in attP40/20XUAS-IVS-CsChrimson:tdTomato in VK00022; fru4-40/fruF |
| Figure 6—figure supplement 2 | C | ♂, P1a-GAL4, UAS-CsChrimson | |
Additional files
-
Source data 1
This file contains data points represented in all figure panels, as well as p-values for all statistical tests shown in the figures.
- https://cdn.elifesciences.org/articles/52701/elife-52701-data1-v1.xlsx
-
Supplementary file 1
Key Resources Table.
- https://cdn.elifesciences.org/articles/52701/elife-52701-supp1-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/52701/elife-52701-transrepform-v1.docx





