The switch-like expression of heme-regulated kinase 1 mediates neuronal proteostasis following proteasome inhibition
Figures
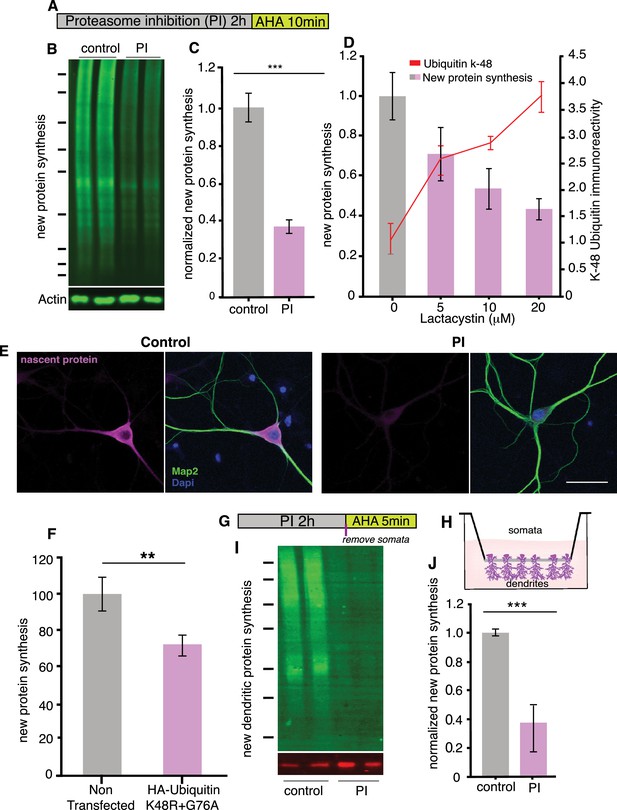
Proteasome inhibition leads to a coordinate inhibition of protein synthesis in neurons.
(A) Scheme indicating experimental protocol: cultured hippocampal neurons were exposed to a proteasome inhibitor (PI; MG132 or lactacystin- see supplementary figs) for 2 hr and then the non-canonical amino acid AHA was added for 10 min to label newly synthesized proteins (see Materials and methods). (B) Representative BONCAT western blot using an anti-biotin antibody to detect newly synthesized proteins (labeled with AHA and then clicked with a biotin alkyne tag). Shown are two biological replicates each from a control sample (not treated with a PI) and a PI-treated sample, respectively. (C) Analysis of experiment shown in B. Protein synthesis in hippocampal neurons was significantly reduced following proteasome inhibition. p<0.001, unpaired t-test, three experiments. Error bars = SD. (D) Analysis of newly synthesized proteins (puromycylation; bar graphs) and K48 ubiquitin chains (line graph) after treatment of cultured neurons with 5, 10 or 20 μM Lactacystin. t-Test (multiple comparisons) n = 4 experiments for each concentration. For protein synthesis, 0 vs 5 μM p<0.05, 0 vs 10 μM p <0.01, 0 vs 20 μM p<0.01, For K48 ubiquitin, 0 vs 5 μM p<0.01, 0 vs 10 μM p <0.001, 0 vs 20 μM p<0.01. (E) Metabolic labeling (magenta; using puromycylation, see Materials and methods) of the global nascent protein pool in hippocampal cell bodies and dendrites following proteasome inhibition, nucleus (DAPI, blue) and dendrites (MAP2, green) can also be visualized in the images. Scale bar = 50 microns. (F) Analysis of protein synthesis (using puromycylation) of the global nascent protein pool in hippocampal neurons transfected with HA-Ubiquitin K48R+G76A –see Figure 1—figure supplement 2, (unpaired t-tests p<0.01, three experiments, n = 110 mock n = 105 ubi-K48R+G76A-transfected neurons). (G) Scheme of experiment to determine whether dendritic protein synthesis is also altered, neurons were treated with PI and labeled with AHA for the last 5 min. (H) Scheme of the experimental set up, neurons were cultured on a special membrane that allows dendrites and axons, but not cell bodies, to grow through pores. The proteasome was inhibited, then the cell bodies were scraped away from the top of the membrane and metabolic labeling was conducted on the dendritic fraction. (I) Inhibition of the proteasome results in a coordinate inhibition of dendritic protein synthesis. Representative BONCAT western blot showing the metabolically labeled newly synthesized dendritic protein in green. (J) Analysis of experiment shown in (I). Protein synthesis in dendrites was significantly reduced by over 60% following proteasome blockade, (p<0.001, unpaired t-test, four experiments) Error bars = SEM. Molecular weight markers in B and G from top-to-bottom are 250, 150, 100, 75, 50, 37, and 25 kD.
-
Figure 1—source data 1
Source data for Figure 1A and G.
- https://cdn.elifesciences.org/articles/52714/elife-52714-fig1-data1-v2.pdf
-
Figure 1—source data 2
Source data for Figure 1C, D, F, J.
- https://cdn.elifesciences.org/articles/52714/elife-52714-fig1-data2-v2.xlsx
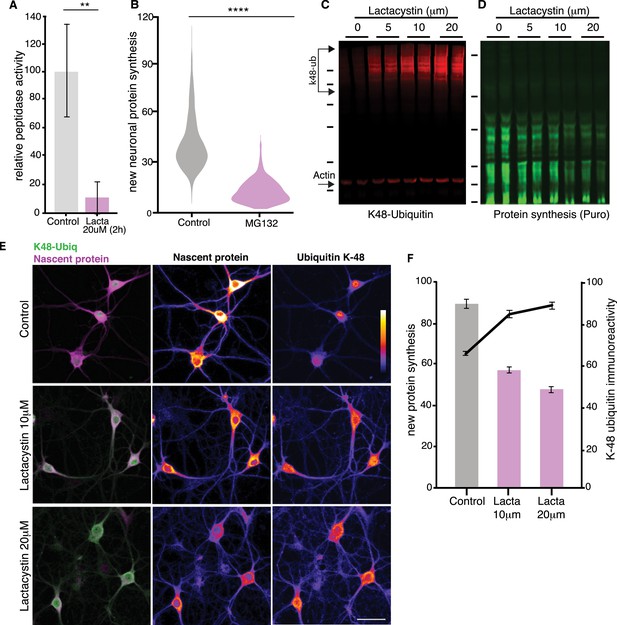
Additional experiments examining the effects of proteasomal inhibition on neuronal protein synthesis.
(A) Proteasome peptidase kinetic activity assay showing that a 2 hr Lactacystin treatment inhibits the proteasome by ~80% (unpaired t-test, p≤0.01, control, treated, kinetic slopes are represented, see Materials and methods, two experiments). (B) Analysis of experiments shown in Figure 1E. Protein synthesis in hippocampal neurons was reduced by over 30% following proteasome inhibition. (p<0.0001, unpaired t-test, ctrl n = 1768, PI n = 1137 neurons, five experiments, two dishes per experiment and condition). (C) Representative western blot of K48 ubiquitin chains. Shown are two biological replicates from a control sample and neurons treated with increasing concentrations of lactacystin (2 hr treatment). Actin is shown as a loading control. Molecular weight markers from top-to-bottom are 250, 150, 100, 75, 50, and 37 kD. (D) Representative western blot of puromycylated newly synthesized proteins (the same membrane shown in C was incubated with an anti-puromycin antibody). Note that the increasing K48-ubiquitin signal (C) is paralleled by a decrease in puromycin signal. Molecular weight markers from top-to-bottom are 75, 50, 37, 25, 20, 15 kD. (E) Metabolic labeling (using puromycylation, see Materials and methods) of the global nascent protein pool and K48-ubiquitin chains in hippocampal cell bodies and dendrites following proteasome inhibition using increasing concentrations of lactacystin (2 hr treatment). In the first column the merged channels for K48-ubiquitin (green) and puromycin (magenta) are shown, followed by color-lookup images for individual channels. Scale bar = 50 μm. (F) Analysis of experiments shown in E. (unpaired t-test, for protein synthesis; control vs 10 μM lactacystin, p≤0.0001, control vs 20 μM lactacystin, p≤0.0001, for k48-ubiquitin; control vs 10 μM lactacystin, p≤0.0001, control vs 20 μM lactacystin, p≤0.0001, control n = 316, lacta 10 μM n = 394, lacta 20 μM n = 378 neurons, three experiments). Error bars = SEM.
-
Figure 1—figure supplement 1—source data 1
Source data for Figure 1—figure supplement 1A, B, F.
- https://cdn.elifesciences.org/articles/52714/elife-52714-fig1-figsupp1-data1-v2.xlsx
-
Figure 1—figure supplement 1—source data 2
Source data for Figure 1—figure supplement 1C, D.
- https://cdn.elifesciences.org/articles/52714/elife-52714-fig1-figsupp1-data2-v2.pdf
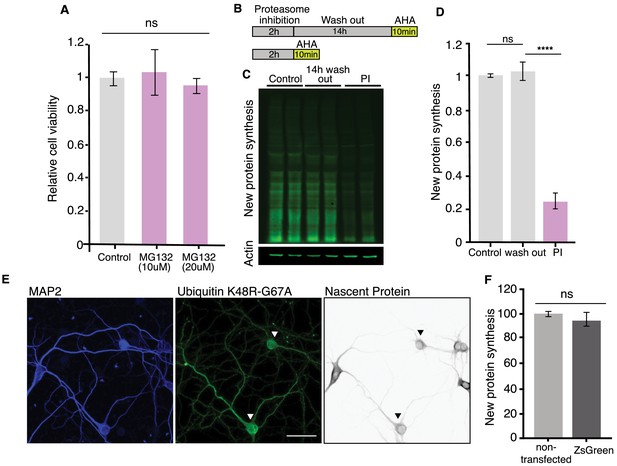
Neuronal health assessment after PI, and control experiments for ubiquitin over-expression.
(A) Proteasome inhibition by MG-132 (10 or 20 µM for 2 hr) does not alter cell viability as measured by a cell counting assay (see Materials and methods) (p>0.05, unpaired t-test, control n = 20, PI n = 20). (B) Scheme indicating experimental protocol: cultured hippocampal neurons were exposed to a proteasome inhibitor (MG132) for 2 hr followed by a 14 hr wash (in the absence of the PI) and then AHA (or puromycin) was added for 10 min to label newly synthesized proteins before or after the washout period (see Materials and methods). (C) Representative puromycylation western blot showing the recovery of protein synthesis after 2 hr treatment with MG132 and then a 14 hr wash. Two biological replicates are shown. Actin is shown as a loading control. (D) Analysis of experiment shown in C. Protein synthesis in hippocampal neurons was significantly reduced compared to that observed after the 14 hr wash, which did not differ from control levels of protein synthesis. Control vs. 14 hr wash, not significant, p>0.05; 14 hr wash vs. PI, p≤0.0001, four experiments. All analyses were unpaired t-tests. Error bars = SD. (E) Representative images of transfected neurons overexpressing HA-Ubiq K48R-G76A (arrowheads) (second panel, green), Map2 (blue) is shown in the first panel, nascent protein synthesis (puromycin labeling) is shown in the third panel (grey), protein synthesis is decreased in the neurons expressing HA-Ubiquitin K48R-G76A compared with non-transfected neurons. Scale bar 50 μm. (F) Analysis of the effect that overexpression of a control protein (ZsGreen) has on neuronal protein synthesis. (unpaired t-tests p>0.05, three experiments mock n = 224, ZsGreen n = 312).
-
Figure 1—figure supplement 2—source data 1
Source data for Figure 1—figure supplement 2A, D, F.
- https://cdn.elifesciences.org/articles/52714/elife-52714-fig1-figsupp2-data1-v2.xlsx
-
Figure 1—figure supplement 2—source data 2
Source data for Figure 1—figure supplement 2C.
- https://cdn.elifesciences.org/articles/52714/elife-52714-fig1-figsupp2-data2-v2.pdf
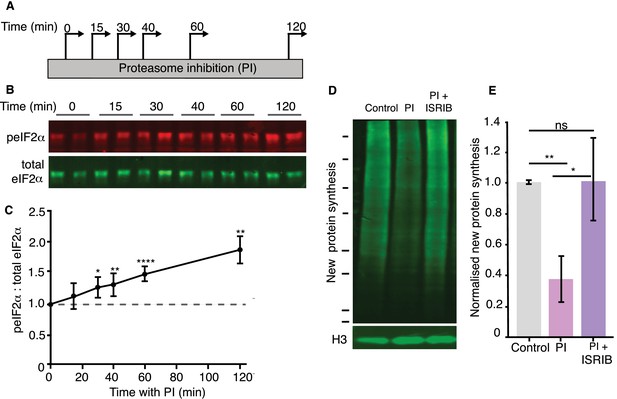
Reduced protein synthesis is mediated by eIF2α phosphorylation.
(A) Scheme indicating experimental protocol: cultured hippocampal neurons were treated with a proteasome inhibitor (PI) and collected at the indicated time points. (B) Representative western blot showing the levels of phosphorylated (detected using a phospho-specific anti-eIF2α antibody) and total eIF2α under control conditions (t = 0) or following the indicated minutes of proteasome inhibition. (C) Analysis of experiments shown in B. PI treatment led to a significant increase in peIF2α levels. (unpaired t-test control vs each time point, Control vs 15 min p>0.05, control vs 30 min p<0.05, control vs 40 min p≤0.01, control vs 60 min p≤0.0001, control vs 120 min p≤0.01. (3 experiments). Error bars = SD. (D) Representative BONCAT western blot showing the metabolically labeled newly synthesized protein in green. Treatment with the small molecule inhibitor ISRIB rescued the PI-induced decrease in protein synthesis (2 hr of treatment). Molecular weight markers from top-to-bottom are 250, 150, 100, 75, 50, 37, 25, and 20 kD. Histone 3is shown as a loading control. (E) Analysis of experiment shown in D. The PI-induced protein synthesis inhibition (control vs. PI: p≤0.05) was rescued by ISRIB treatment (control vs. ISRIB, p>0.05, unpaired t-test), four experiments, Error bars = SD.
-
Figure 2—source data 1
Source data for Figure 2A-B, D.
- https://cdn.elifesciences.org/articles/52714/elife-52714-fig2-data1-v2.pdf
-
Figure 2—source data 2
Source data for Figure 2C, E.
- https://cdn.elifesciences.org/articles/52714/elife-52714-fig2-data2-v2.xlsx
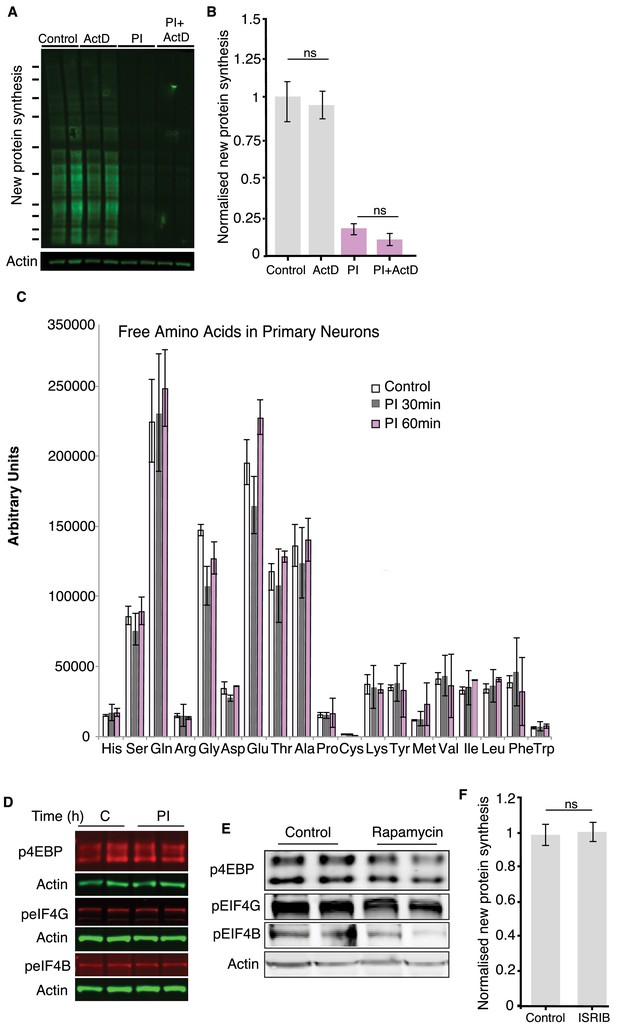
Effect of transcription inhibition on protein synthesis after proteasome inhibition and phosphorylation of other translation factors.
(A) Representative western blot showing that blocking transcription with actinomycin D (10 µM, applied for 2 hr) did not reduce protein synthesis under control conditions or affect the PI-induced decrease in protein synthesis. Actin is shown as a loading control. Two biological replicates per condition are shown. (B) Analysis of experiments shown in A. ActD treatment did not significantly alter protein synthesis under control conditions (p>0.05) nor did it significantly alter the PI-induced decrease in protein synthesis (p>0.05), three experiments. (C) Free amino acid content of primary neurons, measured after 30 and 60 min of proteasome inhibition. All experiments were performed in primary rat hippocampal neurons. (D) Western blot analysis of indicated translation initiation factors and binding proteins showing that proteasome inhibition by MG132 (20 μM; 2 hr) did not alter the phosphorylation levels of 4EBP1/2, eIF4B or eIF4G. Actin is shown as a loading control. (E) Neurons were treated with Rapamycin for 2 hr to confirm the specificity of the used phospho-antibodies. These antibodies are widely used and their specificity has been demonstrated previously. (F) The small molecule inhibitor ISRIB did not significantly alter protein synthesis under basal conditions, control vs. ISRIB, (p>0.05, non-significant, unpaired t-test, four experiments).
-
Figure 2—figure supplement 1—source data 1
Source data for Figure 2—figure supplement 1B, C, F.
- https://cdn.elifesciences.org/articles/52714/elife-52714-fig2-figsupp1-data1-v2.xlsx
-
Figure 2—figure supplement 1—source data 2
Source data for Figure 2—figure supplement 1A, D.
- https://cdn.elifesciences.org/articles/52714/elife-52714-fig2-figsupp1-data2-v2.pptx
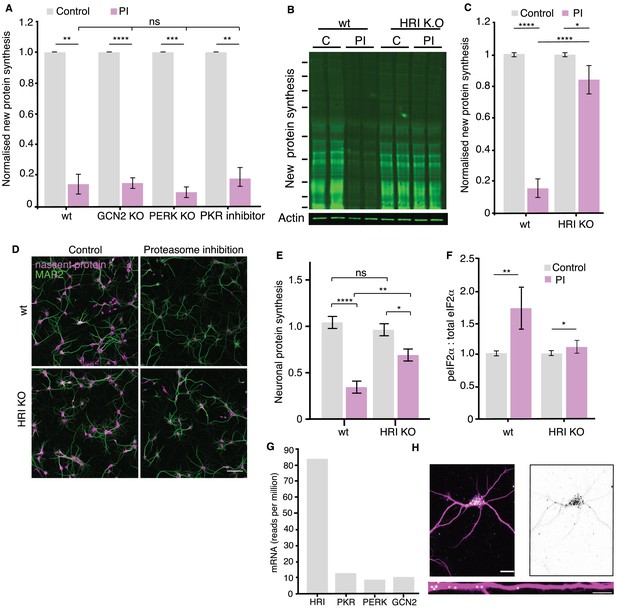
HRI kinase is responsible for the proteasome-inhibition induced increases in eIF2α phosphorylation.
(A) Genetic deletion (KO) or inhibition of the eIF2α kinases GCN2, PERK or PKR did not rescue the inhibition of protein synthesis elicited by proteasome inhibition. (wt vs. kinase KO or inhibition: GCN2, unpaired t-test p≤0.0001, PERK, unpaired t-test p≤0.001 and PKR p≤0.01, respectively, n = 2 experiments, two biological replicates per KO). Error bars = SD. (B) Representative puromycylation western blot showing that genetic deletion of HRI kinase rescues the inhibition of protein synthesis. Molecular weight markers from top-to-bottom are 250, 150, 100, 75, 50, 37, 25, 20, and 15 kD. (C) Analysis of the experiment shown in B. The amount of PI-induced protein synthesis inhibition was significantly reduced in the HRI KO (unpaired t-test on wt PI vs KO. PI p≤0.0001, wt control vs. PI p≤0.0001, KO. control vs. PI p≤0.05, n = 5 experiments, error bars SD. (D) Metabolic labeling (magenta; using puromycylation, see Materials and methods) of the global nascent protein pool in cultured hippocampal neurons following proteasome inhibition showing that HRI KO neurons exhibit less PI-induced inhibition of protein synthesis. Scale bar = 100 μm. (E) Analysis of experiments like that shown in D. Protein synthesis in hippocampal neurons was significantly reduced in the HRI KO following proteasome inhibition (unpaired t-test on wt control vs. k.o. control p>0.05, wt PI vs k.o. PI p≤0.01, wt control vs. PI p≤0.0001, KO control vs. PI p≤0.05. n = 3 experiments, wt control n = 1104, wt-PI n = 955, ko control = 1457, ko-PI n = 1052 neurons). (F) Analysis of eIF2α phosphorylation in response to proteasome inhibition. PI treatment led to a significant increase in peIF2α levels. (unpaired t-test on wt control vs. PI: p≤0.01, t-test on HRI KO control vs. PI: p≤0.05. two experiments –see Figure 3—figure supplement 1E. (G) RNA-seq data from hippocampal slices indicate that HRI mRNA is the most abundant amongst the 4 eIF2α kinases (H) Representative fluorescence in situ hybridization image detecting HRI mRNA in neuronal somata and dendrites. Scale bar = 20 μm for somata and 10 µm for dendrites-See Figure 3—figure supplement 2A–D.
-
Figure 3—source data 1
Source data for Figure 3B.
- https://cdn.elifesciences.org/articles/52714/elife-52714-fig3-data1-v2.pdf
-
Figure 3—source data 2
Source data for Figure 3A, C, E, F.
- https://cdn.elifesciences.org/articles/52714/elife-52714-fig3-data2-v2.xlsx
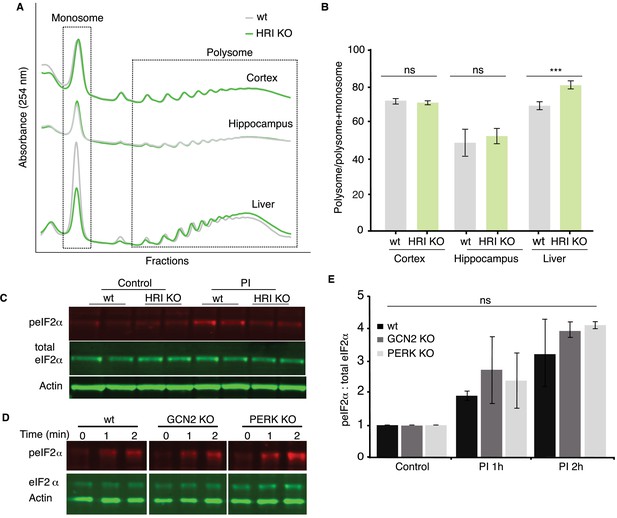
Polysome and eIF2α phosphorylation data from eIF2α kinase knock-outs.
(A) Representative polysome profiles from WT or HRI KO cortex, hippocampus or liver. (B) Quantification of the experiment shown in A and additional similar experiments. Shown is the fraction of profiling signal detected in the polysome fraction, expressed as a ratio of polysome: polysome + monosome. The absence of HRI (HRI KO) did not change the fraction of signal detected in the polysome in either cortex or hippocampus but did reduce the polysome signal (and enhanced the monosome signal) in liver. WT vs. HRI KO in cortex and hippocampus p>0.05, both not significant, WT vs. HRI KO in liver p≤0.001, n = 6 technical replicates and three biological replicates for all samples. All analyses used unpaired t-tests. Error bars = SEM. (C) Representative western blot showing the levels of peIF2α and total eIF2α following proteasome inhibition in cortical cultures prepared from WT or HRI KO mice. (D) Western blot analysis of eIF2α phosphorylation and total levels following 1 or 2 hr of proteasome inhibition (MG132, 10 μM) in WT, GCN2 KO or PERK KO cultured neurons. Actin is shown as a loading control. (E) Analysis of experiment in D and additional similar experiments. The PI-dependent increase in peIF2α was not significantly altered in either the GCN KO or the PERK KO. WT vs. GCN2 KO at 1 (n = 2) and 2 hr (n = 2) of PI treatment: p>0.05 and p>0.05, not significant. WT vs. PERK KO at 1 (n = 2) and 2 hr (n = 2) of PI treatment: p>0.05 and p>0.05, not significant. All unpaired t-tests, error bars = SD.
-
Figure 3—figure supplement 1—source data 1
Source data for Figure 3—figure supplement 1B,E.
- https://cdn.elifesciences.org/articles/52714/elife-52714-fig3-figsupp1-data1-v2.xlsx
-
Figure 3—figure supplement 1—source data 2
Source data for Figure 3—figure supplement 1C,E.
- https://cdn.elifesciences.org/articles/52714/elife-52714-fig3-figsupp1-data2-v2.pdf
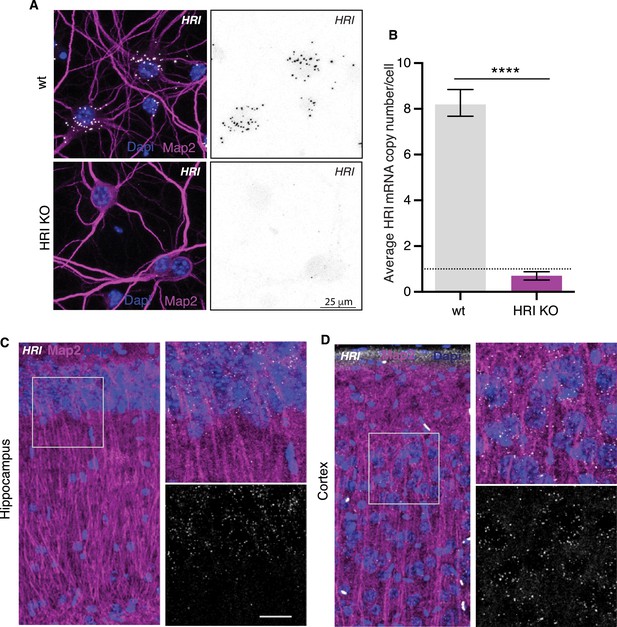
HRI mRNA detection by fluorescence in situ hybridization.
(A) Representative fluorescence in situ hybridization images of cultured neurons detecting HRI mRNA in neuronal somata and dendrites of wt and HRI KO mice. HRI mRNA is detected in neurons (white puncta left panel, and black puncta right panel). Scale bar = 25 μm. (B) Analysis of the experiment shown in A (wt n = 176, KO n = 160, three experiments). The number of HRI mRNA puncta detected in the HRI KO neurons was significantly reduced relative to control (t-Test p<0.0001) (C) Representative fluorescence in situ hybridization image detecting HRI mRNA (white puncta) in hippocampus and cortex of wt mouse. Scale bar = 20 μm. (D) MAP2 (magenta) and Dapi (blue) are shown in all images.
-
Figure 3—figure supplement 2—source data 1
Source data for Figure 3—figure supplement 2B.
- https://cdn.elifesciences.org/articles/52714/elife-52714-fig3-figsupp2-data1-v2.xlsx
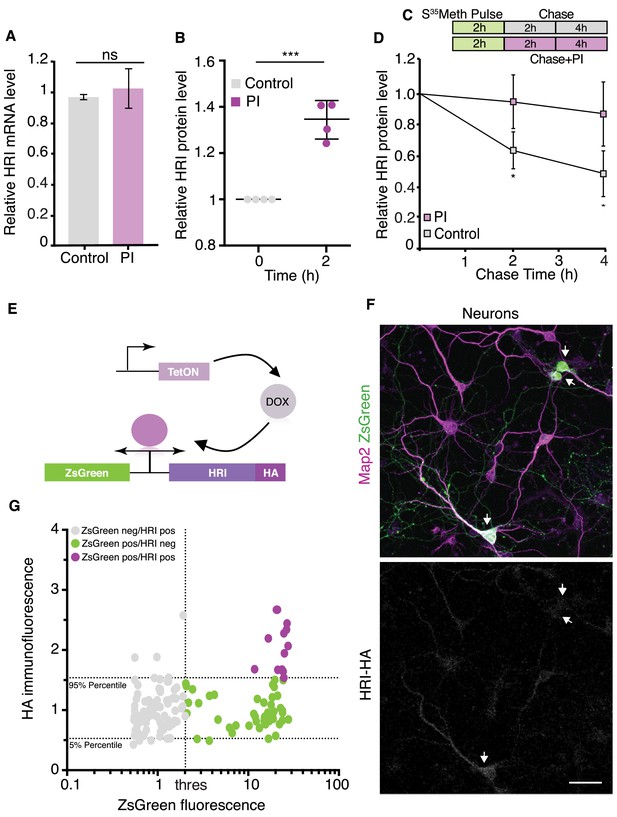
HRI exhibits low expression under basal conditions and increased expression following proteasome inhibition.
(A) Analysis for ddPCR experiments showing there is no change in HRI mRNA level following proteasome inhibition, control vs. PI, (unpaired t-test, p>0.05 n=3, error bars = SD). Samples were normalized to ribosomal RNA. (B) Quantification of immunoprecipitated HRI protein levels under control conditions or after proteasome inhibition (PI). After PI (2 hr) a significant increase in HRI protein was detected in PI samples vs. control, experiments are normalized to the untreated condition (unpaired t-test p≤0.0001, n = 4 experiments, error bars = SD). (C) Scheme showing the experimental procedure, cultured neurons were labeled with S35-Met for 2 hr, washed and collected after 2 and 4 hr + / - PI. (D) Analysis of immunoprecipitated and radiolabeled HRI. Under basal conditions the HRI half-life is ≈4 hr, its degradation is blocked by PI (unpaired t-test, 2 hr control vs PI p≤0.05, 4 hr control vs PI p≤0.05, four experiments, error bars = SD). (E) Scheme showing the doxycycline-inducible expression of the HA-tagged HRI protein. (F) Representative images of transfected neurons (arrowheads) with the bi-directional reporter resulting in robust expression of ZsGreen (green) but near absent expression of HRI (white) in the same population of cells. Also shown are somata and dendrites (labeled with an anti-MAP2 antibody). Scale bar = 50 μm. (G) Analysis of experiments in F, showing the correlation between ZsGreen fluorescence and HA (HRI) immunolabeling in individual neurons. Dotted horizontal lines indicate the area containing 90% of the HA immunofluorescence values of ZsGreen-negative (non-transfected) neurons (5–95% percentiles), the dotted vertical line indicates the threshold between ZsGreen-negative and ZsGreen positive cells. Grey dots represent ZsGreen-negative cells, magenta dots show neurons positive for ZsGreen but negative for HA, and green dots represent neurons positive for ZsGreen and above the HA 95% percentile of the ZsGreen-negative population (n = 149). See Figure 4—figure supplement 2A.
-
Figure 4—source data 1
Source data for Figure 4A, B, D, G.
- https://cdn.elifesciences.org/articles/52714/elife-52714-fig4-data1-v2.xlsx
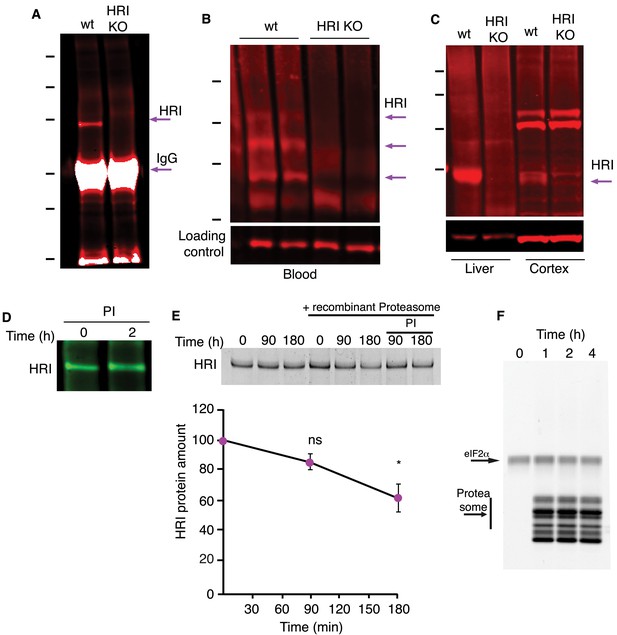
Additional data on HRI expression following proteasome inhibition.
(A) Validation of the HRI antibody in cultured neurons showing a distinct band present at 75 kD, (MW of HRI ≈ 75kD) after IP and the absence of the band in cultured neurons prepared from an HRI knock-out mouse. Molecular weight markers from top-to-bottom are 150, 100, 75, 50, 37 and 25 kD. (B) HRI is expressed at significant levels in blood. Arrows point to three different apparent HRI variants present in WT and absent in HRI KO. Molecular weight markers from top-to-bottom are 150, 100, 75 and 50 kD. (C) HRI expression in liver and cortex. Molecular weight markers from top-to-bottom are 250, 150, 100, and 75 kD. (D) Western blot demonstrating the modest but significant stimulation of HRI protein expression following a 2hr MG132 (10 μM) treatment. The analysis of this representative experiment and other experiments is shown in Figure 4B. (E) In vitro degradation experiment in which recombinant HRI was added to recombinant 20S proteasome for the indicated times. The graph indicates the recombinant proteasome-dependent degradation of HRI, obtained by subtracting the amount of HRI degradation that occurred with the addition of the recombinant proteasome together with a proteasome inhibitor (‘proteasome-independent degradation’). Plot shows analysis of the HRI protein degradation by the recombinant 20S proteasome normalized to control reactions without proteasome. Control vs. 90 min, p>0.05, 0 vs 180 min p≤0.05, two experiments. Error bars = SD. (F) Control experiment for that shown in (E) showing that eIF2α, as one example, is not a proteasome substrate in this assay.
-
Figure 4—figure supplement 1—source data 1
Source data for Figure 4—figure supplement 1E.
- https://cdn.elifesciences.org/articles/52714/elife-52714-fig4-figsupp1-data1-v2.xlsx
-
Figure 4—figure supplement 1—source data 2
Source data for Figure 4—figure supplement 1A, B, C, D, E, F.
- https://cdn.elifesciences.org/articles/52714/elife-52714-fig4-figsupp1-data2-v2.pdf
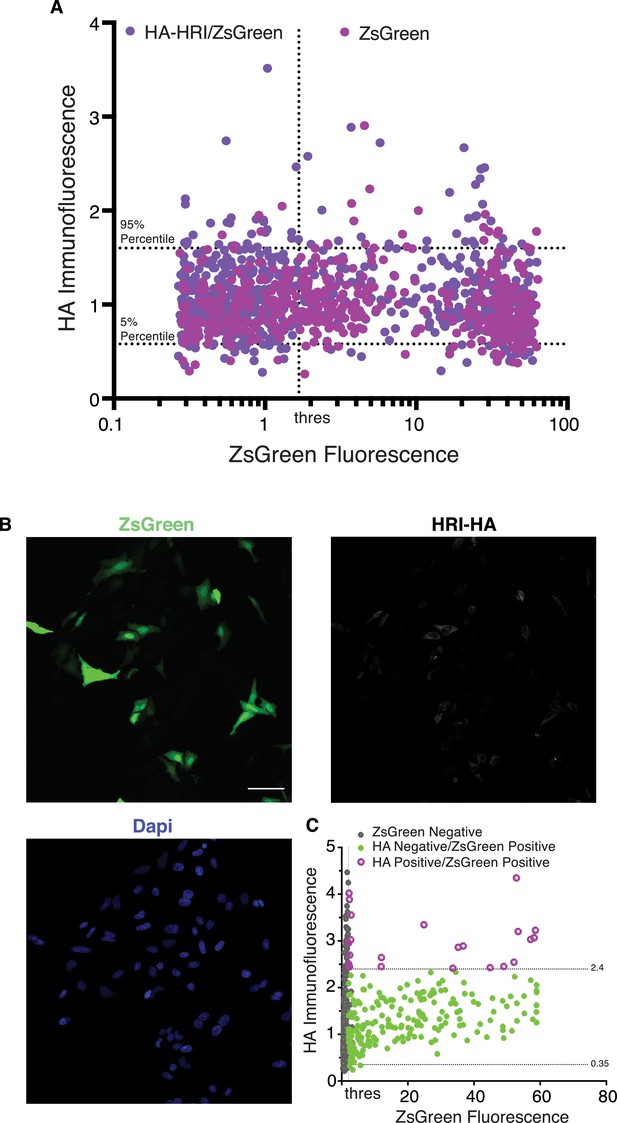
Correlation of HRI expression and ZsGreen in primary neurons and HeLa cells.
(A) Additional experiments for the one shown in the Figure 4E–G, including background controls with neurons transfected with a bi-directional inducible plasmid expressing ZsGreen but lacking HRI-HA. Correlation between ZsGreen fluorescence and HA (HRI) immunofluorescence in individual neurons. Horizontal grey lines correspond to the 95% and 5% percentile for normalized HA values of the cells lacking HRI-HA in the multiple cloning site (thus containing 90% of the HA values from the negative/background control). The vertical grey line represents a manual threshold to delineate ZsGreen positive and ZsGreen negative neurons. As expected ZsGreen can be expressed from both constructs. The distribution of HA immunoreactivity values in the construct intended to express HRI-HA does not differ substantially from the distribution of values in the background control suggesting a very low – if any – expression of HRI. (B) Transfection of the bi-directional reporter in HeLa cells (for scheme see Figure 4E) results in robust expression of ZsGreen but sparse expression of HRI in the same population of cells. Scale bar = 100 μm. (C) Analysis of experiments in B, showing the correlation between ZsGreen fluorescence and HA (HRI) immunolabeling in individual cells. Dotted horizontal lines correspond to the 95% and 5% percentile for normalized HA values of the ZsGreen negative cells (thus containing 90% of the HA values of the ZsGreen negative cells), the dotted vertical line indicates the threshold between ZsGreen-negative and ZsGreen positive cells. Grey dots represent ZsGreen-negative cells, magenta dots show cells positive for ZsGreen and HA, and green dots represent cells positive for ZsGreen and HA negative. HRI Expression was not positively correlated with ZsGreen expression (two experiments, n = 867 cells).
-
Figure 4—figure supplement 2—source data 1
Source data for Figure 4—figure supplement 2A, C.
- https://cdn.elifesciences.org/articles/52714/elife-52714-fig4-figsupp2-data1-v2.xlsx
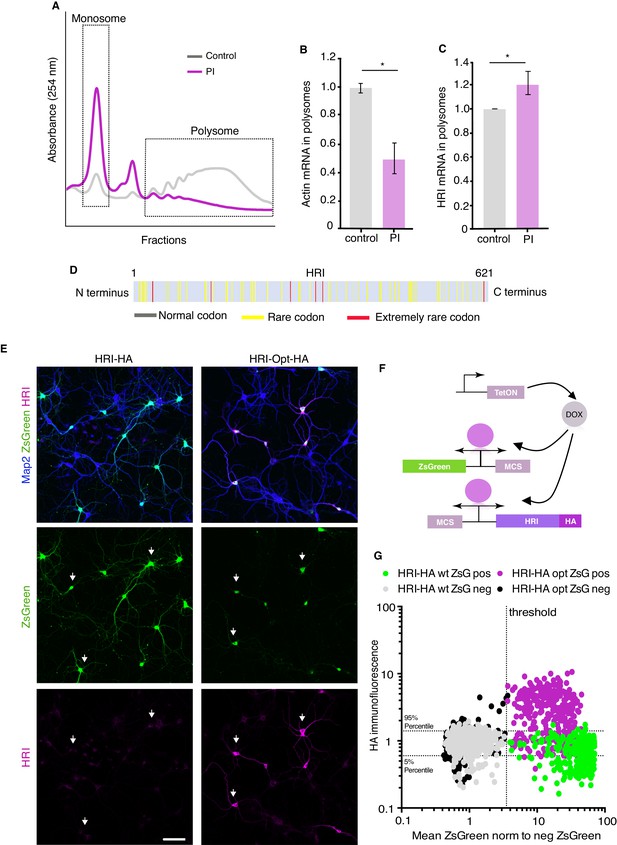
HRI exhibits a codon-dependent paradoxical shift to enhanced translation following proteasome inhibition.
(A) Representative polysome profile showing the effect of proteasome inhibition on translation. PI led to a dramatic shift to the monosome fraction, reflecting reduced global translation. (B) Quantification of ddPCR experiments examining the abundance of actin mRNA in monosomes and polysomes, normalized to control levels. Following PI, actin mRNA exhibited the typical shift to the monosome fraction (unpaired t-test, p≤0.05, n = 2 experiments (three technical replicates per experiment). (C) Quantification of ddPCR experiments examining the abundance of HRI mRNA in monosomes and polysomes, normalized to control levels. Unlike the global RNA population, HRI mRNA exhibited a significant shift to the polysome fraction following PI, (unpaired t-test, p≤0.05, n = 2 experiments three technical replicates). Error bars = SD. (D) Scheme of the HRI protein showing an abundance of many rare and extremely rare codons, consistent with HRI’s extremely low level of translation under basal conditions in neurons. HRI uses more rare codons than the 86.78% of the genes expressed in the brain (see Materials and methods). (E) Representative images showing the increased expression of codon-optimized HRI-HA in comparison with HRI-wt (Scale bar = 100 μm). (F) Scheme showing the plasmids used for the transfection shown in E, the plasmid shown in the Figure 4E was split in two parts, one expressing ZsGreen and another one expressing HRI, the plasmids were transfected in 1:5 ratio (ZsGreen:HRI) (G) Analysis of the experiments in E, showing the correlation between ZsGreen fluorescence and HA immunolabeling in individual neurons from dishes co-transfected with ZsGreen and HRI-HA either as wt sequence (HRI-HA) or codon optimized (HRIopt-HA). Dotted horizontal lines indicate the area containing 90% of the HA immunofluorescence values of ZsGreen-negative (non-transfected) neurons of the HRI-HA transfections (5–95% percentiles), the dotted vertical line indicates the threshold between ZsGreen-negative and ZsGreen positive cells. Grey and black dots are represent ZsGreen-negative (non-transfected) neurons from dishes co-transfected with HRI-HA and HRI-opt-HA, respectively. ZsGreen-positive neurons from dishes co-transfected with HRI-HA are shown in green and from dishes co-transfected with HRIopt-HA are represented in magenta. Neurons positive for ZsGreen and HA are located in the upper right quadrant (n = 2484 neurons).
-
Figure 5—source data 1
Source data for Figure 5B, C, G.
- https://cdn.elifesciences.org/articles/52714/elife-52714-fig5-data1-v2.xlsx
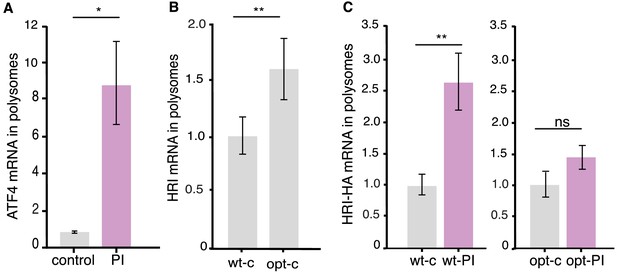
Additional data on HRI’s codon-dependent paradoxical shift to enhanced translation following proteasome inhibition.
(A) Control experiments showing the atypical ATF4 mRNA behavior in polysomes after PI (MG132 20 μM, 2 hr). Following PI, ATF4 mRNA exhibited a noncanonical shift to the polysome fraction, similar to the behavior observed for HRI mRNA (Figure 5C). (Control vs PI p≤0.05 both unpaired t-test, n = 2 per condition, each replicate has 12 transfected dishes, and three technical replicates). Error bars = SD. (B) Analysis of polysome experiment comparing WT HRI (HRIwt) to a codon-optimized HRI (HRIopt). HRIopt exhibited a significantly (p≤0.01) higher occupancy in polysomes under baseline conditions, indicating enhanced translational efficiency (note normalization is to the wt construct). (n = 2 experiments, each replicate has 12 transfected dishes, and three technical replicates). Error bars = SD. (C) Quantification of ddPCR experiments examining the abundance of wild-type HRI (HRI-wt) and a codon-optimized HRI (HRI-opt) mRNA in monosomes and polysomes. Compared to HRI-wt, under control conditions HRI-opt exhibited a higher occupancy in polysomes (B) and upon proteasome inhibition exhibited a blunted shift to the polysome fraction (note that normalization is to each respective control), (unpaired t-test, wt-c vs wt-PI, p≤0.01, t-test wt-opt vs wt-opt-PI p>0.05, n = 2 each replicate has 12 transfected dishes, and three technical replicates). Error bars = SD.
-
Figure 5—figure supplement 1—source data 1
Source data for Figure 5—figure supplement 1A, B, C.
- https://cdn.elifesciences.org/articles/52714/elife-52714-fig5-figsupp1-data1-v2.xlsx
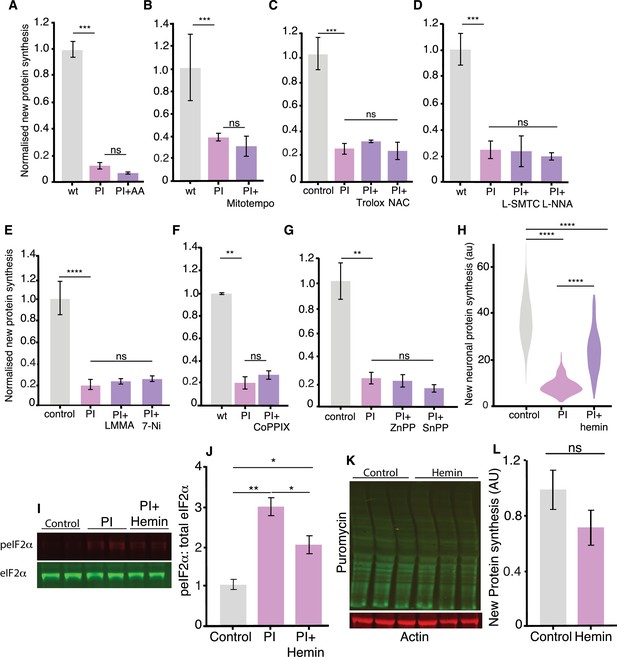
Experiments addressing the mechanism of HRI activation.
Analysis of experiments examining the effect of quenching reactive oxygen species (ROS) (A–C), blocking nitric oxide synthase (D, E) or heme oxygenase (F, G) activity on the PI-induced decrease in protein synthesis (measured using puromycylation- see Materials and methods). None of these treatments resulted in a significant alteration of the PI-induced decrease in protein synthesis: in A; PI vs. control, p≤0.001; PI vs. PI + AA p>0.05; in B PI vs. control, p≤0.001, PI vs. PI+ Mitotempo p≥0.05; in C, PI vs. control, p≤0.001 PI vs. PI+ Trolox and PI+ NAC p≥0.05; in D PI vs. control, p≤0.001, PI vs. PI +SMTC and PI +LNNA p≥0.05; in E PI vs. control, p≤0.0001 PI vs. PI+ LMMA and PI+ 7-Ni p≥0.05; in F, PI vs. control, p≤0.01; PI vs. PI +CoPPIX p≥0.05; in G PI vs. control, p≤0.01 PI +ZnPPIX and PI vs. PI+ SnPPIX p≥0.05 Unpaired t-test for all analyses (n = 2 experiments per condition). Error bars = SD. (H) Analysis of experiments examining neuronal protein synthesis in situ using puromycylation showing that hemin significantly rescues the inhibition of protein synthesis in neurons PI (n = 244) vs. control (n = 243), p≤0.0001; PI vs. PI +hemin (n = 285) p≤0.0001, Unpaired t-test for all analyses (n = 2 experiments, two dishes were imaged per experiment per condition). (I) Western blot showing the of the phosphorylation levels of eIF2α after proteasome inhibition in the presence of hemin. (J) Quantification of the experiments shown in I. (K) Western blot of protein synthesis in the presence of hemin, actin is shown as loading control. (L) Quantification of the experiments shown in K normalized with actin, normalized to the control condition.
-
Figure 5—figure supplement 2—source data 1
Source data for Figure 5—figure supplement 2A, B, C, D, E, F, G, H, L.
- https://cdn.elifesciences.org/articles/52714/elife-52714-fig5-figsupp2-data1-v2.xlsx
-
Figure 5—figure supplement 2—source data 2
Source data for Figure 5—figure supplement 2I,K.
- https://cdn.elifesciences.org/articles/52714/elife-52714-fig5-figsupp2-data2-v2.pdf
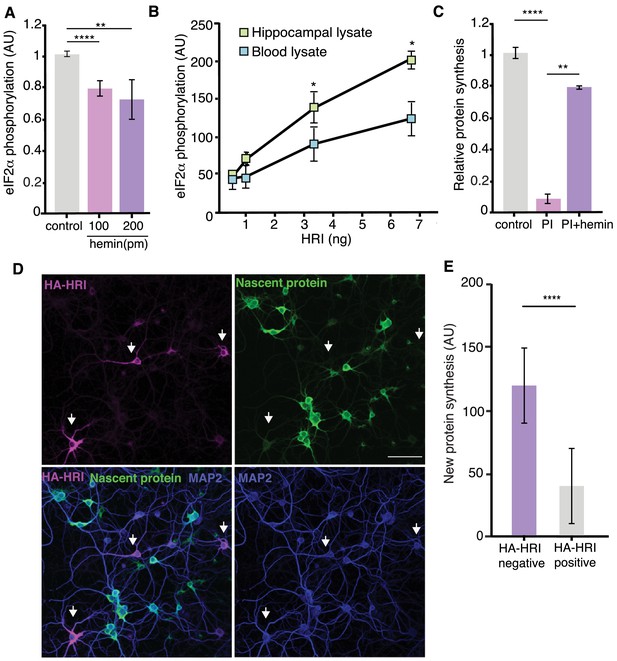
HRI activity is constitutive owing to a low heme content in neurons.
(A) In vitro eIF2α phosphorylation assay by HRI; 100 and 200 pm of hemin were added to the reaction mixture (500 ng eIF2α and 3,3 ng of HRI), inhibiting the activity of the kinase (unpaired t-test control vs hemin 100pm p≤0.0001, control vs hemin 200pm p≤0.01). (B) In vitro eIF2α phosphorylation assay by HRI; a constant amount of eIF2α was added for each reaction combined with increasing amounts of HRI as indicated (See Materials and methods). Increasing amounts of HRI protein lead to significantly higher levels of eIF2α phosphorylation in hippocampal lysates, compared to blood lysates (the lysates were cleared of protein by heating 10 min at 60°C) (unpaired t-test 0.05 ng HRI Blood vs hippocampus p≥0.05, 0.1 ng HRI Blood vs hippocampus p≥0.05, 0.33 ng HRI Blood vs hippocampus p≤0.05, 66 ng HRI Blood vs hippocampus p≤0.05, n = 3 experiments). (C) Western blot analysis of experiments testing whether hemin (heme complexed with Fe3+) can mitigate the effects of PI. Hemin significantly reduced the PI-induced protein synthesis inhibition. (PI vs. control, p≤0.0001; PI vs. PI +hemin, p≤0.01 Error bars = SD, three experiments) (D) Representative images showing that HA-HRIexpressing neurons (arrow) (green; anti-HA labeling) exhibit lower levels of protein synthesis (magenta) when compared to neighboring neurons, which do not express HA-HRI. Also shown are somata and dendrites (labeled with an anti-MAP2 antibody) and nuclei (labeled with DAPI). Scale bar = 50 μm. (E) Quantification of somatic protein synthesis levels in neurons expressing or lacking HA-HRI; HA-HRI positive neurons exhibited significantly lower levels of protein synthesis, (p≤0.0001, HA negative neurons n = 852 and HA positive neurons n = 253, unpaired t-test).
-
Figure 6—source data 1
Source data for Figure 6A, B, C, E.
- https://cdn.elifesciences.org/articles/52714/elife-52714-fig6-data1-v2.xlsx
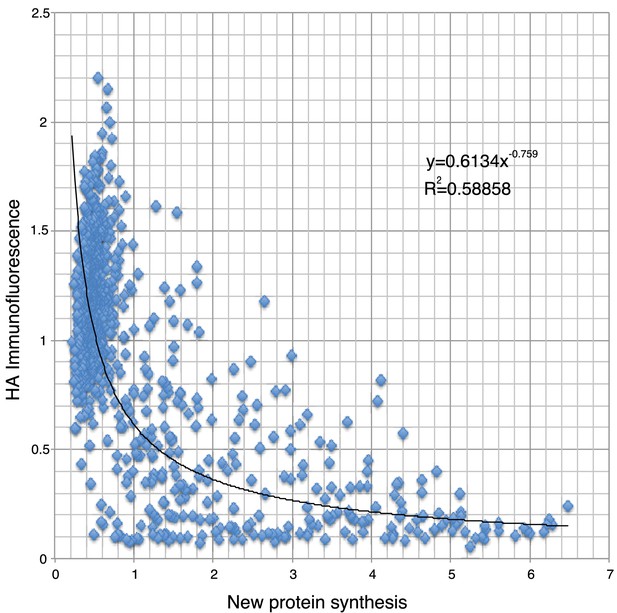
Analysis of the data shown in Figure 6.
Correlation of the level of HA-HRI expression (using a transfected construct) and the corresponding nascent protein signal (obtained using brief puromycin labeling) in the same neuron. As expected, the correlation between the expression of HRI kinase and the inhibition in protein synthesis was not linear, the best fit corresponds to a power line. This is consistent with the assumption that one molecule of HRI will lead to the phosphorylation of multiple molecules of substrate. Given this dependence, small increases in HRI expression will exert the greatest impact on protein synthesis within a low to intermediate range: when expression is not too low to impact eIF2α phosphorylation, but also not too high because then most protein synthesis is already shut off. Consequently, it is possible that a 30% increase in HRI expression will decrease protein synthesis by more than 50%.

Western Immunoblot for Lamin B1.
Neurons were scraped from the upper and lower part of the membrane and the nuclear protein Lamin B1 was detected by Western Blot.
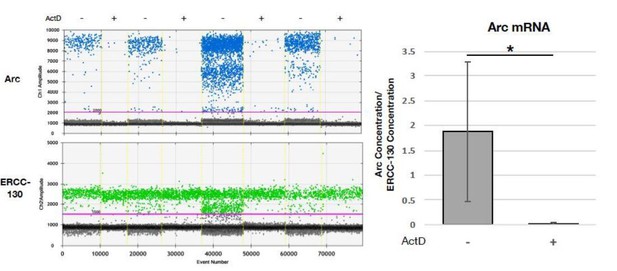
Testing actinomycin D’s effectiveness in blocking transcription using ddPCR of Arc mRNA.
Neurons were treated with ActD for 2h and mRNA levels of Arc, an immediate early gene, in non-treated and treated neurons were measured. In the left graph the ddPCR counts for Arc mRNA and ERCC (used for normalization) are shown. Treatment with actinomycin D (indicated by “+”) completely prevented the transcription of Arc mRNA. In the right graph, the average of the experiments normalized to the ERCC standards are shown.
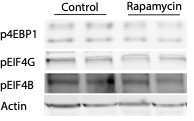
Western Immunoblot for phosphorylated translation factors.
Neurons were treated with Rapamycin (10 µm) for 2h and the phosphorylation of 4EBP1, EIF4G and EIF4B was detected by Western Blot using the phospho-specific antibodies described in the paper. A decrease in the phosphorylation of the proteins in response to Rapamycin treatment was observed.
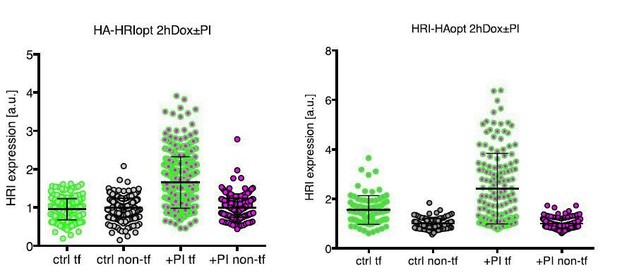
Expression of transfected HRI in primary neurons -/+ proteasome inhibition, after short induction with Doxycycline (2h).
Left panel shows the expression of HRI tagged on the N- terminus, and the right panel shows the expression of HRI tagged on the C-terminus. HRI expression was measured by Immunofluorescence using the HA tag. tf = transfected, PI = proteasome inhibitor.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information | |
|---|---|---|---|---|---|
| Gene (Rat or Mouse) | -HRI -HRI-opt -ZsGreen | GeneScript Clone ID: OMu18622C | Entrez Gene: NM_013557.2 | ||
| Strain, strain background (Escherichia coli) | BL21 (DE3) | Sigma-Aldrich | CMC0016 | Electrocompetent cells | |
| Genetic reagent (Rat or Mouse) | -Primary Neurons | Other | Animals obtained from our animal house | ||
| Cell line (Human) | -HeLa (RRID:CVCL_0030) | -ATCC | |||
| Transfected construct (mouse, Rat) | -HA-HRI -HRI-HA -HRI-HA-opt -ZsGreen -ZsGreen-HRI-HA -Ubiquitin K48R | This paper | Constructs used to over-express HRI or ubiquitin genes in primary neurons. The constructs of HRI with the different tags can be obtained from Erin Schuman’s Laboratory (MPI-BR) | ||
| Biological sample (Mouse) | -Blood (mouse wt and HRI K.O) -Liver (mouse wt and HRI K.O) -Brain (mouse wt and HRI K.O) | DOI: 10.1093/emboj/20.23.6909 | |||
| Antibody | -Rabbit polyclonal anti-peIF2α | Invitrogen | 44728G RRID:AB_1500038 | (1:1000) WB | |
| Antibody | -Mouse monoclonal eIF2α | Cell signalling | 2103 | (1:1000) WB | |
| Antibody | -Rabbit polyclonal peIF4B | Cell signalling | 5399 | (1:2000) WB | |
| Antibody | -Rabbit Monoclonal p4EBP1 | Cell signalling | 2855 RRID:AB_560835 | (1:1000) WB | |
| Antibody | -Rabbit polyclonal peIF4G | Cell signalling | 2441 | (1:25000) WB | |
| Antibody | -Rabbit polyclonal HRI | Millipore | 07–728 RRID:AB_441964 | (1:1000) WB | |
| Aantibody | -Rabbit polyclonal anti-actin | abcam | Ab8227 RRID:AB_2305186 | (1:5000) WB | |
| Antibody | -Rabbit polyclonal anti-H3 | Abcam | AB18521 RRID:AB_732917 | (1:10000) WB | |
| Antibody | -Rabbit polyclonal anti-biotin | SIGMA | 31852 | (1:10000) WB | |
| Antibody | -Mouse monoclonal anti-puromycin | Kerafast | 3RH11 | (WB, 1:1000, IF 1:3500) | |
| Antibody | -Guinea pig polyclonal anti-MAP2 | Synaptic Systems | 188004 | (IF 1:1000) | |
| Antibody | -Rabbit polyclonal anti- HA-tag | Rockland | 600-401-384 | (IF 1:2000) | |
| Antibody | -Goat polyclonal anti-mouse or anti-rabbit IR680 or IR800 | Licor | -Goat anti-mouse or anti-rabbit IR680 or IR800 | (WB 1:5.000,) | |
| Antibody | -Goat polyclonal anti-guinea pig Dylight405 | Dianova | 106-475-008 RRID:AB_2337434 | (IF 1:1000) | |
| Antibody | -Goat polyclonal anti-guinea pig-Alexa488 | Dianova | 106-546-003 RRID:AB_2337441 | (IF 1:1000) | |
| Antibody | -Alexa488, polyclonal goat anti-rabbit Alexa647 or -Alexa546 | ThermoFisher | R37116, A32733, A-21085 | (IF all 1:1000) | |
| Peptide, recombinant protein | -eIF2a -HRI | Abcam | ab95932 ab131665 | ||
| Peptide, recombinant protein | −20S proteasome | Enzo | BML-PW8720-0050 | ||
| Commercial assay or kit | -Free heme measurement kit | Sigma | MAK316 | ||
| Chemical compound, drug | -Puromycin -Doxycycline (Doxo) -MG132 -Lactacystin −7-Nitroindazole (7-NA) -S-methyl-L-thiocitrulline (L-SMTC) -N-acetyl-cysteine (NAC) -Ascorbic Acid (AA) -MitoTempo -Trolox -Actinomycin-D (ActD) -FePPIX (hemin) -CoPPIX | Sigma | -Puromycin (P8833) -Doxycycline (D9891) -MG132 (M8699) -Lactacystin (L6785) −7-Nitroindazole (N7778) -S-methyl-L-thiocitrulline (M5171) -N-acetyl-cysteine (A9165) -Ascorbic Acid (A1968) -MitoTempo (SML0737) -Trolox (648471) -Actinomycin-D (A1410) -FePPIX (hemin) (H9039) -CoPPIX (C1900) | ||
| Chemical compound, drug | -SnMPIX -ZnPPIX -NG mono methyl L-Arginine (L-NMMA) -The PKR inhibitor | Cayman | -SnMPIX (19071) -ZnPPIX (14483) -L-NMMA (10005031) -The PKR inhibitor (15323) | ||
| Genetic reagent (Mus musculus) | -PERK K.O. -GCN2 K.O. | Jackson | -PERK K.O. (line #009340) RRID:IMSR_JAX:009340 -GCN2 K.O. (line #008240) RRID:IMSR_JAX:008240 | ||
| Genetic reagent (Mus musculus) | -HRI K.O. Obtained from the laboratory of Dr, Jane Chen (Harvard-MIT) | PMID:11726526 (Han et al., 2001) | |||
| Software, algorithm | https://github.molgen.mpg.de/MPIBR-Bioinformatics/CodonUsage | https://github.molgen.mpg.de/MPIBR-Bioinformatics/CodonUsage | The kinase gene list was generated by a keyword search in gene description. Analysis and plotting scripts can be found in this repository | ||
| Sequence-based reagent | HRI Taqman Assay | IDT | Rn.PT.58.5930825 | Used for ddPCR | |
| Sequence-based reagent | Rn18s rRNA 18 s Taqman Assay | Thermo-fisher | Mm0427757_s1 | Used for ddPCR | |
| Sequence-based reagent | Actin beta Taqman Assay | IDT | Rn.PT.39a.22214838.g | Used for ddPCR | |
| Sequence-based reagent | Atf4 Taqman Assay | IDT | Rn.PT.58.13690870.g | Used for ddPCR | |
| Sequence-based reagent | HRI-HA | IDT | PCR primer one for HRI-hemagglutinin tag | Used for ddPCR CAGCTACTGCAGAGCGAACT | |
| Sequence-based reagent | HRI-HA | IDT | PCR primer two for HRI-hemagglutinin tag | Used for ddPCR GCGTAATCTGGAACATCGTATGG | |
| Sequence-based reagent | HRI-HA | IDT | Probe | Used for ddPCR /56-FAM/AGCCTCCTT/ZEN/TCGCAGGACAAAGGGCTG/3IABkFQ/ | |
| Sequence-based reagent | HRI-HA-opt | IDT | PCR primer 1 HRI-hemagglutinin tag optimized | Used for ddPCR TCCAGAGCGAGCTGTTCCAG | |
| Sequence-based reagent | HRI-HA-opt | IDT | PCR primer 2 HRI-hemagglutinin tag optimized | Used for ddPCR GCATAGTCAGGCACATCATAAGGG | |
| Sequence-based reagent | HRI-HA-opt | IDT | Probe HRI-hemagglutinin tag optimized | Used for ddPCR /56-FAM/ACCACCGGC/ZEN/AACGTGAACCTGACCC/3IABkFQ/ | |
Additional files
-
Supplementary file 1
List of rare codons in HRI mRNA.
- https://cdn.elifesciences.org/articles/52714/elife-52714-supp1-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/52714/elife-52714-transrepform-v2.pdf





