Selective egg cell polyspermy bypasses the triploid block
Figures
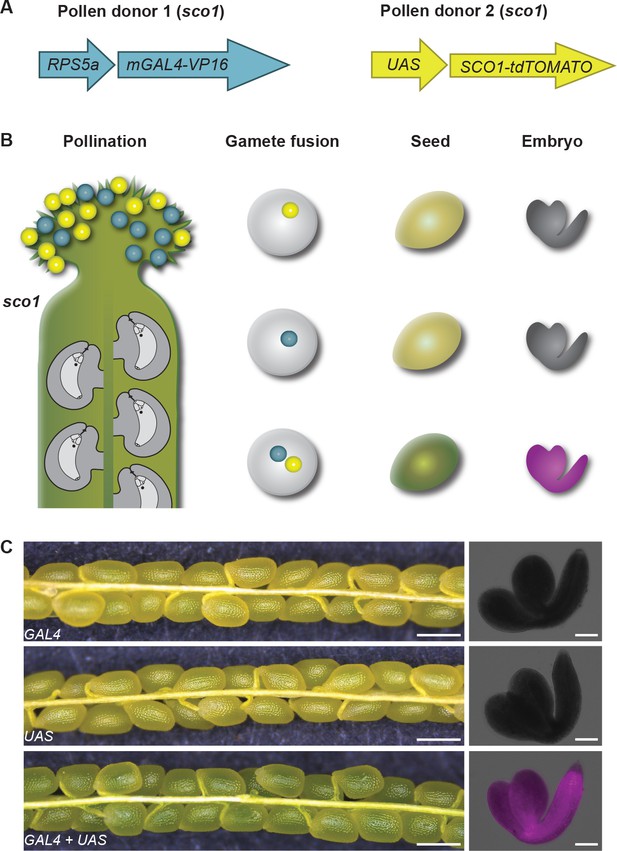
Establishment of a detection assay for polyspermy-derived embryos.
(A) Illustration of HIPODSCO1. The assay is based on the UAS-GAL4 two-component system whereby a synthetic transcription factor mGAL4 expressed under the control of the ubiquitous RPS5a promoter activates the tdTOMATO-tagged SCO1 gene. These two components were combined with the sco1 mutant to generate pollen donor 1 and 2 (PD1 and PD2), respectively. (B) Pollen of PD1 and PD2 (blue, yellow) are applied to the stigma of a sco1 gynoecium (green). Gamete fusion involving two sperm from two different pollen donors leads to transactivation of the SCO1 gene resulting in dark green seeds and fluorescence of tdTOMATO in the embryo, while monospermy-derived seeds remain pale green with no fluorescence. (C) Silique and seed analysis of sco1 mutants containing only pRPS5a::mGAL4-VP16, (upper panel), only pUAS::SCO1-tdTOMATO (middle panel), and both pRPS5a::mGAL4-VP16 and pUAS::SCO1-tdTOMATO (lower panel). Scale bars, 500 μm and 100 μm in left and right panel, respectively.
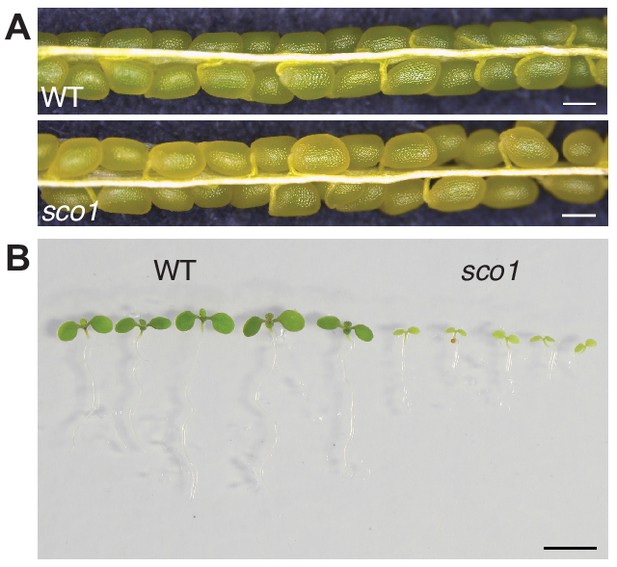
sco1 exhibited yellowish seeds and cotyledons compared to wild-type.
(A) Seeds of wild-type (upper) and sco1 T-DNA insertion mutant (lower) in silique 7 DAP. (B) 6-day-old seedlings of wild-type (left) and sco1 mutant (right).
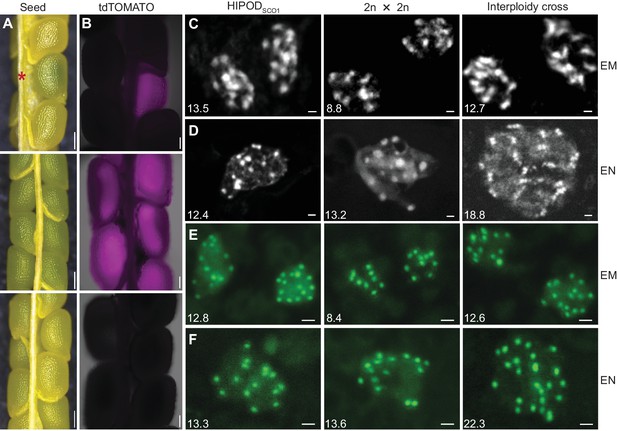
HIPODSCO1 identifies developing seeds harboring polyspermy-derived embryos.
(A and B) Bright- and fluorescence light images of seeds from different crosses seven days after pollination (DAP). Upper panel (HIPODSCO1): sco1 × pRPS5a::mGAL4-VP16/+ sco1 (PD1) × pUAS::SCO1-tdTOMATO/+ sco1 (PD2); middle panel: pUAS::SCO1-tdTOMATO/+ sco1 × pRPS5a::mGAL4-VP16/+ sco1; lower panel: sco1 × sco1. Asterisk indicates polyspermy-induced complementation of a sco1 seed. (C and D) DAPI-stained chromosome spreads of embryo (EM) (C) and endosperm (EN) (D) resulting from different crosses. Left panel: HIPODSCO1-rescued embryo segregating from cross between sco1 × pRPS5a::mGAL4-VP16/+ sco1 (PD1) × pUAS::SCO1-tdTOMATO/+ sco1 (PD2); middle panel: pUAS::SCO1-tdTOMATO/+ sco1 × pRPS5a::mGAL4-VP16/+ sco1; right panel: sco1 × wild type (4n). (E and F) Chromosome counting through centromere-targeted CENH3-GFP of embryo (E) and endosperm (F) resulting from different crosses. Left panel: rescued embryo segregating from cross between sco1 × pRPS5a::mGAL4-VP16/+ p35S::CENH3-GFP sco1 (PD1 with CENH3-GFP) × pUAS::SCO1-tdTOMATO/+ sco1 (PD2); middle panel: pUAS::SCO1-tdTOMATO/+ sco1 × pRPS5a::mGAL4-VP16/+ p35S::CENH3-GFP sco1 ; right panel: wild type (4n) × pRPS5a::mGAL4-VP16/+ p35S::CENH3-GFP sco1 . The numbers in parenthesis indicate the average counted chromosomes from all analyzed cells, from left to right, (C) n = 11, 11, 9, (D) n = 20, 6, 9, (E), n = 82, 57, 78, (F), n = 12, 26, 6. Scale bars, 200 μm (A), 100 μm (B), 1 μm (C–E).
-
Figure 2—source data 1
Chromosome counting through centromere-targeted CENH3-GFP in embryo and endosperm resulting from different crosses.
- https://cdn.elifesciences.org/articles/52976/elife-52976-fig2-data1-v1.xlsx
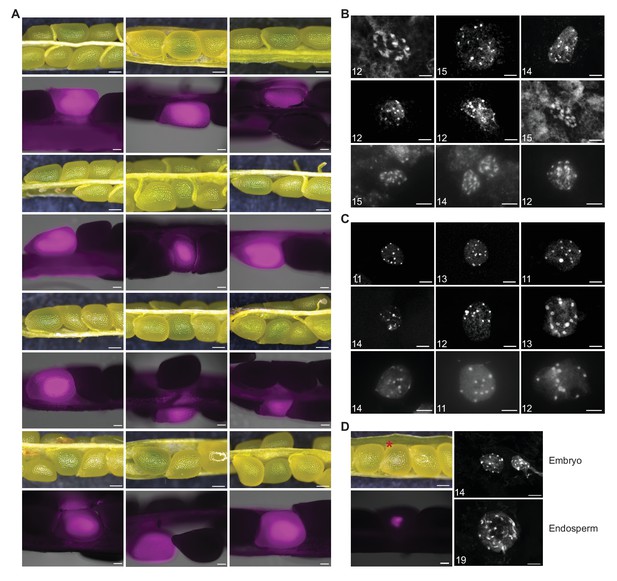
Identification and characterization of rescued green seeds and one underdeveloped seed recovered from HIPODSCO1.
(A) Silique segregating a green seed from 7 DAP HIPODSCO1 (upper) and corresponding tdTOMATO signal (lower). The last three siliques from HIPODSCO1 with p35S::CENH3-GFP in PD1. Scale bars, 200 μm (upper), 100 μm (lower). (B and C). Chromosome spread of the embryo (B) and endosperm (C) isolated from green seeds recovered from HIPODSCO1. The number in the left corner labels the visible chromosome numbers in one cell. The pictures in the lower row in (B) and (C) were captured with a Leica DMI6000b epifluorescence inverted microscope (100 × objective) with manually increased brightness and contrast. Scale bar, 5 μm. (D) Silique (upper left) segregating one single abnormal polyspermy-derived seed (asterisk) recovered from HIPODSCO1, the respective tdTOMATO fluorescence analysis (lower left), and DAPI stained chromosome spreads of embryo and endosperm (right panel) from the asterisk- labeled seed Scale bars, 200 μm (upper left), 100 μm (lower left), 5 μm (right panel). The number in the left corner labels the visible chromosome numbers in one cell.
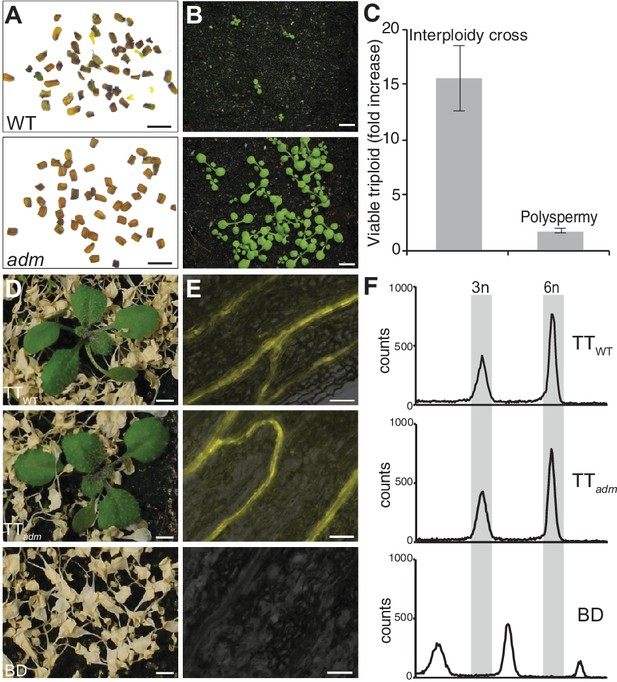
Polyspermy-induced polyploidization is partially insensitive of adm-mediated triploid block repression.
(A and B) Mature seed (A) and corresponding 9 day old seedling (B) from one silique of wild-type pollinated with diploid wild-type pollen and diploid adm-1 pollen. (C) Effect of the triploid block repressor adm on interploidy cross- recovered triploids (2n × 4n) and polyspermy- derived triploids. Shown is the ratio of triploid plants recovered from two crosses involving either adm or wild-type pollen donors (adm/WT). The data are means ± SEM (n = 3 experiments). (D) Herbicide-treated offspring of triparental triploid (TT) plants recovered from HIPOD with (TTWT) or without (TTadm) ADMETOS segregating pollen donors. Lower panel, herbicide-sensitive offspring of biparental diploid wild type (BD). (E and F) YFP fluorescence (E) and flow-cytometric analysis (F) of TTWT, TTadm and BD plants corresponding to the categories shown in (D). Scale bars, 1 mm (A), 5 mm (B, D), 50 μm (E).
-
Figure 3—source data 1
Comparison of viable triploids recovered from polyspermy and interploidy crosses (2n × 4n).
- https://cdn.elifesciences.org/articles/52976/elife-52976-fig3-data1-v1.xlsx
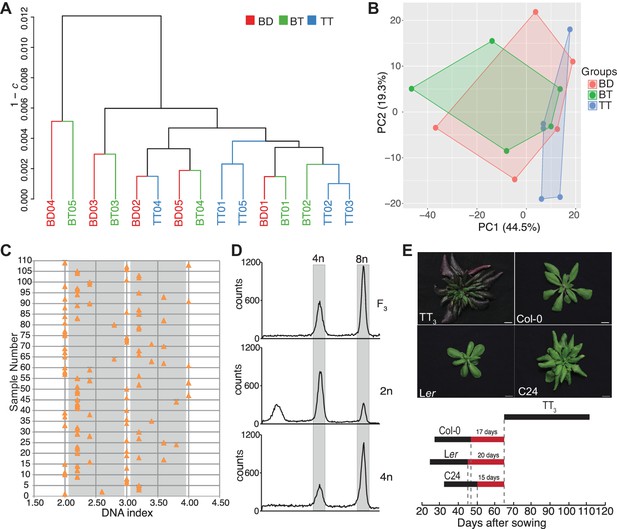
Characterization of triparental triploid plants.
(A) Dendrogram from hierarchical clustering of the 3 × 5 biological replicates of biparental diploid (BD), biparental triploid (BT), and triparental triploid (TT) recovered from polyspermy plant samples. The 15 × 15 matrix of Spearman’s correlation coefficients c was computed from the fifteen regularized log transformed expression profiles. The dendrogram was computed by average linkage clustering (UPGMA) of the 15 × 15 dissimilarity matrix with elements 1 – c. (B) Two-dimensional principle component analysis (PCA) of the fifteen 21,450-dimensional regularized log transformed expression profiles. The points represent the biological replicates, while the shapes resulting from connecting biological replicates from the same genotype highlight the similarity between the transcriptome profiles. (C) Distribution of ploidy level in the aneuploidy swarms produced by a triparental triploid plant. 2.0 in DNA index represents near diploids, 3.0, near triploids, and 4.0, near tetraploids. The gray areas indicate the intermediate ploidies. Each orange triangle represents an F2 plant derived from a triparental triploid. (D) Flow cytometric analysis of tetraploid progeny plants in F3 generation. 2n and 4n represent diploid and tetraploid controls. (E) Analysis of flowering time window of triparental triploid plants recovered from a three accession cross (TT3), Col-0, Ler and C24 during bolting stage. Scale bar, 1 cm. The black bold line represents the flowering period of different accessions. The three red bold lines bordered by the gray dashed lines label the day gaps between flowering termination of the parents and flowering initiation in the TT3.
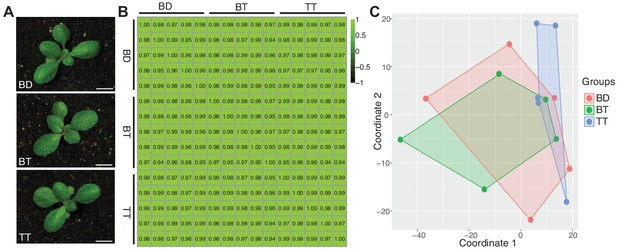
Transcriptome analysis of plants from different origin and ploidy.
(A) RNA was isolated from biparental diploids (BD), biparental triploids (BT) and triparental triploids (TT) 18 DAS. Scale bar, 5 mm. (B and C) Pairwise Spearman’s correlation coefficients (B) and two-dimensional multidimensional scaling (MDS) analysis (C) of the fifteen 21,450-dimensional regularized log transformed mRNA expression profiles. The points represent the single biological replicates, while the shapes resulting from connecting biological replicates from the same genotype highlight the similarity between the transcriptome profiles.
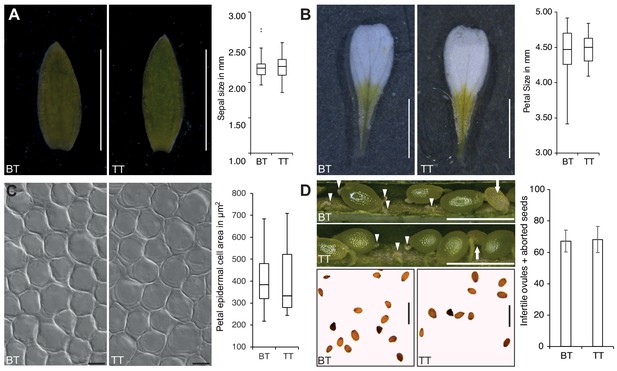
Polyspermy-induced triploid plants are similar to triploid plants originating from a regular monospermic fertilization mode.
(A) Sepal size comparison of biparental triploids (BT; n = 56) and triparental triploids (TT; n = 107). The brightness in (A) was equally increased by using Adobe Photoshop CC. (B) Petal size of BT (n = 121) and TT (n = 104). (C) Petal epidermal cell area of BT (n = 112) and TT (n = 93). (D) Silique analysis of BT (n = 19) and TT (n = 39). Siliques are showing dark green seeds, sterile ovules (white arrowhead) and abnormal seeds (white arrow) (upper panel). Seed morphology of BT and TT plants (lower panel). Box plots show median, quartiles, maximum and minimum. Scale bars, 2 mm (A, B), 10 µm (C), 1 mm (D). TT data was already published before (Nakel et al., 2017).
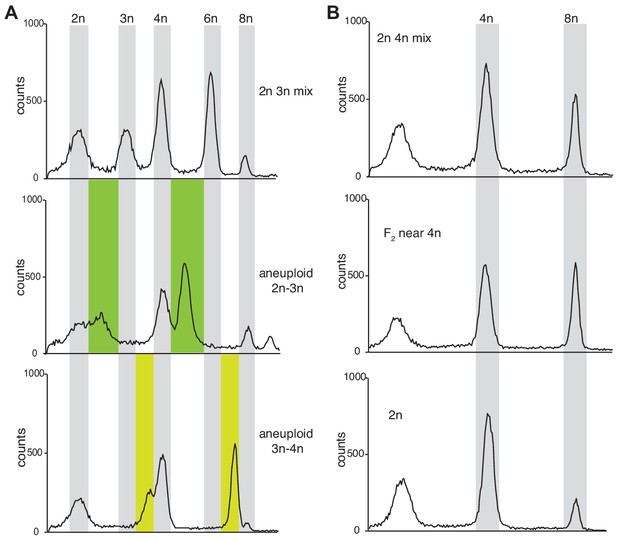
Ploidy assessment of offspring derived from triparental triploids.
(A) Euploid peak distribution was assessed by measuring a mixture of extracted nuclei from 2n and 3n plants that gives rise to five prominent peaks representing 2n, 3n, 4n, 6n, and 8n (upper panel). Leaf samples were taken from triparental offspring (F2) and mixed with 2n Col-0 as internal standard. Aneuploid samples exhibited additional peaks that were grouped into two distinct categories; i) aneuploid 2n-3n which exhibit additional peaks in the green region (middle panel) and ii) aneuploid 3n-4n which exhibit additional peaks in the yellow region (lower panel). (B) Near-tetraploid plant samples derived from triparental offspring (F2) were identified by changes in peak height. 2n Col-0 control plant exhibits a small 8n peak (lower panel). 2n Col-0 mixed with 4n Col-0 control samples exhibit a pronounced 8n peak (upper panel) comparable to 2n Col-0 mixed with near-tetraploid samples (middle panel).
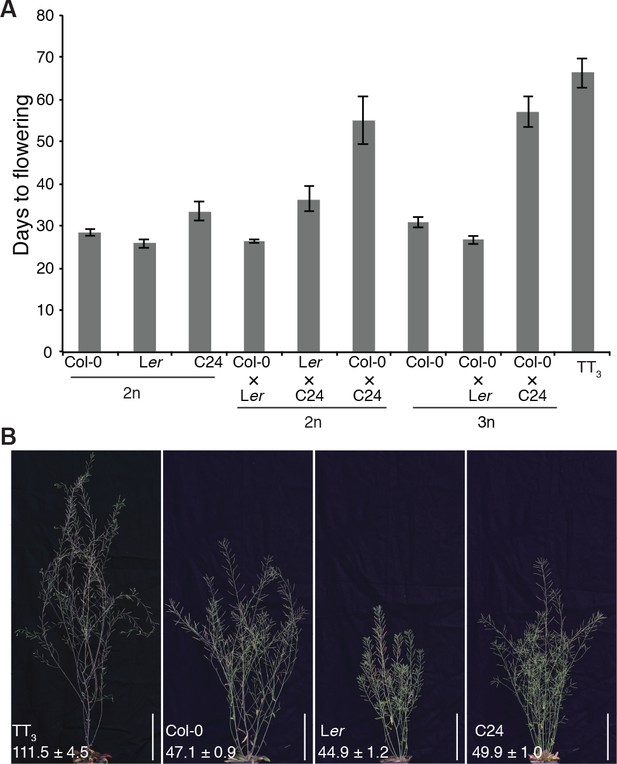
Flowering initiation and termination comparison.
(A) Flowering time comparison among plants from different ploidy and hybrids among the three accessions. The 2n hybrid of Col-0 × Ler, Ler × C24, and Col-0 × C24 were produced by crossing the respective diploid parents. The 3n hybrid of Col-0 × Ler and Col-0 × C24 were recovered from the cross of 2n Col-0 with 4n male Ler and C24. 3n Col-0 was acquired by crossing 2n with 4n Col-0 plants. All the data are means ± SD, from left to right, n = 13, 13, 12, 19, 15, 9, 4, 8, 14, 3. (B) Triparental triploid plant recovered from three accession cross (TT3), Col-0, Ler and C24 at flowering end stage corresponding to 44 DAS for Col-0, Ler and C24, and 102 DAS for TT3. The number in parenthesis indicates the average DAS of flowering termination for each genotype. All the data are means ± SEM, (n = 3 independent experiments for Col-0, Ler, and C24, n = 2 for TT3). Scale bar, 10 cm.
Additional files
-
Supplementary file 1
List of primers used for PCR.
- https://cdn.elifesciences.org/articles/52976/elife-52976-supp1-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/52976/elife-52976-transrepform-v1.docx





