Losing Dnmt3a dependent methylation in inhibitory neurons impairs neural function by a mechanism impacting Rett syndrome
Figures
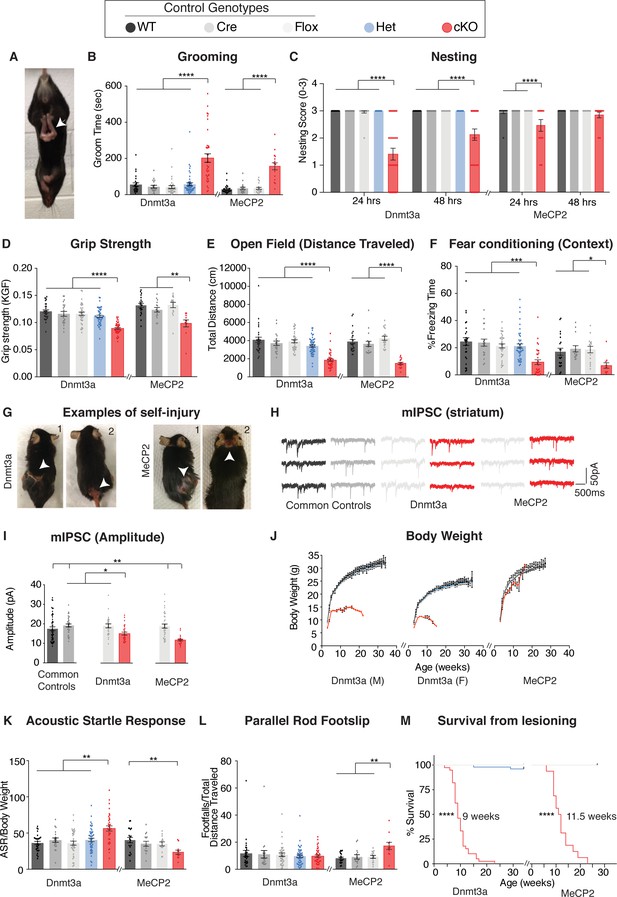
Dnmt3a cKO and Mecp2 cKO mice show overlapping as well as distinct neurological deficits.
(A) Mice that lack Dnmt3a or MeCP2 in inhibitory neurons present with hindlimb spasticity. (B) Obsessive grooming is increased in both cKO models. (C) Nest building, (D) grip strength, (E) open field, (F) fear conditioning tests revealed impairments in both cKO lines. (G) Self-injury in Dnmt3a cKO and Mecp2 cKO mice necessitated humane euthanasia. (H) Example traces of miniature inhibitory postsynaptic currents (mIPSCs) recorded in the dorsal striatum. Both cKO models show similar alterations in (I) amplitude. (J) Weekly body weight records for Dnmt3a cKO and Mecp2 cKO mice showed only Dnmt3a cKO mice (here separated by sex- see Materials and methods) were runted. (K) Dnmt3a cKO and Mecp2 cKO mice showed opposite alterations in acoustic startle response. (L) Only Mecp2 cKO mice displayed impairment on the parallel rod. M) Dnmt3a cKO mice had to undergo earlier euthanasia than Mecp2 cKO mice due to the severity of their self-lesioning. n = 11–52 (behavior), n = 5–9 mice per genotype with 24–50 neurons total (electrophysiology). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. See Supplementary file 1 for full statistics.
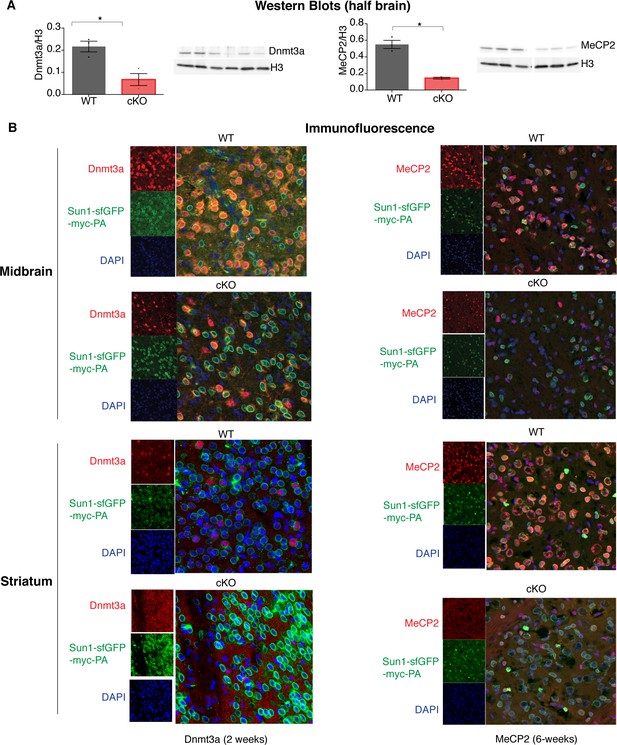
Protein levels of Dnmt3a and MeCP2 are reduced in inhibitory neurons of conditional knockout mice.
(A) Western blot of half brain hemisphere from Dnmt3a or Mecp2 cKO mice demonstrating loss of each protein. (B) Immunofluorescence (IF) images of WT, Dnmt3a cKO or Mecp2 cKO mice probing for Dnmt3a or MeCP2 (red), the Sun1-sfGFP-myc-PA fusion protein that marks the nuclear envelope and is dependent on Cre expression (green), and DAPI to mark genomic DNA (blue). n = 3 mice per genotype (western), n = 3 mice (IF) representative images for two brain regions shown.
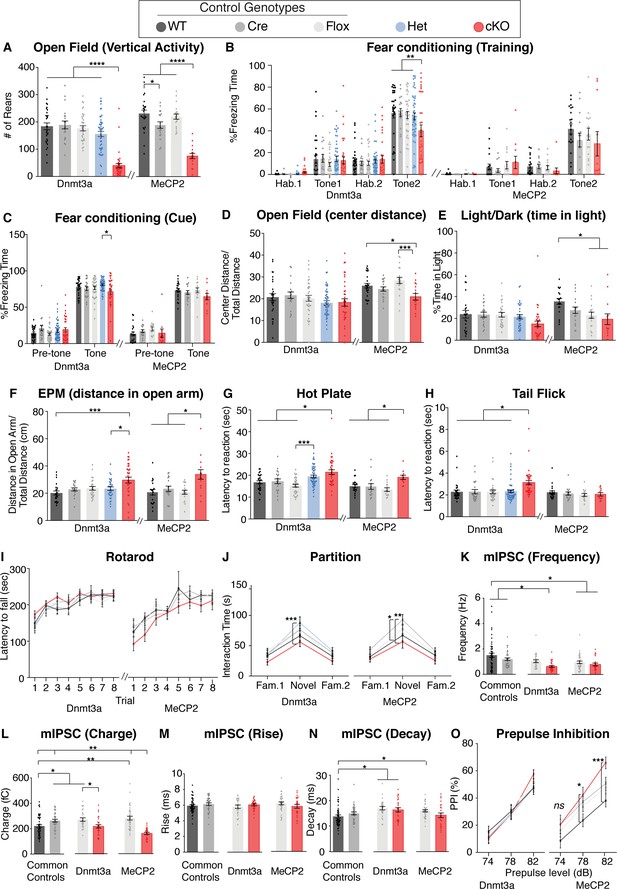
Supplemental behavioral and physiological data for Dnmt3a cKO and Mecp2 cKO mice.
(A) Both mouse lines showed decreased rearing in the open field test. (B) Dnmt3a cKO mice showed impaired fear learning, whereas Mecp2 cKO mice did not differ from control mice. (C) Cue memory was normal in both cKO mice. (D–F) Tests for anxiety-like behaviors (open field, light dark or elevated plus maze, respectively). (G) Hot plate and (H) tail flick testing for nociceptive pain in both cKO mice. (I) Rotarod test for motor learning and coordination and (J) the partition test for social interaction. (K–N) Frequency, charge, rise and decay measures from mIPSCs from the striatum. (O) Only Mecp2 cKO had a trend for increased pre-pulse inhibition. n = 11–50 per genotype (behavior), n = 5–9 mice per genotype with 24–50 neurons total (electrophysiology), *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. See Supplementary file 1 for full numbers and statistics.
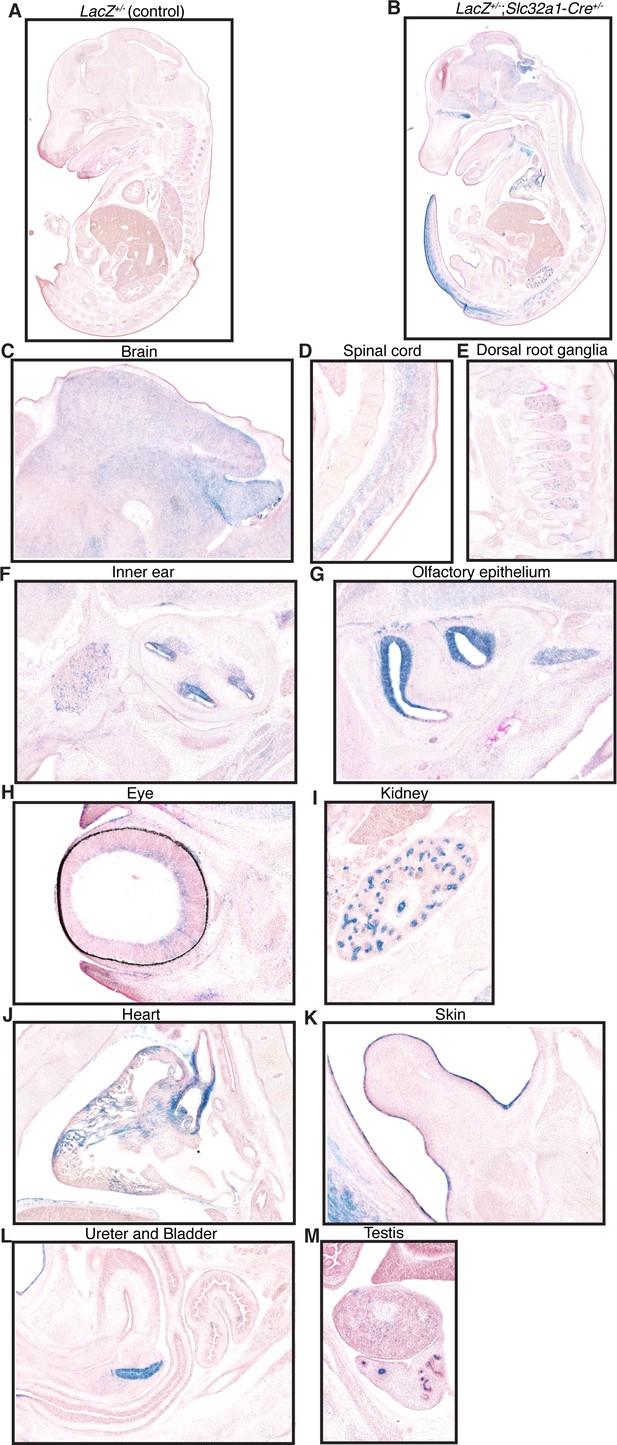
X-gal staining of LacZ+/-;Slc32a1-Cre+/- and control E14.5 embryos shows widespread expression of Cre transgene in the nervous system and select expression in peripheral tissues.
(A) X-gal staining of LacZ+/- control and (B) LacZ+/-;Slc32a1-Cre+/- reporter mice demonstrating specific staining of tissues with expression of Cre recombinase under the Slc32a1 promoter. (C–H) Images showing positive X-gal staining in nervous system tissues consistent across three biological replicates: (C) brain (D) spinal cord (E) dorsal root ganglia (F) inner ear (G) olfactory epithelium (H) and eye. (I–L) Images showing positive X-gal staining in non-neural tissues consistent across three biological replicates: (I) kidney (J) heart (K) skin (only in select regions: surrounding genital tubercle and tail and area on the face/nose) (L) ureter and bladder. Additional positive X-gal staining was noted in the (M) testis in the only embryo where the testis were visible, as well as rare, scattered cells of the intestine and pancreas (not shown). The observation of expression of Slc32a1-Cre in the testis is consistent with our noted germline recombination of floxed genes if F1, Dnmt3aflox/+;Slc32a1-Cre+/- males are used to generate F2 offspring; a breeding scheme we deliberately avoided to generate our experimental mice (see Materials and methods for more details).
Forepaw stereotypies are apparent in Dnmt3a cKO mice.
Video shows an example of forepaw stereotypies in a Dnmt3a cKO male mouse at 8 week of age.
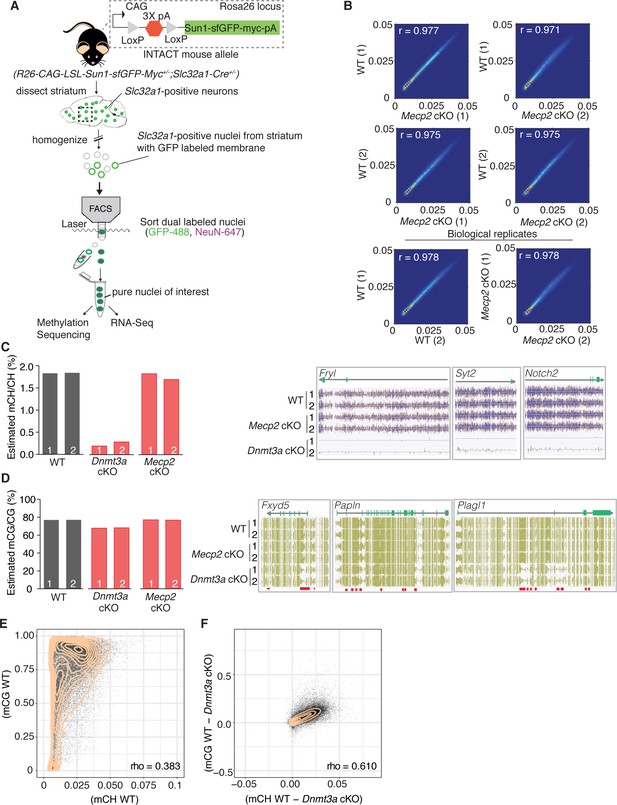
Methylation in striatal inhibitory neurons remains stable without MeCP2, but without Dnmt3a there is global loss of mCH and some loss of mCG.
(A) Schematic of INTACT method used to isolate inhibitory neurons from the striata of WT, Dnmt3a cKO, or Mecp2 cKO mice that also conditionally express the INTACT allele. (B) Spearman correlation of methylation profiles from inhibitory neurons sorted from WT vs. Mecp2 cKO striatum, showing that methylation is stable in the absence of MeCP2. (C) Bar graph showing the global mCH level in each biological replicate (left). Example genes from DNA methylome sequencing tracks showing mCH signal in two biological replicates per genotype (right). (D) Bar graph showing the global mCG level in each biological replicate (left). Example genes from DNA methylome sequencing tracks showing mCG signal (right). Red bars indicate DMRs. (E) Genome wide correlation between mCH and mCG in wild-type mice showing poor correlation. (F) Genome wide correlation of Dnmt3a dependent mCG and mCH showing good correlation to indicate that mCH and mCG written by Dnmt3a are coupled. Correlation values for E and F are Pearson correlations designated as ‘rho’. n = 2 mice per genotype (see Materials and methods for specific genotype information). See Supplementary file 1 for replicate statistics.
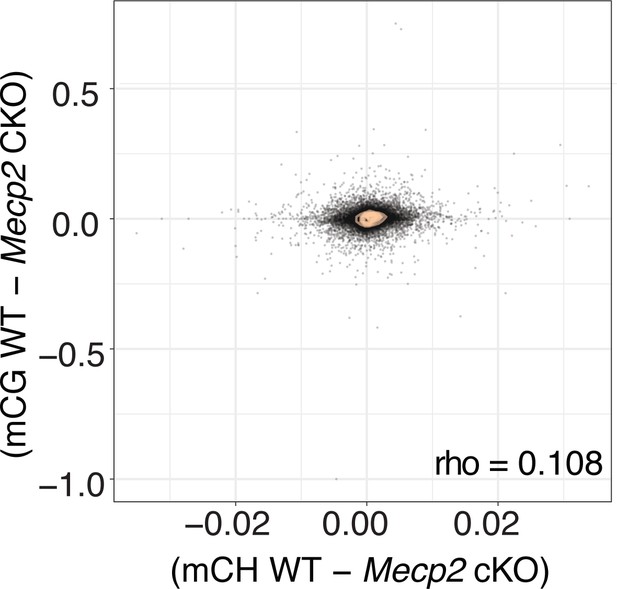
Plot of the difference in genome wide mCH versus mCG methylation between the WT and MeCP2 cKO mice.
Person correlation designated as rho. Consistent with stable methylation in the absence of MeCP2, the difference centers at 0.
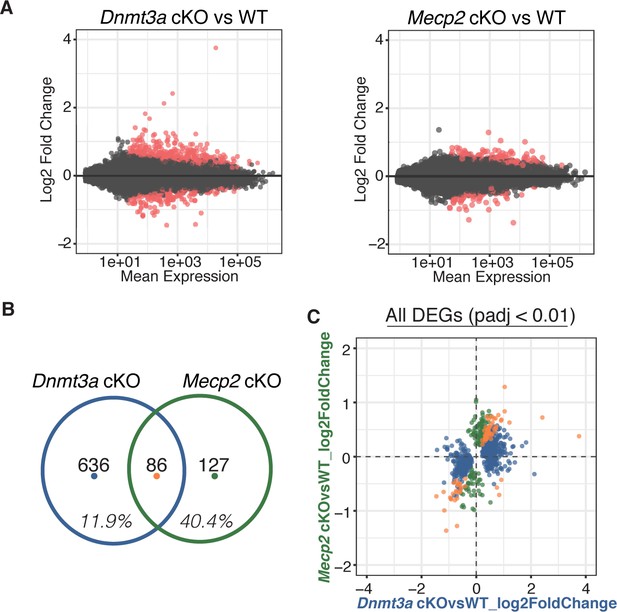
RNA-seq from sorted striatal inhibitory neurons of WT and cKO mice reveal MeCP2 is a restricted reader for Dnmt3a dependent methylation.
(A) RNA-seq data of sorted WT vs Dnmt3a cKO or Mecp2 cKO striatal inhibitory neurons that also express the INTACT allele. Red dots represent genes with altered expression in the knockout cells (padj <0.01). (B) Differentially expressed genes (DEGs) that overlap between knockout models. Inhibitory neurons that lack MeCP2 share ~40% of the same DEGs as the same neurons that lack Dnmt3a. Only ~12% of DEGs in inhibitory neurons that lack Dnmt3a are shared with the same neurons that lack MeCP2. (C) Plot of log2 fold-change for DEGs in Dnmt3a cKO and Mecp2 cKO models. DEGs that are only significantly misregulated in the Dnmt3a cKO model, only significantly misregulated in the Mecp2 cKO model, or common to both models are colored in blue, green, or orange, respectively. The plot shows that the DEGs common to both models have similar degree and direction of change.
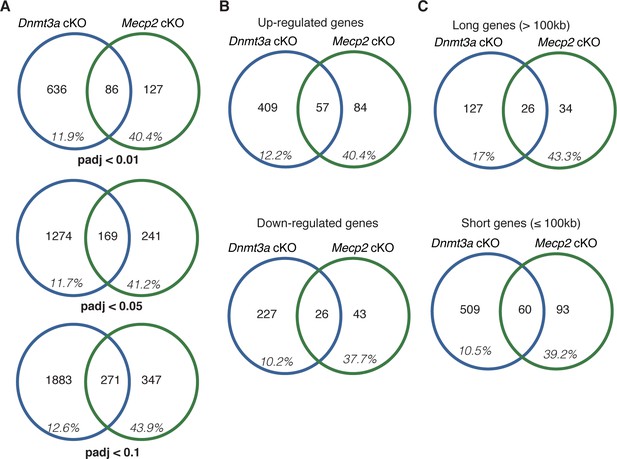
The percentage of DEGs that overlap between cKO mouse models broken down as a function of (A) p-value, (B) direction of change (down- or up-regulated), (C) gene length.
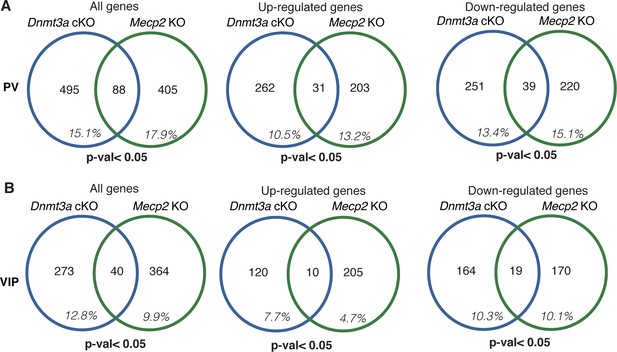
The percentage of DEGs that overlap between parvalbumin (PV) and vasoactive intestinal polypeptide (VIP) neurons in the mouse cortex from Dnmt3a cKO (Nestin-Cre) and Mecp2 KO mouse models.
The data are a re-analysis of single-nuclear RNA sequencing (Stroud et al., 2017). The percentages are shown as all genes and genes broken down by direction of change (down- or up-regulated) for (A) PV and (B) VIP neurons. Consistent with our data there are few DEGs significantly misregulated in Dnmt3a cKO mice that are also significantly misregulated in the same neurons that lack MeCP2. Notably, our data show a significantly higher overlap of genes (40%) misregualted in Mecp2 cKO that are also misregulated in the Dnmt3a cKO.
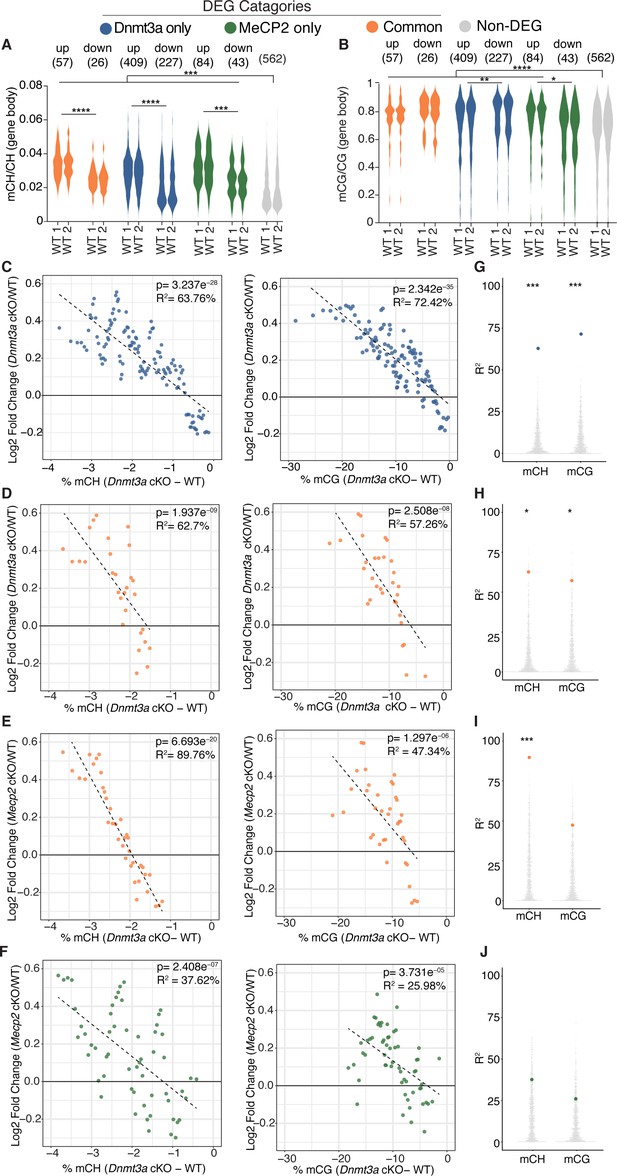
Integrative gene expression and methylation analysis shows mCH and mCG loss contribute to Dnmt3a cKO DEGs, and reveals a strong mCH contribution to RTT.
(A) Gene-body mCH levels in different categories of DEGs in WT mice are plotted, highlighting that misregulated genes have higher mCH than genes that are unchanged and the significant differences between mCH levels on up- and down-regulated genes. (B) Gene-body mCG levels in different categories of DEGs in WT mice are plotted, demonstrating that misregulated genes (with the exception of MeCP2 down-regulated genes) have higher mCG than genes that are unchanged and the significant differences between mCG levels on up- and down-regulated genes. (C) Running average plot of log2fold change in gene expression for genes significantly misregulated only in the Dnmt3a cKO model versus the change in mCH methylation observed in the Dnmt3a cKO model (‘Dnmt3a dependent mCH methylation’) (left). Running average plot of log2fold change in gene expression for these same genes versus the change in mCG methylation observed in the Dnmt3a cKO (‘Dnmt3a dependent mCG methylation’) (right). (D) Running average plots of log2fold change in gene expression in the Dnmt3a cKO model for genes commonly misregulated in both cKO models versus Dnmt3a dependent mCH (right) and mCG (left). (E) Running average plots of log2fold change in gene expression in the MeCP2 cKO for genes commonly misregulated in both cKO models versus Dnmt3a dependent mCH (right) and mCG (left). (F) Running average plots of log2fold change in gene expression for genes that are only significantly misregulated in the MeCP2 cKO model. (G–J) R2 values from analysis in panels C-F shown as blue, orange or green dots, respectively, plotted over 1000 random repetitions of the analysis with each repetition containing the same number of non-DEGs (padj >0.01). The results of random repetitions are shown as gray dots. All plots were made with DEGs padj <0.01. n = 2 mice per genotype for methylation data. n = 4 mice per genotype (RNA-seq). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
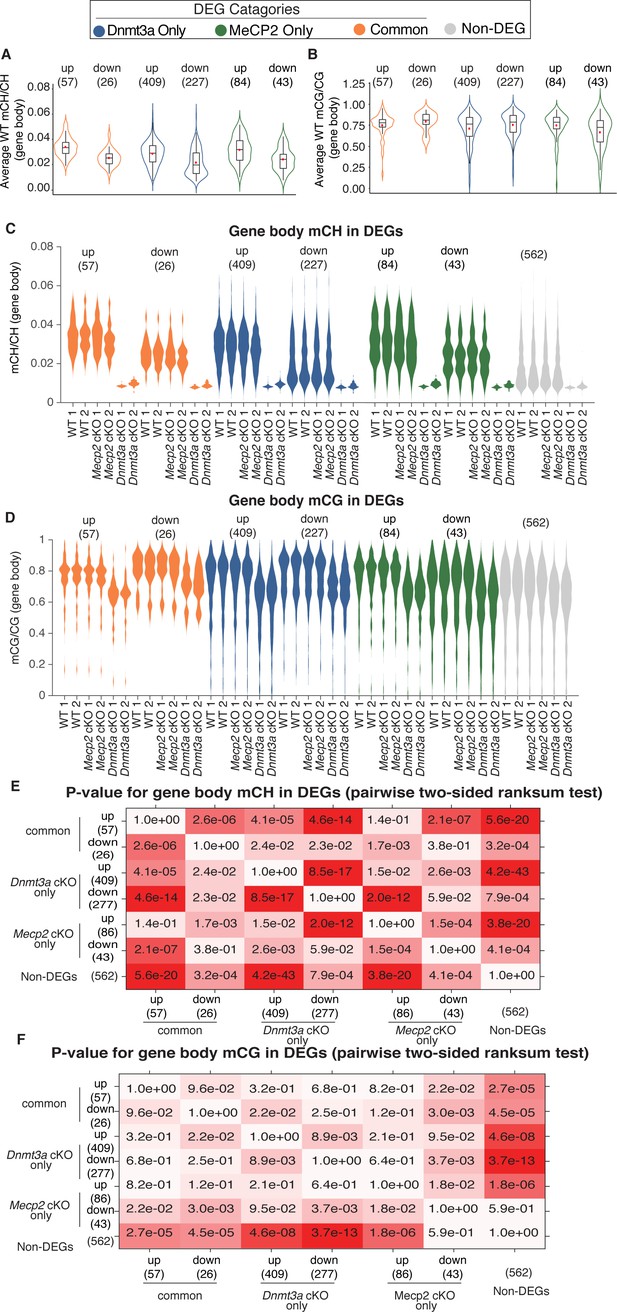
Methylation levels on categories of DEG genes in WT, Dnmt3a cKO, and Mecp2 cKO mice with statistical comparisons.
(A) Averaged gene-body mCH levels (average of two replicates) in different categories of DEGs in WT mice, highlighting the direction of difference in mCH levels between up- and down-regulated DEGs. The box plots within each segment highlights the median of the distribution, while the red point represents the mean of the distribution. (B) Averaged gene-body mCG levels (average of two replicates) in different categories of DEGs in WT mice, highlighting the direction of difference in mCG levels between up- and down-regulated DEGs. The box plots within each segment highlights the median of the distribution, while the red point represents the mean of the distribution. (C) Gene-body mCH levels in different categories of DEGs in WT, Dnmt3a cKO and Mecp2 cKO mice. (D) Gene-body mCG levels in different categories of DEGs in WT, Dnmt3a cKO and Mecp2 cKO mice. (E) P-value matrix for comparison of mCH levels of different categories of genes. The boxes are colored such that more significant p-values are darker shades of red. (F) P-value matrix for comparison of mCG levels of different categories of genes. As in C, the boxes are colored such that more significant p-values are darker shades of red.
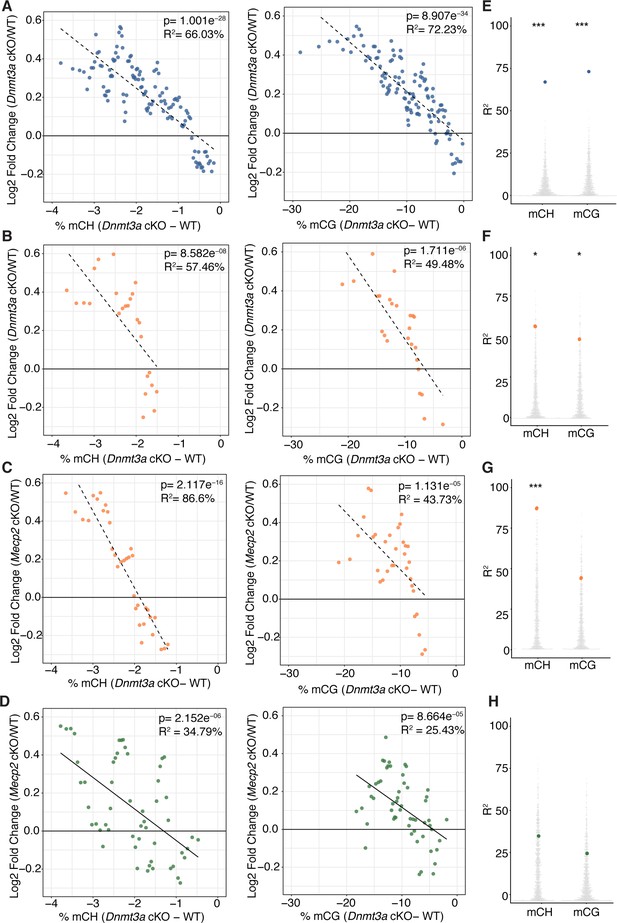
Integrative gene expression and methylation analysis excluding interneuron specific DEGs determined in PSEA analysis shows the same trends and statistical significance as in Figure 4.
(A–H) Running average and validation plots from Figure 4 re-plotted after removing DEGs that may represent changes in gene expression in the minor population of co-purified inhibitory interneurons. All plots were made with DEGs padj <0.01. n = 2 mice per genotype for methylation data. n = 4 mice per genotype (RNA-seq). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Tables
Summary of behavioral and physiological test results in cKO models.
| Symptom Category | Test/observation | Result |
|---|---|---|
| General Health | Reduced body weight | Dnmt3a |
| Prematurely moribund (humane end due to lesioning) | both | |
| Spasticity | Hind limb clasping | both |
| Repetitive Behavior | Forepaw stereotypies | Dnmt3a |
| Perseverative grooming and self-injury | both | |
| Nociceptive pain | Delayed response to hot plate | both |
| Delayed response to tail flick | Dnmt3a | |
| Apraxia | Poor nest building | both |
| Muscle Strength | Decreased grip strength | both |
| Motor | Rotarod | none |
| Parallel rod footslip- more footslips | Mecp2 | |
| Open field – decreased distance traveled | both | |
| Open field – decreased vertical activity | both | |
| Anxiety | Open field – time spent in center | none |
| Elevated plus maze- increased time in open arms | Mecp2 | |
| Light/dark box | none | |
| Learning and Memory | Conditioned fear – decreased contextual fear response | both |
| Conditioned fear – cued | none | |
| Conditioned fear – decreased response to tone #2 during fear learning (training) | Dnmt3a | |
| Sensory processing | Increased acoustic startle response | Dnmt3a |
| Paired pulse inhibition | none | |
| Social Behavior | Partition | none |
| Inhibitory signaling | Altered miniature inhibitory postsynaptic currents | both |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional Information |
|---|---|---|---|---|
| Antibody | Rabbit polyclonal anti-Dnmt3a | Santa Cruz Biotechnology | Cat. #: 20703; RRID:AB_2093990 | (1:1000) |
| Antibody | Rabbit polyclonal anti-MeCP2 | Huda Zoghbi lab (in house) | 0535 | (1:10,000) |
| Antibody | Rabbit polyclonal anti-histone H3 | Abcam | Cat. #: 1791; RRID:AB_302613 | (1:20,000) |
| Antibody | goat anti-rabbit-HRP | BioRad | Cat. #: 170–5046;RRID:AB_11125757 | (1:10,000) |
| Antibody | Mouse monoclonal anti-Dnmt3a | Novus | Cat. #: NB120-13888; RRID:AB_789607 | (1:250) |
| Antibody | Rabbit polyclonal anti-Myc | Sigma | Cat. #: C3956; RRID:AB_439680 | (1:200) |
| Antibody | Rabbit monoclonal anti-MeCP2 | Cell Signaling Technology | Cat. #: 3456; RRID:AB_2143849 | (1:500) |
| Antibody | Chicken polyclonal anti-GFP | Abcam | Cat. #: 13970; RRID:AB_300798 | (1:800) |
| Antibody | Goat anti-mouse Alexa Fluor 555 | Invitrogen | Cat. #: A21127; RRID:AB_141596 | (1:1000) |
| Antibody | Goat polyclonal anti-rabbit Dylight 488 | Bethyl Laboratories | Cat. #: A120-201D2; RRID:AB_10634085 | (1:750) |
| Antibody | Goat anti-chicken Alexa Fluor 488 | Invitrogen | Cat. #: A11039; RRID:AB_142924 | (1:750) |
| Antibody | Donkey anti-rabbit Alexa Fluor 568 | Thermo Fisher Scientific | Cat. #: A10042; RRID:AB_2534017 | (1:1000) |
| Antibody | Mouse monoclonal anti-NeuN | Millipore | Cat. #: MAB377; RRID:AB_2298772 | (1:300) (each batch needs to be empirically tested after labeling with Alexa Fluor 647) |
| Antibody | Rabbit polyclonal anti-GFP 488 | Thermo Fisher Scientific | Cat. #: A-21311; RRID:AB_221477 | (1:1000) |
| Other | cOmplete, EDTA-free Protease Inhibitor Cocktail | Sigma | Cat. #: 5056489001 | |
| Other | Pierce Universal Nuclease for Cell Lysis | Thermo Fisher Scientific | Cat. #: 88701 | |
| Other | Optimal Cutting Temperature medium | VWR | Cat. #: 25608–930 | |
| Other | VECTASHIELD HardSet Antifade Mounting Medium without DAPI | Vector Laboratories | Cat. #: H-1400 | |
| Other | Cryoseal XYL | Fisher Scientific | Cat. #: 22-050-262 | |
| Other | Aqueous Glutaraldehyde EM Grade, 10% 10 ML | Electron Microscopy Sciences | Cat. #: 16100 | Dilute fresh |
| Other | Paraformaldehyde | Sigma | Cat. #: P6148 | Make fresh |
| Other | X-Gal | Gold Biotechnologies | Cat. #: X4281C | |
| Other | RNasin Ribonuclease Inhibitors | Promega | Cat. #: N261A | |
| Other | UltraPure BSA (50 mg/mL) | Thermo Fisher Scientific | Cat. # AM2618 | |
| Other | DPBS, no calcium, no magnesium | Invitrogen | Cat. #: 14190144 | |
| Other | UltraPure DEPC-Treated Water (1L) | Invitrogen | Cat. #: 750023 | |
| Commercial assay, kit | GE Healthcare Amersham ECL Prime Western Blotting Detection Reagent | Fisher Scientific | Cat. #: 45010090 | |
| Commercial assay, kit | APEX Alexa Fluor 647 Antibody Labeling Kit | Thermo Fisher Scientific | Cat. #: A10475 | |
| Commercial assay, kit | Single Cell RNA Purification Kit | Norgen Biotek | Cat. #: 51800 | |
| Commercial assay, kit | NuGEN Ovation RNA-Seq v2 | NuGen | protocol p/n 7102, kit p/n 7102–08 | |
| Commercial assay, kit | Rubicon ThruPlex DNA-Seq | Rubicon Genomics | protocol: QAM-108–002, kit p/n R400428 | |
| Commercial assay, kit | EZ DNA Methylation-Direct Kit | Zymo | Cat. #: D5021 | |
| Other | Unmethylated lambda DNA spike-in | Promega | Cat. #: D1521 | |
| Strain, Strain background (Mus musculus) | Dnmt3aflox/flox mice | Dr. Margaret Goodell, Baylor College of Medicine (Can be purchased from Riken BRC) | Cat. #: RBRC03731; RRID:IMSR_RBRC03731 | |
| Strain, Strain background (Mus musculus) | Slc32a1-Cre+/+ mice | Jackson Laboratory | Cat. #: 017535; RRID:IMSR_JAX:017535 | backcrossed to C57Bl/6J |
| Strain, Strain background (Mus musculus) | Mecp2flox/flox and Mecp2flox/y mice | MMRRC | Cat. #: 011918-UCD; RRID:MMRRC_011918-UCD | backcrossed to C57Bl/6J |
| Strain, Strain background (Mus musculus) | R26-CAG-LSL-Sun1-sfGFP-Myc+/+ mice | Dr. M. Margarita Behrens, The Salk Institute for Biological Studies (can be purchased from Jackson Laboratory) | Cat. #: 021039; RRID:IMSR_JAX:021039 | backcrossed to C57Bl/6J |
| Software, algorithm | GraphPad Prism version 6 | GraphPad Software | www.graphpad.com | |
| Other | pClamp10 | Molecular Devices | https://www.moleculardevices.com | |
| Software, algorithm | Minianalysis 6.0.3 | Synaptosoft Inc | http://www.synaptosoft.com/MiniAnalysis/ | |
| Software, algorithm | STAR aligner version 2.5.3a | GitHub | https://github.com/alexdobin/STAR | |
| Software, algorithm | FeatureCount v1.5.3 | Subread | http://subread.sourceforge.net | |
| Software, algorithm | DESeq2 v1.6.2 | Bioconductor | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
Additional files
-
Supplementary file 1
Numbers and statistics for all mouse behavioral assays and methylation datasets.
- https://cdn.elifesciences.org/articles/52981/elife-52981-supp1-v1.xlsx
-
Supplementary file 2
RNA-seq data and DEGs used in figures.
- https://cdn.elifesciences.org/articles/52981/elife-52981-supp2-v1.xlsx
-
Supplementary file 3
Table of interneuron DEGs identified by PSEA analysis.
- https://cdn.elifesciences.org/articles/52981/elife-52981-supp3-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/52981/elife-52981-transrepform-v1.docx





