High-fat diet enhances starvation-induced hyperactivity via sensitizing hunger-sensing neurons in Drosophila
Figures
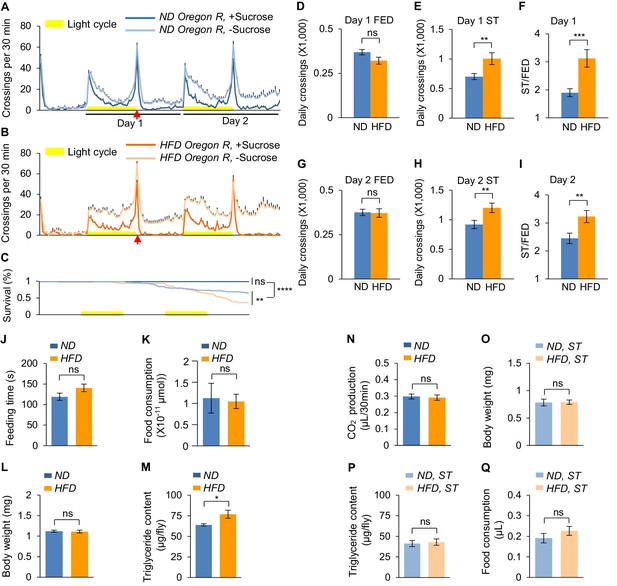
HFD promotesstarvation-induced hyperactivity in adult Drosophila.
(A–B) Wild-type Oregon-R virgin female flies fed with normal fly food (A, ND, blue) or high-fat diet (B, HFD, orange) were assayed in the presence (dark color) and absence (light color) of 5% sucrose using DAMS-based locomotion assay (n = 46–63). Midline crossing activities of indicated flies are shown. Yellow bars represent 12 hr light-on period in this and following figures. (C) Survival curves of different fly groups assayed in A-B. (D–I) Average daily midline crossing activity of fed flies (D and G), starved flies (E and H), and starvation-induced hyperactivity (F and I) of flies assayed in A-B. Data were broken down to Day 1 (D–F) and Day 2 (G–I). In the following figures only Day one data are presented. (J) Total duration of feeding time during 24 hr recording in the FLIC assay (n = 38–47). (K) 1 hr food consumption measured by the BARCODE assay (n = 12–16 biological replicates, each containing four flies). (L) Average body weight of indicated flies fed ad libitum (n = 6 biological replicates, each containing three flies). (M) Triglyceride storage of indicated flies fed ad libitum (n = 6). (N) 1 hr CO2 production of flies fed with ND or HFD (n = 12–14 biological replicates, each containing five flies). (O–Q) Average body weight (O, n = 18 biological replicates, each containing 10 flies), triglyceride storage (P, n = 6), and 100 mM sucrose consumption (Q, n = 29) of indicated flies starved for 24 hr (ST: starvation, the time point marked by arrows in A and B). ns, p>0.05; *p<0.05; ***p<0.001; ****p<0.0001. Student’s t-test and one-way ANOVA followed by post hoc test with Bonferroni correction were used for pair-wise and multiple comparisons, respectively.
-
Figure 1—source data 1
Raw data of the behavioral experiments shown in Figure 1 and Figure 1—figure supplements 1–5.
- https://cdn.elifesciences.org/articles/53103/elife-53103-fig1-data1-v2.xlsx
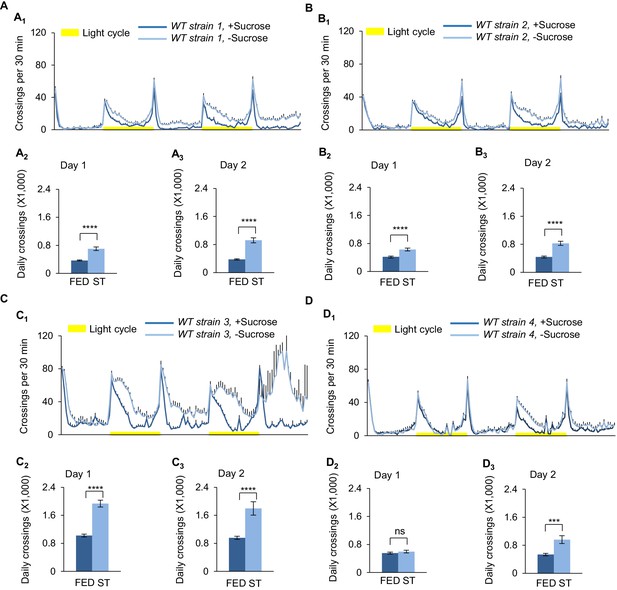
Starvation-induced hyperactivity across different wild-type fly strains.
(A1, B1, C1, D1) Midline crossing activity of different wild-type strains assayed in the presence or absence of 5% sucrose (n = 46–63). (A2, B2, C2, D2) Average daily midline crossing activity in Day 1 of flies assayed in A1-D1. (A3, B3, C3, D3) Average daily midline crossing activity in Day 2 of flies assayed in A1-D1. ns, p>0.05; *p<0.05; **p<0.01; ***p<0.001. Strain 1: Oregon-R; Strain 2: Canton-S; Strain 3: a control strain from VDRC; Strain 4: a control strain from Bloomington. Note that for all these strains, most of the starved flies were dead after two days of starvation. Student’s t-test and one-way ANOVA followed by post hoc test with Bonferroni correction were used for pair-wise and multiple comparisons, respectively.
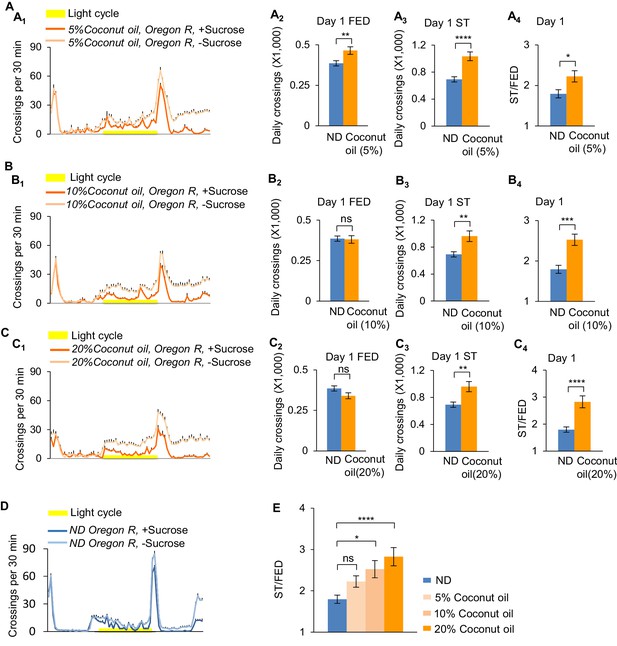
HFD promotes starvation-induced hyperactivity in a dose-dependent manner.
Wild-type Oregon-R virgin female flies fed with normal fly food (ND, blue) or different concentrations of coconut oil (orange) were assayed in the presence (dark color) and absence (light color) of 5% sucrose using DAMS-based locomotion assay (n = 52–63). Midline crossing activities of indicated flies are shown in A1, B1, C1, and D. (A2, B2, C2) Average daily midline crossing activity of fed flies assayed in A1, B1, C1, and D. (A3, B3, C3) Average daily midline crossing activity of s;tarved flies assayed in A1, B1, C1, and D. (A4, B4, C4) Starvation-induced hyperactivity of flies assayed in A1, B1, C1, and D. (E) A summary of the effect of coconut oil feeding. ns, p>0.05; *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. Student’s t-test and one-way ANOVA followed by post hoc test with Bonferroni correction were used for pair-wise and multiple comparisons, respectively.
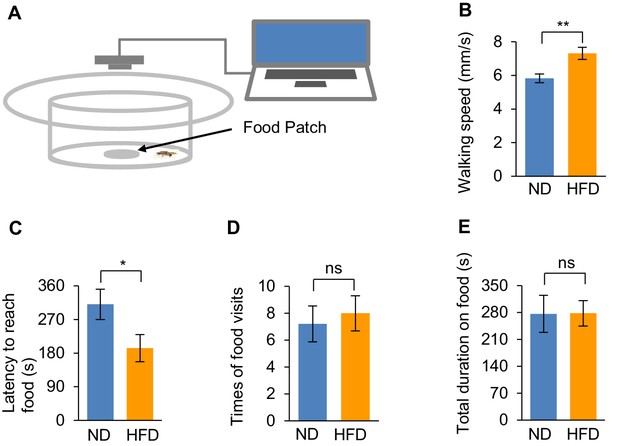
HFD feeding enhanced starvation-induced food seeking.
(A) Schematic diagram of the video recording-based food seeking assay. Briefly, individual flies starved for 24 hr were introduced into a behavioral chamber with a small food patch (5% sucrose) located in the center, and their positions and moving trajectories were recorded by a camera placed on top and analyzed by a custom computer program. (B–C) Average walking speed before locating the food source (B) and latency to reach the food source (C). (D–E) Total number of food visits (D) and total duration on food (E) of starved flies during the 30 min assay. n = 14–19. Student’s t-test was used for pair-wise comparisons.
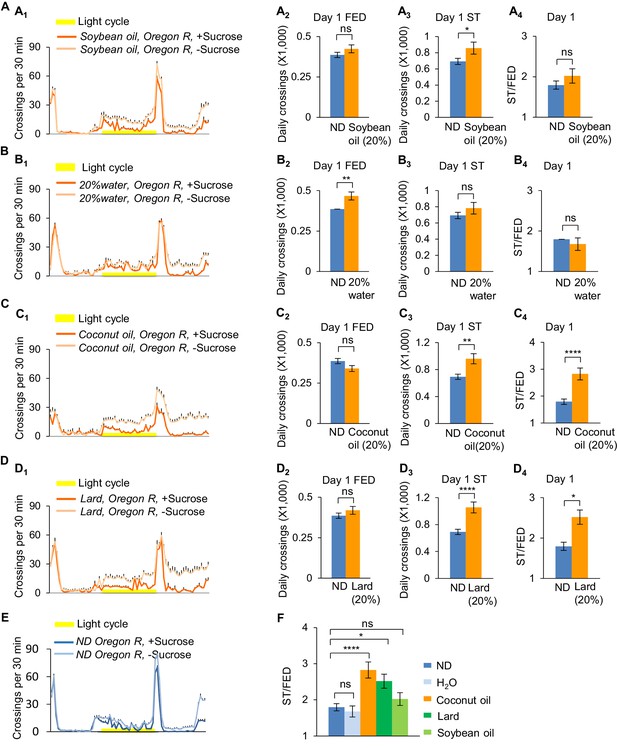
Dietary saturated fat promotes starvation-induced hyperactivity.
Wild-type Oregon-R virgin female flies fed with normal fly food (ND, blue) or different types of lipids (orange) were assayed in the presence (dark color) and absence (light color) of 5% sucrose using DAMS-based locomotion assay (n = 39–63). Midline crossing activities of indicated flies are shown in A1, B1, C1, D1, and E. (A2, B2, C2, D2) Average daily midline crossing activity of fed flies assayed in A1, B1, C1, D1, and E. (A3, B3, C3, D3) Average daily midline crossing activity of starved flies assayed in A1, B1, C1, D1, and E. (A4, B4, C4, D4) Starvation-induced hyperactivity of flies assayed in A1, B1, C1, D1, and E. (F) A summary of the effect of different oil feeding. Note that data shown in E are the same as data shown in Figure 1—figure supplement 2D. ns, p>0.05; *p<0.05; **p<0.01; ****p<0.0001. Student’s t-test and one-way ANOVA followed by post hoc test with Bonferroni correction were used for pair-wise and multiple comparisons, respectively.
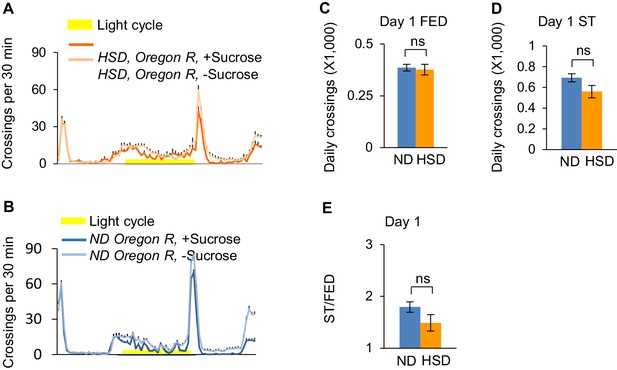
Dietary sugar does not affect starvation-induced hyperactivity.
Wild-type Oregon-R virgin female flies fed with ND (blue) or HSD (orange) were assayed in the presence (dark color) and absence (light color) of 5% sucrose using DAMS-based locomotion assay (n = 41–63). Midline crossing activities of indicated flies are shown in A-B. (C–D) Average daily midline crossing activity of fed flies (C) and starved flies (D) assayed in A-B. (E) Starvation-induced hyperactivity of flies assayed in A-B. ns, p>0.05; *p<0.05; **p<0.01; ****p<0.0001. Student’s t-test and one-way ANOVA followed by post hoc test with Bonferroni correction were used for pair-wise and multiple comparisons, respectively.
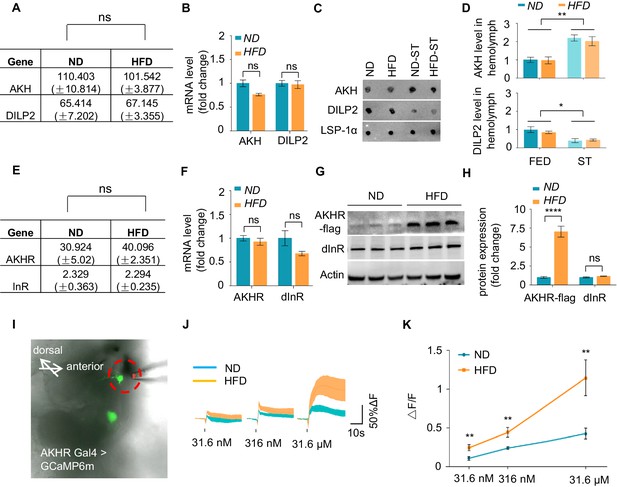
HFD increases neuronal AKHR protein.
(A–B) AKH and DILP2 mRNA levels of wild-type virgin female flies fed with ND or HFD. The fly heads and associated tissues were collected and subjected to RNAseq (A) or quantitative RT-PCR (B) (n = 3 biological replicates, each containing 25 flies). (C–D) AKH and DILP2 protein levels in the hemolymph of wild-type virgin female flies fed with ND or HFD and assayed under both fed and starved conditions (n = 3 biological replicates, each containing 30 flies). Hemolymph samples were collected from the flies and AKH and DILP2 protein levels were analyzed by dot blot with AKH and DILP2 antibodies (C), and the quantification was shown in (D). (E–F) AKHR and InR mRNA levels were analyzed similar to A-B (n = 3 biological replicates, each containing 25 flies). (G–H) AKHR and dInR protein levels of fly heads were analyzed by western blot (n = 3 biological replicates, each containing 25 flies). Note that AKHR-flag knock-in flies were generated by inserting FLAG sequence into the C terminal of AKHR gene via CRISPR/cas9 mediated gene editing. Antibodies against FLAG and dInR were used in the western blot. (I) Schematic diagram of the ex vivo calcium imaging. Green signals indicate AKHR+ neurons expressing GCaMP6 (AKHR:BD/+; nSyb:AD/UAS-GCaMP6m). Dotted red circle indicates the position of the pipette delivering synthetic AKH. (J–K) Representative traces (J) and quantification (K) of peak calcium transients of AKHR+ neurons upon AKH administration (n = 7–10). ns, p>0.05; *p<0.05; ***p<0.001. Student’s t-test and one-way ANOVA followed by post hoc test with Bonferroni correction were used for pair-wise and multiple comparisons, respectively.
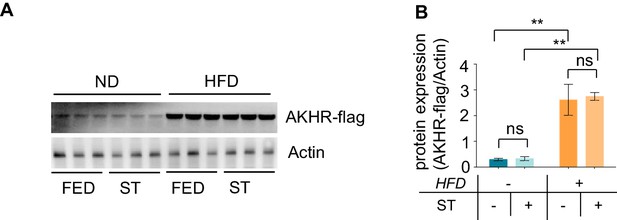
HFD increases neuronal AKHR protein accumulation in both fed and starved conditions.
(A-B) AKHR-flag knock-in flies were fed with ND or HFD for 5 d, and then starved for 18 hr or not. AKHR-flag protein level in the head tissue was analyzed by western blot (A, n = 3 biological replicates, each containing 25 flies), and the quantification was shown in (B). *p < 0.05. Student’s t-test was used for pair-wise comparisons.
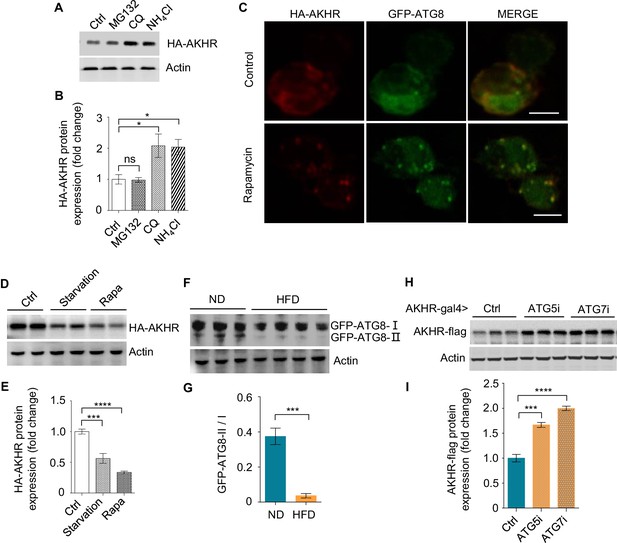
AKHR accumulation is induced by the suppression of autophagy.
(A–B) Quantification of AKHR protein levels in cultured Drosophila S2 cells upon the treatment of proteasome and lysosome inhibitors (n = 3 biological replicates). Cultured cells transiently expressing AKHR-HA were treated with the indicated chemicals. The lysates were analyzed by western blot with HA antibody (A) and quantified in (B). (C) Formation of autophagosome in cultured S2 cells. Cultured cells transiently expressing AKHR-HA and GFP-ATG8 were treated with 200 nM rapamycin for 12 hr and stained with HA antibody. (D–E) Quantification of AKHR protein levels in Cultured Drosophila S2 cells upon starvation and the treatment of rapamycin (n = 3 biological replicates). Cultured S2 cells transiently expressing AKHR-HA were starved or treated with rapamycin, and AKHR-HA was analyzed by western blot. (F–G) Quantification of ATG8 activation in the head tissues of flies in vivo (n = 3–4 biological replicates, each containing 25 flies). GFP-ATG8 was specifically expressed in AKHR+ neurons by using the GAL4-UAS system. Flies were fed with ND or HFD and their head tissues were collected for western blot with GFP antibody. The ratio between ATG8-II and ATG8-I bands indicates the level of autophagy. (H–I) Quantification of AKHR protein levels upon suppressing neuronal autophagy in AKHR+ neurons in vivo (n = 3 biological replicates,, each containing 25 flies). ATG5 and ATG7 were knocked down by the expression of RNAi constructs in AKHR+ neurons. AKHR-flag expression was assayed by western blot. ns, p>0.05; *p<0.05; **p<0.01; ****p<0.0001. Student’s t-test and one-way ANOVA followed by post hoc test with Bonferroni correction were used for pair-wise and multiple comparisons, respectively.
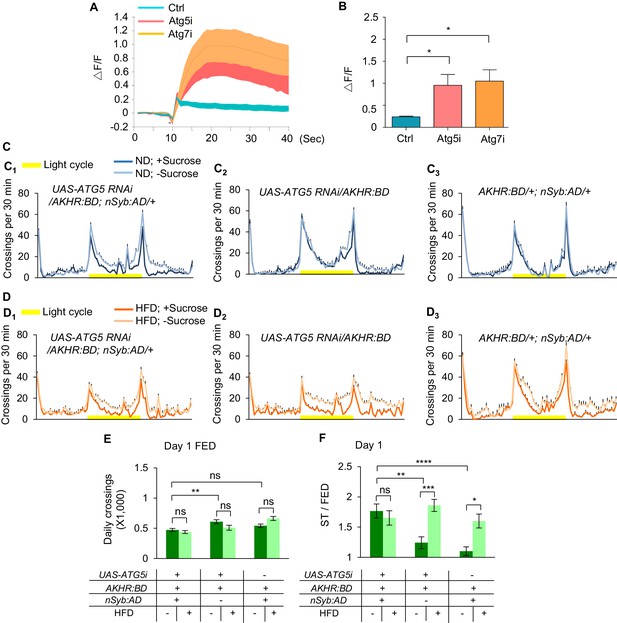
Inhibition of autophagy increases starvation-induced hyperactivity.
(A–B) Representative traces (A) and quantification (B) of peak calcium responses of AKHR+ neurons to 316 nM AKH upon suppressing neuronal autophagy in AKHR+ neurons (n = 7–10). (C–D) Midline crossing activity of indicated genotypes and diet treatments assayed in the presence or absence of 5% sucrose (n = 43–62). (E) Average daily midline crossing activity of fed flies assayed in C-D. (F) Starvation-induced hyperactivity of flies assayed in C-D. ns, p>0.05; *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. One-way and two-wayANOVA followed by post hoc test with Bonferroni correction were used for multiple comparisons when applicable.
-
Figure 4—source data 1
Raw data of the behavioral experiments shown in Figure 4 and Figure 4—figure supplements 1–4.
- https://cdn.elifesciences.org/articles/53103/elife-53103-fig4-data1-v2.xlsx
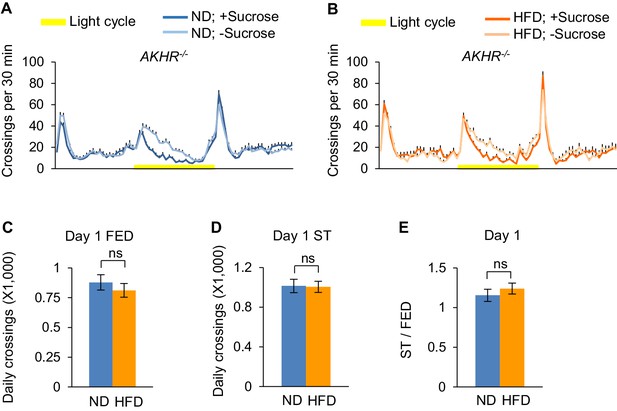
AKHR gene is required for the enhancement of starvation-induced hyperactivity by HFD.
(A–B) Midline crossing activity of ND- and HFD-fed AKHR-/- flies assayed in the presence or absence of 5% sucrose (n = 43–73). (C–D) Average daily midline crossing activity of fed flies (C) and starved flies (D) assayed in A-B. (E) Starvation-induced hyperactivity of flies assayed in A-B. ns, p > 0.05. Student’s t-test was used for pair-wise comparisons.
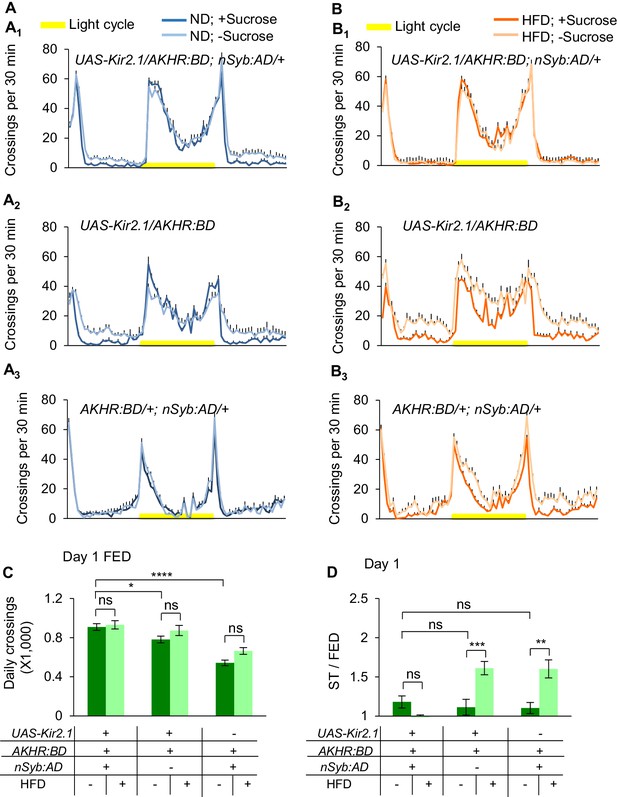
Silencing AKHR+ neurons diminishes the effect of HFD feeding to enhance starvation-induced hyperactivity.
(A–B) Midline crossing activity of indicated genotypes and diet treatments assayed in the presence or absence of 5% sucrose (n = 39–64). (C) Average daily midline crossing activity of fed flies assayed in A-B. (D) Starvation-induced hyperactivity of flies assayed in A-B. ns, p>0.05; *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. One-way and two-way ANOVA followed by post hoc test with Bonferroni correction were used for multiple comparisons when applicable.
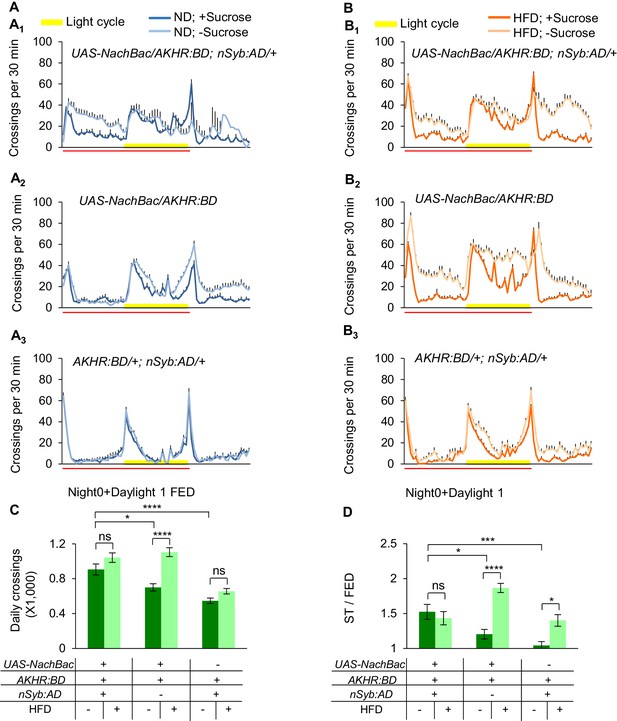
Activation AKHR+ neurons enhances starvation-induced hyperactivity under ND feeding.
(A–B) Midline crossing activity of indicated genotypes and diet treatments assayed in the presence or absence of 5% sucrose (n = 30–61). Red line indicates the time window for quantifying average midline crossing activity. (C) Average daily midline crossing activity of fed flies assayed in A-B. (D) Starvation-induced hyperactivity of flies assayed in A-B. ns, p>0.05; *p<0.05; ***p<0.001; ****p<0.0001. One-way and two-way ANOVA followed by post hoc test with Bonferroni correction were used for multiple comparisons when applicable.
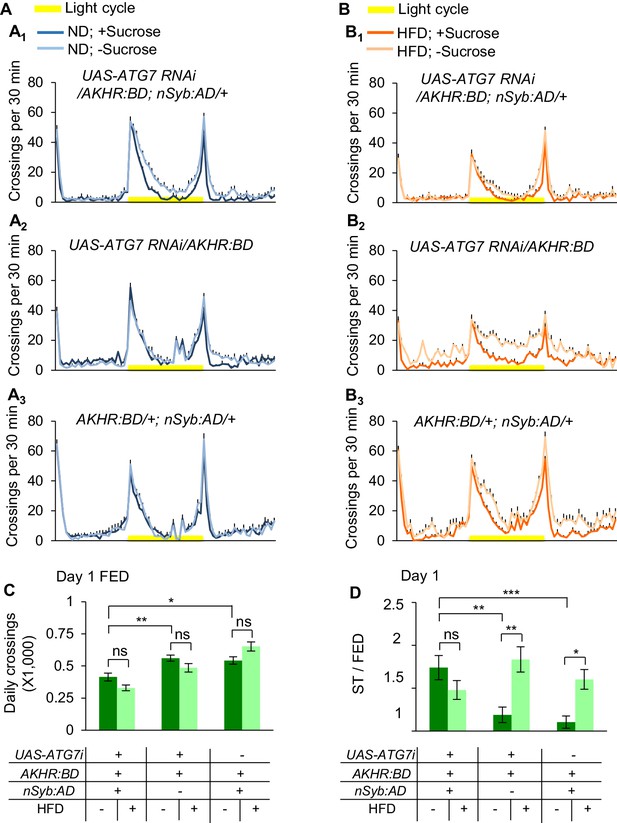
Knocking down ATG7 in AKHR+ neurons promotes starvation-induced hyperactivity.
(A–B) Midline crossing activity of indicated genotypes and diet treatments assayed in the presence or absence of 5% sucrose (n = 30–60). (C) Average daily midline crossing activity of fed flies assayed in A-B. (D) Starvation-induced hyperactivity of flies assayed in A-B. ns, p>0.05; *p<0.05; **p<0.01; ****p<0.0001. One-way and two-way ANOVA followed by post hoc test with Bonferroni correction were used for multiple comparisons when applicable.
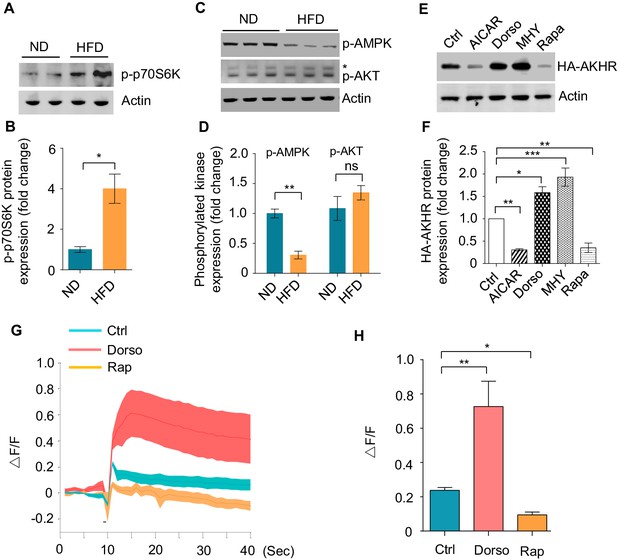
HFD activates TOR signaling.
(A–D) p70s6k/AKT/AMPK phosphorylation under ND vs. HFD feeding conditions. The head tissues of wild-type flies fed with ND or HFD were harvested and subjected to western blot with phosphorylated p70s6k antibody (A–B) and p-AKT/p-AMPK antibodies (C–D) (n = 3 biological replicates, each containing 25 flies). *Nonspecific bands. (E–F) AKHR accumulation upon pharmacological manipulation of AMPK-TOR signaling in cultured Drosophila S2 cells (n = 3). Cultured S2 cells transiently expressing AKHR-HA were treated with the indicated chemicals and then analyzed by western blot with HA antibody. (G–H) Representative traces (G) and quantification (H) of peak calcium responses of AKHR+ neurons to 316 nM AKH, with or without the indicated drug treatment (n = 7–10). ns, p>0.05; *p<0.05; **p<0.01; ***p<0.001. Student’s t-test and one-way ANOVA followed by post hoc test with Bonferroni correction were used for pair-wise and multiple comparisons, respectively.
-
Figure 5—source data 1
Top differentially expressed genes upon HFD feeding.
Wild-type virgin female flies were fed with ND or HFD for 5 d before their head tissues harvested and subjected to RNAseq. Top 40 differentially expressed genes (20 up-regulated and 20 down-regulated) were shown.
- https://cdn.elifesciences.org/articles/53103/elife-53103-fig5-data1-v2.xlsx
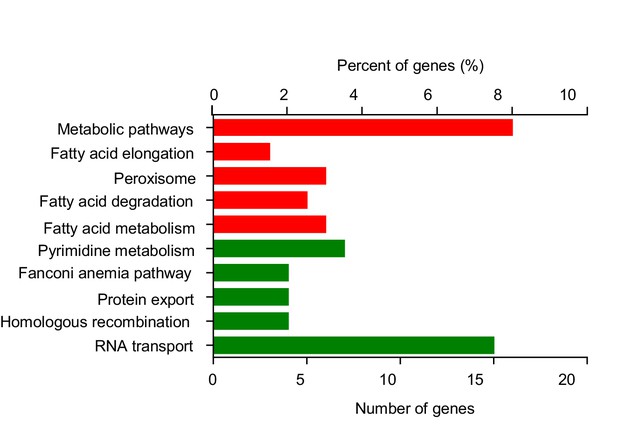
KEGG pathway analysis of differentially expressed genes upon HFD feeding.
Wild-type virgin female flies were fed with ND or HFD for 5 d before their head tissues harvested and subjected to RNAseq. The highly influenced KEGG pathways were shown (red: up-regulated by HFD; green: down-regulated by HFD).
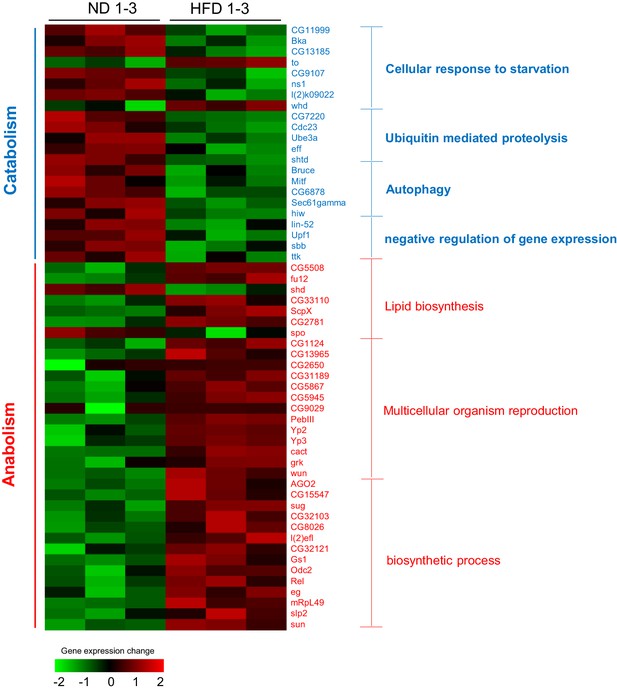
TOR downstream genes are modulated by HFD feeding.
Heat map of differentially expressed genes enriched in TOR signaling pathways. Wild-type virgin female flies were fed with ND or HFD for 5 d before their head tissues harvested and subjected to RNAseq. Green and red blocks represent down-regulated genes and up-regulated genes upon HFD feeding, respectively. Red and blue font colors indicate anabolism and catabolism genes, respectively. Note that most anabolism related genes were up-regulated while most catabolism related genes were down-regulated by HFD.
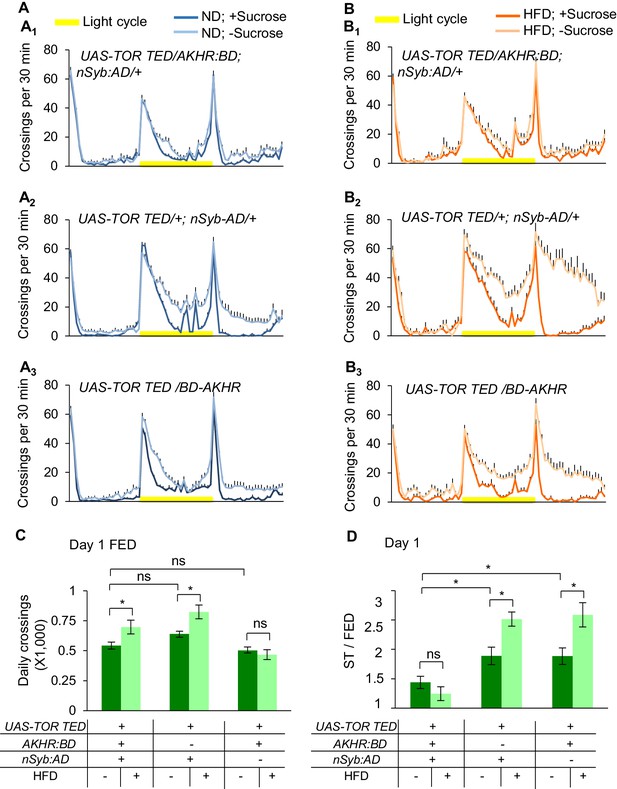
AMPK-TOR signaling modulates starvation-induced hyperactivity.
(A–B) Midline crossing activity of indicated genotypes and diet treatments assayed in the presence or absence of 5% sucrose (n = 35–58). (C) Average daily midline crossing activity of fed flies assayed in A-B. (D) Starvation-induced hyperactivity of flies assayed in A-B. ns, p>0.05; *p<0.05; **p<0.01; ***p<0.001. One-way and two-way ANOVA followed by post hoc test with Bonferroni correction were used for multiple comparisons when applicable.
-
Figure 6—source data 1
Raw data of the behavioral experiments shown in Figure 6 and Figure 6—figure supplements 1–3.
- https://cdn.elifesciences.org/articles/53103/elife-53103-fig6-data1-v2.xlsx
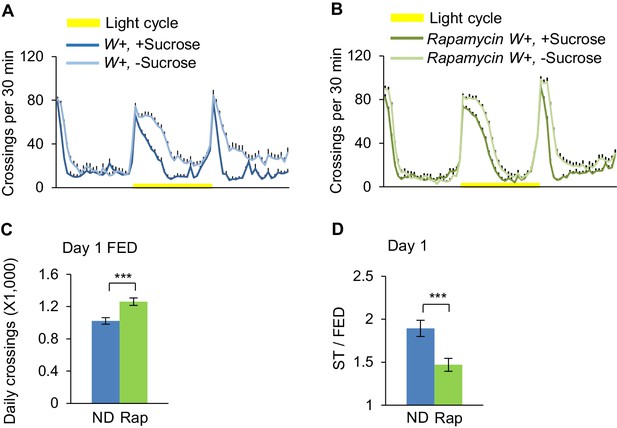
Rapamycin feeding suppresses starvation-induced hyperactivity.
Wild-type virgin female flies fed with ND (blue) or ND plus rapmycin (green) were assayed in the presence (dark color) and absence (light color) of 5% sucrose using DAMS-based locomotion assay (n = 48–64). Midline crossing activities of indicated flies are shown in A-B. (C) Average daily midline crossing activity of fed flies assayed in A-B. (D) Starvation-induced hyperactivity of flies assayed in A-B ns, p>0.05; *p<0.05; **p<0.01; ****p<0.0001. Student’s t-test and one-way ANOVA followed by post hoc test with Bonferroni correction were used for pair-wise and multiple comparisons, respectively.
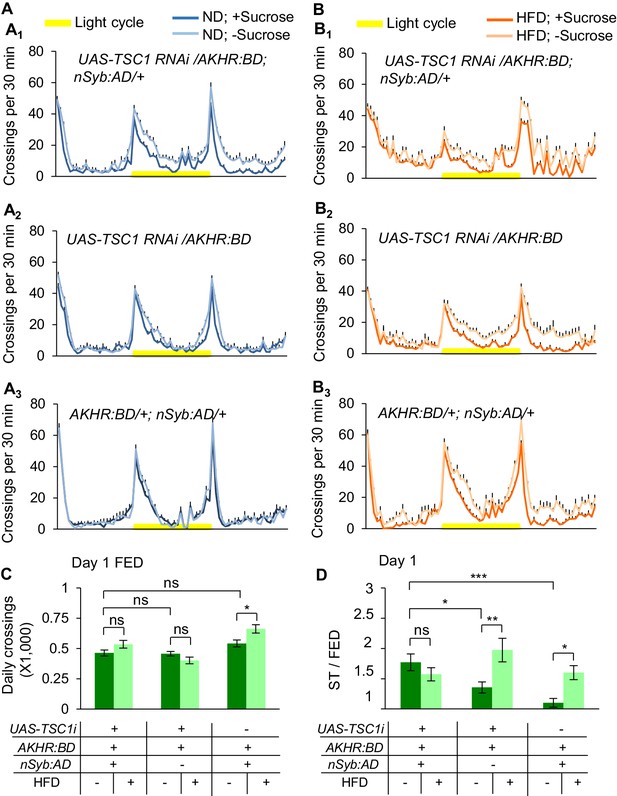
Knocking down TSC1 in AKHR+ neurons promotes starvation-induced hyperactivity.
(A–B) Midline crossing activity of indicated genotypes and diet treatments assayed in the presence or absence of 5% sucrose (n = 36–46). (C) Average daily midline crossing activity of fed flies assayed in A-B. (D) Starvation-induced hyperactivity of flies assayed in A-B. ns, p>0.05; *p<0.05; **p<0.01; ****p<0.0001. Student’s t-test, one-way and two-way ANOVA followed by post hoc test with Bonferroni correction were used for multiple comparisons when applicable.
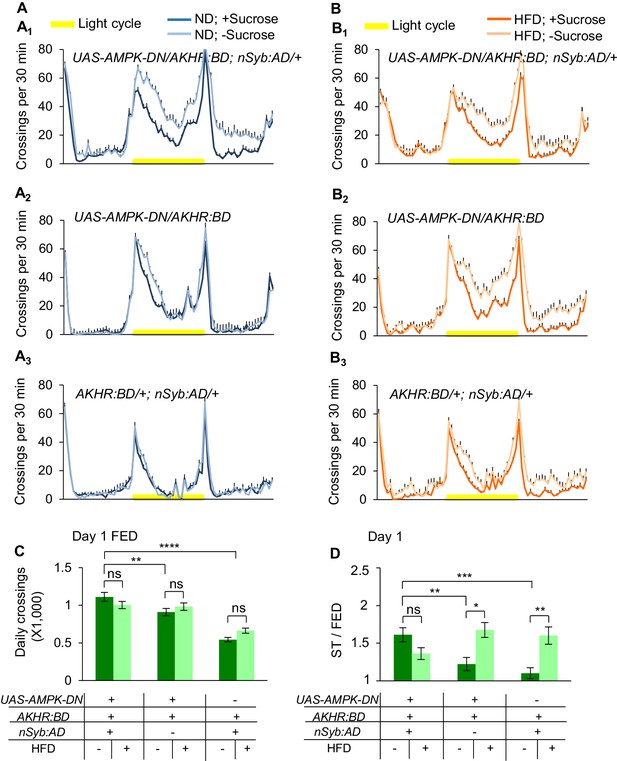
Suppressing AMPK signaling in AKHR+ neurons promotesstarvation-induced hyperactivity.
(A–B) Midline crossing activity of indicated genotypes and diet treatments assayed in the presence or absence of 5% sucrose (n = 30–59). (C) Average daily midline crossing activity of fed flies assayed in A-B. (D) Starvation-induced hyperactivity of flies assayed in A-B. ns, p>0.05; *p<0.05; **p<0.01; ****p<0.0001. Student’s t-test, one-way and two-way ANOVA followed by post hoc test with Bonferroni correction were used for multiple comparisons when applicable.
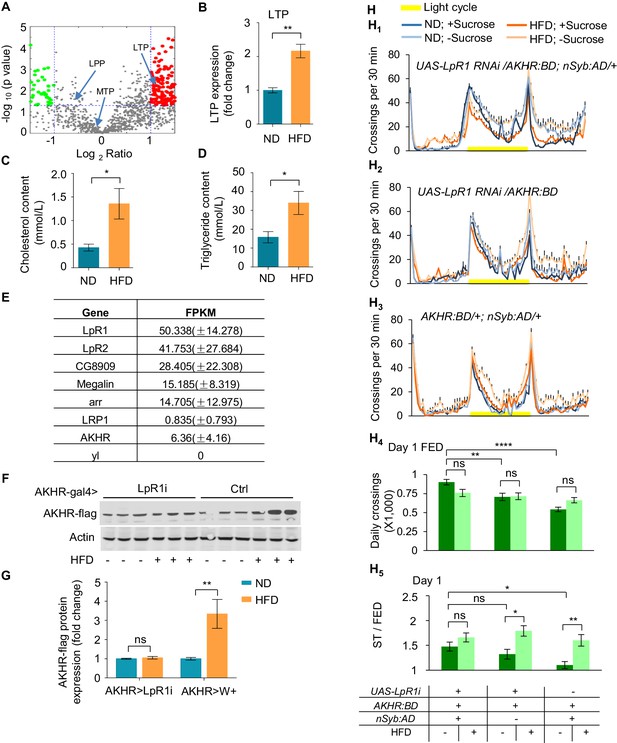
LpR1 is required for HFD-strengthened hyperactivity under starvation.
(A–B) LTP protein levels in the hemolymph. Hemolymph was collected from flies fed with ND or HFD and subjected to LC-MS/MS analysis. Volcano plot (A) shows peptides with differential enrichment (under ND vs. HFD feeding conditions). The horizontal line indicates the significance threshold (p=0.01) and the vertical lines indicate two-fold change. Green and red dots represent down-regulated and up-regulated peptides, respectively. Arrows indicate three lipoproteins (LTP, LPP and MTP). The level of LTP was shown in B (n = 3 biological replicates, each containing ~300 flies). (C–D) The cholesterol level (C) and triglyceride level (D) in the hemolymph harvested from ND vs. HFD fed flies (n = 4–6 biological replicates, each containing 60 flies). (E) Single cell RNAseq shows the gene expression level of candidate lipoprotein receptors in AKHR+ neurons (n = 5 cells). (F–G) AKHR protein levels from the head tissues of the indicated genotypes fed with ND or HFD (n = 3 biological replicates, each containing 25 flies). (H1–H3) Midline crossing activity of indicated genotypes and diet treatments assayed in the presence or absence of 5% sucrose (n = 43–57). (H4) Average daily midline crossing activity of fed flies assayed in H1-H3. (H5) Starvation-induced hyperactivity of flies assayed in H1-H3. ns, p>0.05; *p<0.05; **p<0.01. Student’s t-test, one-way and two-way ANOVA followed by post hoc test with Bonferroni correction were used for multiple comparisons when applicable.
-
Figure 7—source data 1
Raw data of the behavioral experiments and mass spec experiments shown in Figure 7.
- https://cdn.elifesciences.org/articles/53103/elife-53103-fig7-data1-v2.xlsx
-
Figure 7—source data 2
Top differentially expressed peptides upon HFD feeding.
Wild-type virgin female flies were fed with ND or HFD for 5 d before their hemolymph harvested and subjected to LC-MS/MS. Top 40 differentially expressed peptides (20 up-regulated and 20 down-regulated) were shown.
- https://cdn.elifesciences.org/articles/53103/elife-53103-fig7-data2-v2.xlsx
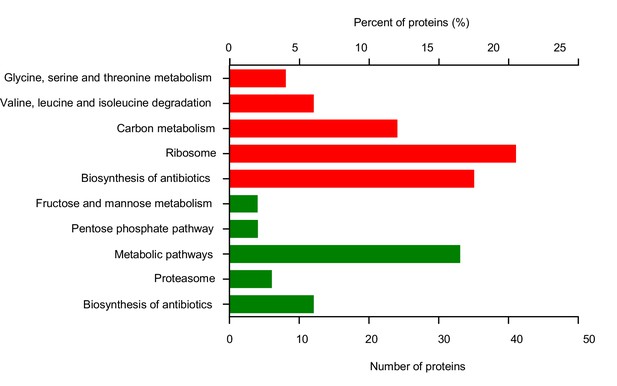
KEGG pathway analysis of differentially expressed peptides upon HFD feeding.
Wild-type virgin female flies were fed with ND or HFD for 5 d before their hemolymph harvested and subjected to LC-MS/MS. The highly influenced KEGG pathways were shown (red: up-regulated by HFD; green: down-regulated by HFD).
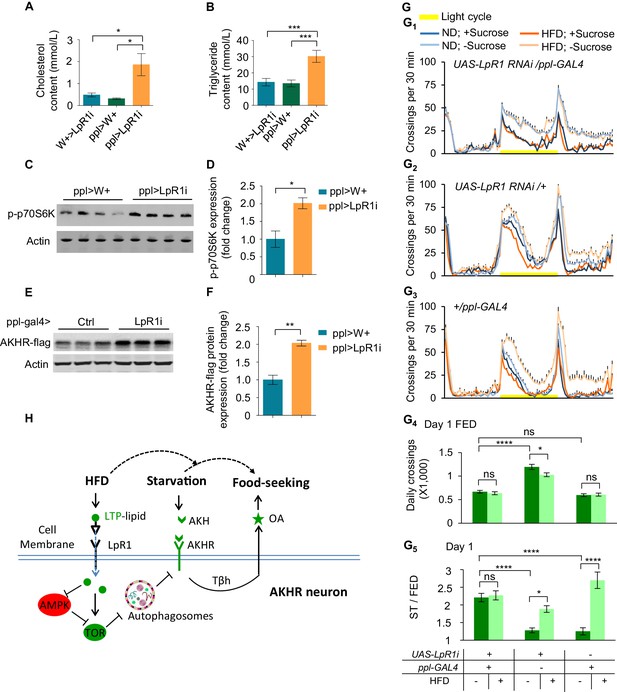
Diet-independent hyperlipidemia promotes the excitability of AKHR+ neurons and starvation-induced hyperactivity.
(A–B) Average levels of cholesterol (A) and triglyceride (B) in the hemolymph of the indicated genotypes (n = 4–11 biological replicates, each containing 60 flies). (C–F) The levels of phosphorylated p70s6k (C–D) and AKHR-flag (E–F) from the head tissues of indicated genotypes (n = 4 biological replicates, each containing 25 flies). (G1-3) Midline crossing activity of indicated genotypes and diet treatments assayed in the presence or absence of 5% sucrose (n = 41–63). (G4) Average daily midline crossing activity of fed flies assayed in G1-G3. (G5) Starvation-induced hyperactivity of flies assayed in G1-G3. ns, p>0.05; *p<0.05; ***p<0.001; ****p<0.0001. Student’s t-test, one-way and two-way ANOVA followed by post hoc test with Bonferroni correction were used for multiple comparisons when applicable. (H) A working model: We propose the following working model based on our results. AKHR protein levels in AKHR+ neurons are well maintained via protein expression and autophagic protein degradation. Under ND feeding, starvation induces the expression and release of AKH, which in turn activates AKHR+ neurons and hence the release of OA. OA signaling promotes starvation-induced hyperactivity in these starved flies. Upon HFD feeding, however, the regulation of starvation-induced hyperactivity is disrupted and greatly enhanced. Mechanistically, LTP delivers excess dietary lipids to AKHR+ neurons via its cognate receptor LpR1. Subsequently, neuronal autophagy is suppressed in AKHR+ neurons via AMPK-TOR signaling, which leads to AKHR accumulation. As a result, AKHR+ neurons become hyper sensitive to AKH and therefore starvation-induced hyperactivity is enhanced.
-
Figure 8—source data 1
Raw data of the behavioral experiments shown in Figure 8.
- https://cdn.elifesciences.org/articles/53103/elife-53103-fig8-data1-v2.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (D. melanogaster) | UAS-TORTED | Bloomington Drosophila Stock Center | Cat. #7013; RRID:BDSC_7013 | FlyBase symbol: P{w+mC=UAS-Tor.TED} |
| Genetic reagent (D. melanogaster) | UAS-AMPK-DN | Bloomington Drosophila Stock Center | Cat. #32112; RRID:BDSC_32112 | FlyBase symbol: P{w+mC=UAS-AMPKalpha.K57A} |
| Genetic reagent (D. melanogaster) | UAS-mCD8GFP | Bloomington Drosophila Stock Center | Cat. #32186; RRID:BDSC_32186 | FlyBase symbol: P{y+t7.7 w+mC = 10XUAS-IVS-mCD8::GFP} |
| Genetic reagent (D. melanogaster) | UAS-mCD8GFP | Bloomington Drosophila Stock Center | Cat. #32186; RRID:BDSC_32186 | FlyBase symbol: P{y+t7.7 w+mC = 10XUAS-IVS-mCD8::GFP} |
| Cell line (include species here) | ppl-GAL4 | Bloomington Drosophila Stock Center | Cat. #58768; RRID:BDSC_58768 | FlyBase symbol: P{w+mC = ppl-GAL4.P} |
| Genetic reagent (D. melanogaster) | UAS-NaChBac | Bloomington Drosophila Stock Center | Cat. # 9469; RRID:BDSC_9469 | FlyBase symbol: P{w+mC = UAS-NaChBac} |
| Genetic reagent (D. melanogaster) | UAS-nSyb-AD-AKHR-BD | Our laboratory PMID:27612383 | Dr. Liming Wang (Zhejiang University) | |
| Genetic reagent (D. melanogaster) | UAS-nSyb-AD | Our laboratory PMID:27612383 | Dr. Liming Wang (Zhejiang University) | |
| Genetic reagent (D. melanogaster) | UAS- AKHR-BD | Our laboratory PMID:27612383 | Dr. Liming Wang (Zhejiang University) | |
| Genetic reagent (D. melanogaster) | UAS- AKHR-/- | Our laboratory PMID:27612383 | Dr. Liming Wang (Zhejiang University) | |
| Genetic reagent (D. melanogaster) | UAS- kir2.1 | Our laboratory PMID:30209352 | Dr. Liming Wang (Zhejiang University) | |
| Genetic reagent (D. melanogaster) | UAS-TSC1 RNAi | Tsinghua Fly Center | Cat. #5074 | |
| Genetic reagent (D. melanogaster) | UAS-ATG7 RNAi | Tsinghua Fly Center | Cat. #2793 | |
| Genetic reagent (D. melanogaster) | UAS-LpR1 RNAi | Tsinghua Fly Center | Cat. #2568 | |
| Genetic reagent (D. melanogaster) | UAS-ATG5 RNAi | Laboratory of Dr. Chao Tong | Dr. Chao Tong (Zhejiang University) | |
| Genetic reagent (D. melanogaster) | AKHR-FLAG | This paper | Dr. Liming Wang (Zhejiang University) | |
| Cell line (D. melanogaster) | S2 | Thermofisher | Cat. #R69007 | |
| Antibody | Anti-phospho-Akt (Ser473) (Rabbit polyclonal) | Cell Signaling Technology | Cat. #9271; RRID:AB_329825 | WB (1:1000) |
| Antibody | Anti-phospho-AMPK (Thr172) (Rabbit polyclonal) | Cell Signaling Technology | Cat. #2535; RRID:AB_331250 | WB (1:1000) |
| Antibody | Anti-GFP (Rabbit polyclonal) | Cell Signaling Technology | Cat. #2555; RRID:AB_10692764 | WB (1:1000) |
| Antibody | Anti-FLAG (Rabbit polyclonal) | Cell Signaling Technology | Cat. #86861; RRID:AB_2800094 | WB (1:1000) |
| Antibody | Anti-HA (Rabbit polyclonal) | Cell Signaling Technology | Cat. #3724; RRID:AB_1549585 | WB (1:1000) |
| Antibody | Anti-HA (Mouse monoclonal) | Cell Signaling Technology | Cat. #2367, RRID:AB_10691311 | IF(1:200); WB (1:1000) |
| Antibody | Anti-actin (Rabbit polyclonal) | Sigma-Aldrich | Cat. #A5316; RRID:AB_476743 | WB (1:5000) |
| Antibody | Anti-AKH (Rabbit polyclonal) | biorbyt | Cat. #orb97730 | DOT BLOT (1:1000) |
| Antibody | Anti-Dilp2 (Rabbit polyclonal) | Laboratory of Dr. Zhefeng Gong | DOT BLOT (1:1000) Dr. Zhefeng Gong (Zhejiang Universtiy) | |
| Recombinant DNA reagent | AKHR-HA (plasmid) | This paper | Dr. Liming Wang (Zhejiang University) | |
| Recombinant DNA reagent | GFP-ATG8 (plasmid) | This paper | Dr. Liming Wang (Zhejiang University) | |
| Peptide, recombinant protein | AKH | PMID:2117437 | pGlu-Leu-Thr-Phe-Ser-Pro-Asp-Trp-NH2 | |
| Chemical compound, drug | MG132 | Beyotime Biotechnology | Cat. #S1748 | |
| Chemical compound, drug | AICAR | Beyotime Biotechnology | Cat. #S1516 | |
| Chemical compound, drug | MHY (MHY1485) | Sigma-Aldrich | Cat. #SML0810 | |
| Chemical compound, drug | dorsomorphin | Sigma-Aldrich | Cat. #P5499 | |
| Chemical compound, drug | Chloroquine (CQ) | Sigma-Aldrich | Cat. #C6628 | |
| Chemical compound, drug | Rapamycin | Sangon Biotech | Cat. #A606203 | |
| Software, algorithm | GraphPad Prism 6 | GraphPad Software | www.graphpad.com |





