An H3K9 methylation-dependent protein interaction regulates the non-enzymatic functions of a putative histone demethylase
Figures
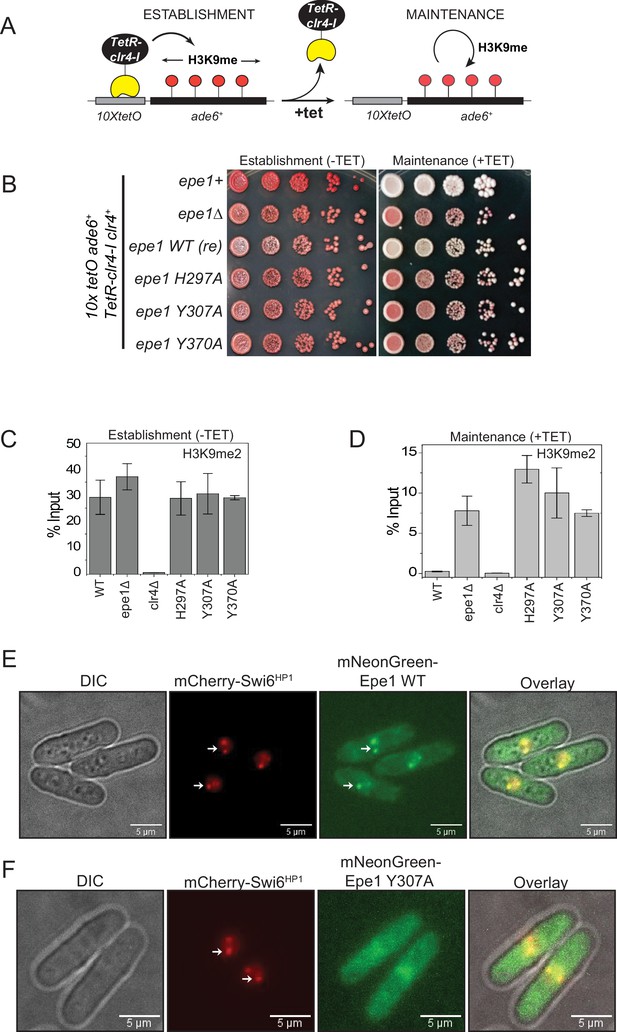
Point mutation within the catalytic JmjC domain of Epe1 affects protein localization at sites of constitutive heterochromatin.
(A) Reporter system to measure epigenetic inheritance. TetR-Clr4-I binding (−tetracycline) leads to ectopic establishment of H3K9 methylation. Addition of tetracycline promotes TetR-Clr4-I dissociation, enabling us to measure epigenetic inheritance of H3K9 methylation. (B) Color-based assay to detect establishment and maintenance of epigenetic states. The establishment of epigenetic silencing (−tetracycline) leads to the appearance of red colonies. Epigenetic inheritance, indicated by red or sectored colonies, (+tetracycline) is critically dependent on Epe1 activity. Point mutations within the JmjC domain of Epe1 disrupt its anti-silencing function in cells, leading to the appearance of red or sectored colonies. (C) ChIP-qPCR measurements of H3K9me2 levels at the ectopic site (10X tetO ade6+) during establishment (−tetracycline) in different Epe1 mutant backgrounds (N = 2). Error bars represent standard deviations. (D) ChIP-qPCR measurements of H3K9me2 levels at the ectopic site (10X tetO ade6+) during maintenance (+tetracycline) in different Epe1 mutant backgrounds (N = 2). Error bars represent standard deviations. (E) Live cell imaging of Epe1 and Swi6HP1. Four images in each case correspond to DIC, 488 excitation, 560 excitation, and overlay of the three emission channels. mNeonGreen-Epe1 and mCherry- Swi6HP1 form co-localized foci in green and red emission channels, respectively (see white arrows). (F) mNeonGreen-Epe1 Y307A fails to form any foci and instead exhibits a diffuse signal that permeates the nucleus. mCherry- Swi6HP1 forms foci corresponding to sites of constitutive heterochromatin.
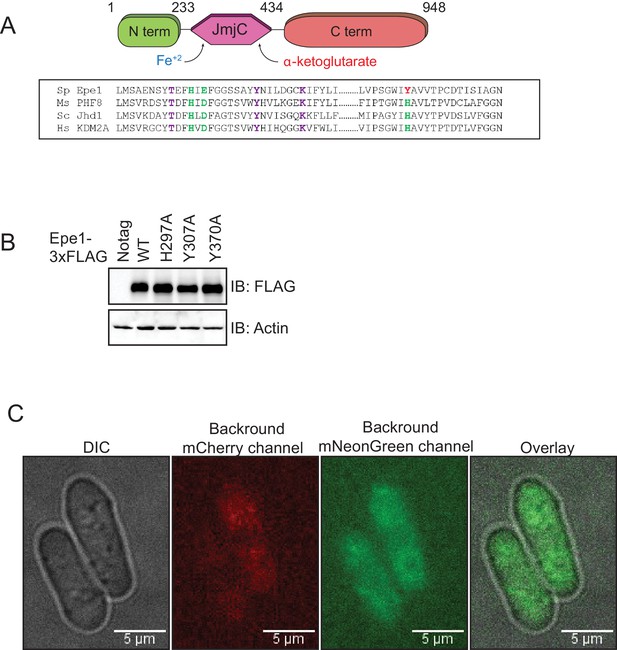
Epe1 mutants are expressed at similar levels to that of the wild-type protein.
(A) Sequence alignment of related JmjC domain-containing proteins reveals the identity of conserved residues in Epe1 that are involved in iron (Fe2+) and α-ketoglutarate binding. Iron-binding residues are colored in green and α-ketoglutarate-binding residues are colored in purple. (B) Western blots comparing expression levels of Epe1 proteins. Wild-type Epe1-3X FLAG and Epe1 mutants exhibit similar levels of expression from the endogenous promoter. Actin levels are shown as a loading control. (C) Images of control cells expressing no mCherry or mNeonGreen proteins reveal the extent of cytoplasmic background in each emission channel.
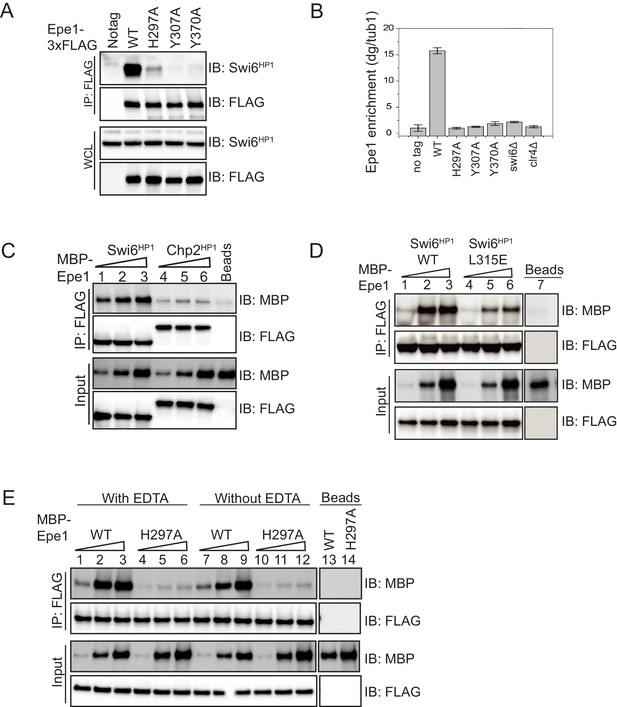
Mutations within the JmjC domain perturb a direct interaction between Epe1 and Swi6HP1.
(A) Western blots of co-immunoprecipitation (co-IP) measurements to test the interaction between Epe1-3XFLAG and Swi6HP1. Epe1 is detected using a FLAG antibody and Swi6HP1 is detected using a primary antibody. The interaction between the two proteins is preserved in wild-type cells and is completely eliminated in all Epe1 JmjC mutants. (B) ChIP-qPCR measurements of Epe1 occupancy at sites of constitutive heterochromatin (dg pericentromeric repeats) (N = 2). Error bars represent standard deviations. Epe1 enrichment is reduced to near background levels (swi6Δ and clr4Δ) in loss of function mutants of Epe1. (C) Western blots of in vitro binding assays using recombinant Epe1 protein. Increasing amounts of wild-type MBP-Epe1 protein are added while maintaining a fixed amount of 3X-FLAG-Swi6HP1 or 3X-FLAG-Chp2 HP1 on beads. Epe1 exhibits a binding preference for Swi6HP1over Chp2 HP1. (D) Increasing amounts of wild-type MBP-Epe1 protein are added while maintaining a fixed amount of 3X-FLAG- Swi6HP1 or a CSD domain mutant, 3X-FLAG Swi6HP1 L315E on beads. Western blots revealed that a point mutation in the conserved Swi6 HP1CSD domain (L315E) leads to reduced levels of interaction between recombinant Epe1 and Swi6HP1 L315E. (E) Increasing amounts of wild-type MBP-Epe1 and MBP-Epe1 H297A are added while maintaining a fixed amount of 3XFLAG- Swi6HP1 on beads. Experiments were performed in the presence and absence of EDTA to measure co-factor independent interactions between the two proteins. The binding recombinant Epe1 H297A binding to Swi6HP1 is significantly reduced relative to the wild-type protein.
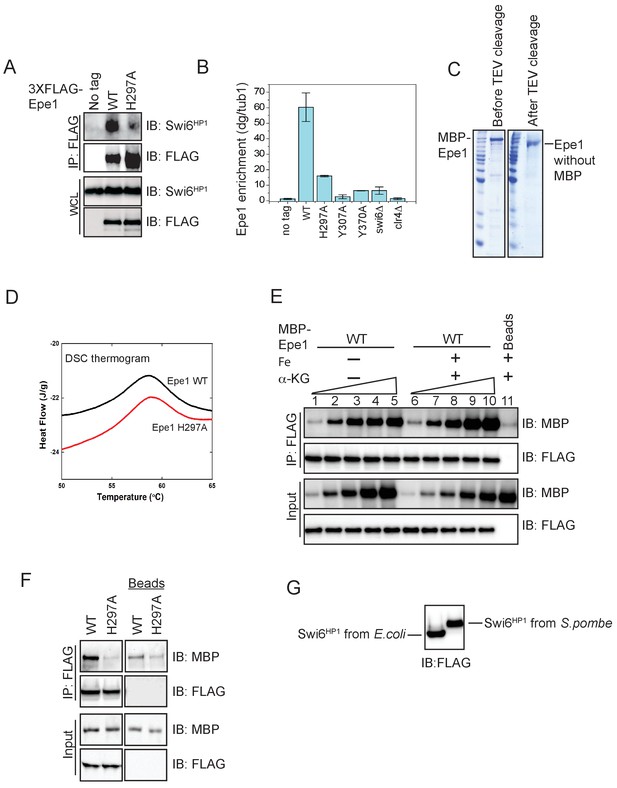
Characterization of recombinant Epe1 wild-type and Epe1 mutant proteins.
(A) Western blot of a co-immunoprecipitation experiment in cells expressing Epe1 that is fused to a FLAG epitope tag at the N-terminus. N-terminal FLAG fusions of Epe1 and Epe1 H297A exhibit the same pattern of interaction with Swi6HP1 (the wild-type protein exhibits an interaction and the Epe1 mutant exhibits reduced binding) as their C-terminal counterparts. (B) ChIP-qPCR measurements of Epe1 occupancy at the pericentromeric dg repeats. The use of additional crosslinkers improves crosslinking efficiency and traps transient interactions. The wild-type Epe1 protein is enriched at sites of heterochromatin formation, whereas Epe1 JmjC mutants do not exhibit any significant enrichment. (C) Recombinant Epe1 protein purification from insect cells before and after cleavage of the MBP tag. Epe1 purified from Sf9 insect cells remains soluble even after MBP tag cleavage. (D) Differential scanning calorimetry (DSC) assays demonstrate that wild-type Epe1 and Epe1 H297A proteins exhibit similar denaturation temperatures, implying that the proteins are equally stable in vitro. The difference in peak intensities reflects slightly different protein amounts in the DSC. (E) In vitro binding assays between Epe1 and Swi6HP1 in the presence of Fe (II) and α-ketoglutarate as co-factors. The presence of Fe (II) and α-ketoglutarate did not alter the binding between Epe1 and Swi6HP1 compared to reactions performed in the absence of any co-factors. (F) In vitro binding experiments using recombinant wild-type and Epe1 H297A proteins with Swi6HP1 purified from fission yeast cells. 3X FLAG- Swi6HP1 was expressed at endogenous levels in fission yeast and purified under high salt conditions. The fission yeast Swi6HP1 protein exhibits the same binding preference for wild-type Epe1 as E. coli Swi6HP1. (G) Swi6HP1 purified from S. pombe exhibits a mass shift compared to Swi6HP1 purified from E. coli because of protein phosphorylation. Therefore, our purification approach preserves the endogenously phosphorylated state of Swi6HP1.
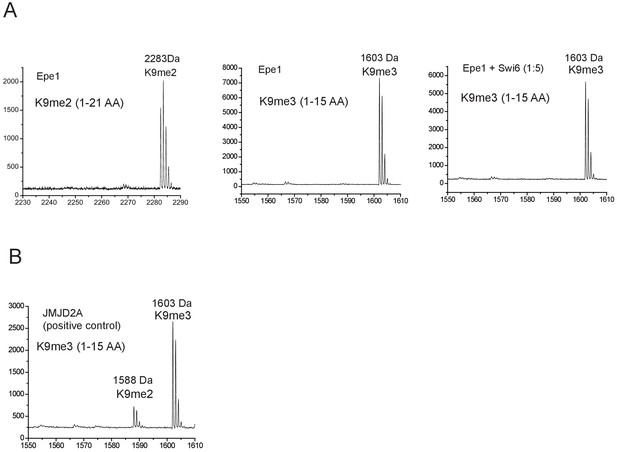
Epe1 exhibits no enzymatic activity in the presence or absence of Swi6HP1.
(A) Mass spectrometry analysis of modified H3 peptides following an in vitro demethylase assay. The H3K9me2 tail peptide spans 1-21 amino acids with an intact mass of 2283 Da. The H3K9me3 tail peptide spans 1-15 amino acids with an intact mass of 1603 Da. Demethylase assays were carried out in the presence and absence of Swi6HP1. (B) The positive control active demethylase, JMJD2A produces a 15kDa mass shift corresponding to the removal of a single methyl group. JMJD2A preferentially demethylates H3K9me3 peptides.
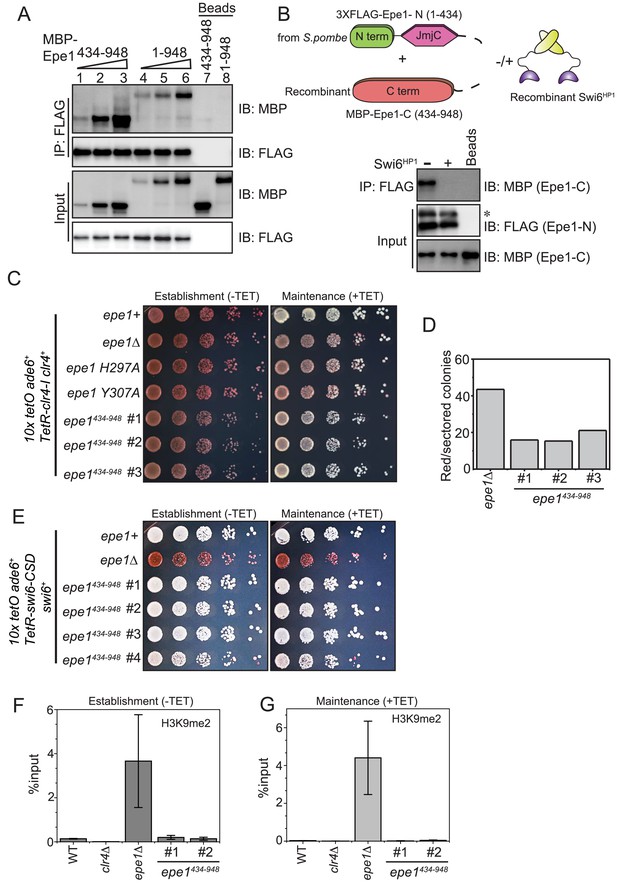
Swi6HP1 interacts with the C-terminus of Epe1 through a region that is proximal to the JmjC domain.
(A) Western blots of in vitro binding assays to validate the interaction between Swi6HP1 and the Epe1 C-terminus. Increasing amounts of MBP-Epe1434-948 protein are added while maintaining a fixed amount of 3X-FLAG-Swi6HP1 on beads. MBP-Epe1434-948 exhibits an increase in its interaction with Swi6 HP1 compared to full-length Epe1. (B) Western blots of an in vitro binding assay between FLAG-Epe1-N (1–434 amino acids) purified from S. pombe and recombinant MBP-Epe1434-948 in the presence and absence of recombinant Swi6HP1. The addition of Swi6HP1 disrupts a direct trans interaction between the N- and C-terminal halves of Epe1. The asterisk in the figure denotes the non-specific FLAG antibody band. (C) Phenotype assays of epigenetic inheritance as a measure of Epe1 activity. Cells expressing Epe1434-948 were plated on −tetracycline and +tetracycline medium. Cells are initially red during establishment. Despite the absence of the putative catalytic JmjC domain, cells expressing Epe1434-948 partly turn white on +tetracycline medium. (D) Quantification of red or sectored colonies comparing epe1Δ and Epe1434-948 expressing cells. Epe1434-948 expressing cells have 30% fewer sectored colonies compared to epe1Δ cells. (E) A dominant interaction between the Epe1 C-terminus and Swi6HP1 opposes heterochromatin establishment. TetR-Swi6-CSD was expressed in cells that harbor 10X TetO binding sites upstream of an ade6+ reporter gene. The expression of the Epe1 C-terminus alone in the absence of its putative catalytic JmjC domain disrupts heterochromatin establishment causing cells to remain white under –tetracycline and +tetracycline conditions. (F) ChIP-qPCR measurements of H3K9me2 levels at the ectopic site (10X tetO ade6+) during establishment (−tetracycline) in a wild-type, epe1Δ and two independent clones expressing Epe1434-948 (N = 2). Error bars represent standard deviations. (G) ChIP-qPCR measurements of H3K9me2 levels at the ectopic site (10X tetO ade6+) during maintenance (+tetracycline) in wild-type, epe1Δ and two independent clones expressing Epe1434-948 (N = 2). Error bars represent standard deviations.
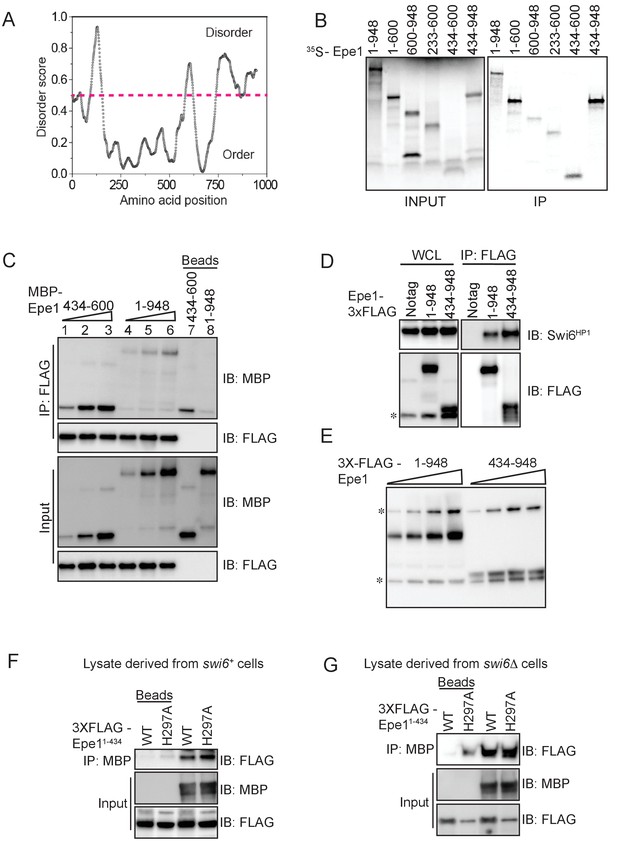
In vitro assays to map the site of interaction between Epe1 and Swi6HP1.
(A) Using a computational disorder prediction program (http://www.pondr.com, VSL2) and a user-specified cut-off, we defined ordered and disordered regions within Epe1. The JmjC domain spans 233-434 amino acids and emerges as one of two ordered domains in Epe1. The second ordered region within the C-terminus of Epe1 has no structural similarity to existing proteins. (B) An in vitro translation assay using rabbit reticulocyte lysates to generate S35 labeled fragments of Epe1. Swi6HP1 was immobilized on beads and incubated with IVT extracts. Two Epe1 regions ranging from 1-600 amino acids and 434-948 amino acids emerge as the primary interacting fragments. (C) Increasing amounts of MBP-Epe1434-600 protein is added while maintaining a fixed amount of 3X-FLAG- Swi6HP1on beads. MBP-Epe1434-600 exhibits an increase in binding to Swi6HP1 compared to full-length Epe1. (D) Western blots of co-immunoprecipitation experiments to detect an interaction between Epe1434-948 and Swi6HP1. (E) Western blots of whole-cell extracts of cells expressing full-length Epe1 or Epe1434-948 fused to a FLAG epitope tag. The Epe1434-948 protein is expressed at levels that are at least 4-5 fold lower than full-length Epe1 which in part might explain its limited efficacy in cells. The asterisk in the figure denotes the non-specific FLAG antibody band. (F) A recombinant C-terminal fragment of Epe1, MBP-Epe1434-948 was incubated with fission yeast cell extracts expressing the N-terminal half of the protein, FLAG-Epe1-N (1-434 amino acids) or FLAG-Epe1-N (1-434 amino acids) H297A. Extracts were derived from swi6+ (G) A recombinant C-terminal fragment of Epe1, MBP-Epe1434-948 was incubated with fission yeast cell extracts expressing the N-terminal half of the protein, FLAG-Epe1-N (1-434 amino acids) or FLAG-Epe1-N (1-434 amino acids) H297A. Extracts were derived from swi6Δ cells.
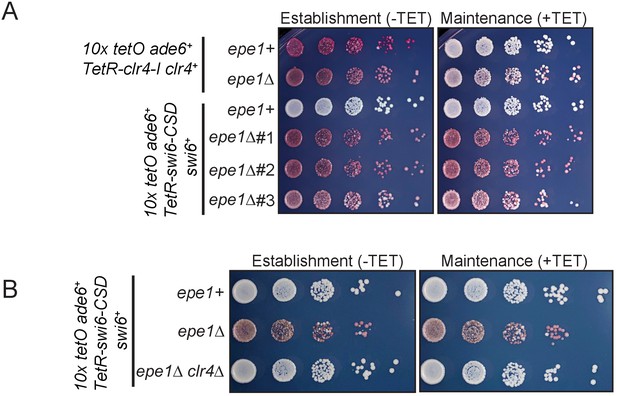
TetR-Swi6 CSD mediated heterochromatin establishment occurs only in the absence of Epe1 and is dependent on H3K9 methylation.
(A) TetR-Swi6-CSD was expressed in cells that have 10X TetO binding sites placed upstream of an ade6+ reporter gene. Cells establish heterochromatin only in an epe1Δ background. epe1+ cells completely suppress TetR-Swi6-CSD initiated epigenetic silencing. (B) TetR-Swi6-CSD mediated heterochromatin establishment is critically dependent on H3K9 methylation. epe1Δ clr4Δ cells expressing TetR-Swi6-CSD fail to establish epigenetic silencing.
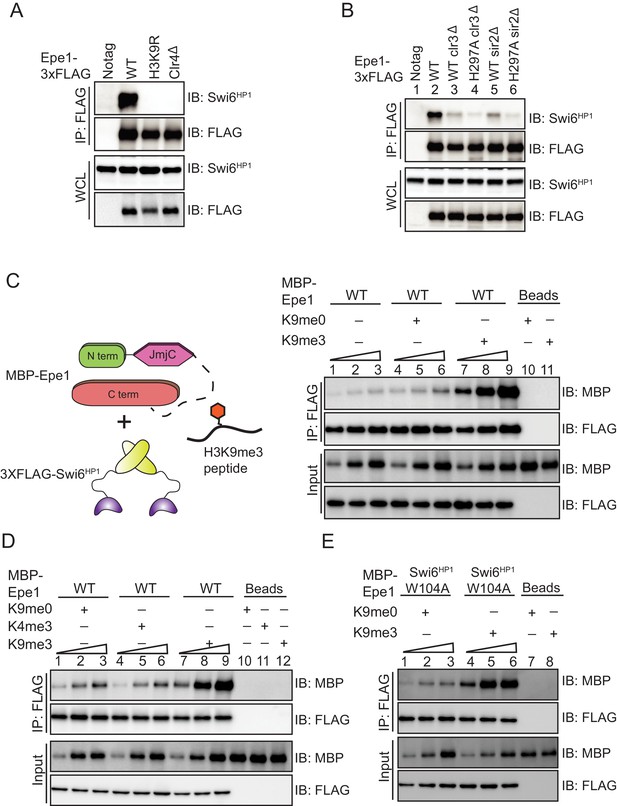
H3K9 methylation stimulates complex formation between Epe1 and Swi6HP1.
(A) Western blots of co-immunoprecipitation measurements reveal an interaction between Epe1-3XFLAG and Swi6HP1 in the case of wild-type cells, which is completely absent in H3K9 methylation deficient cells (clr4Δ and H3K9R). (B) Co-immunoprecipitation measurements reveal that deletion of histone deacetylases Clr3 or Sir2 which disrupt heterochromatin formation, also leads to a concomitant loss in the interaction between Epe1-3XFLAG and Swi6 HP1. (C) Western blots of in vitro binding assays between recombinant Epe1 and Swi6 HP1 in the presence of histone H3 tail peptides (1–15 amino acids). Increasing amounts of wild-type MBP-Epe1 are added while maintaining a fixed amount of 3XFLAG- Swi6HP1 on beads. Experiments were performed in the presence of an unmethylated H3 peptide (H3K9me0) (lanes 4–6) or an H3K9 tri-methylated peptide (H3K9me3) (lanes 7–9). Epe1 exhibits a significant increase in its ability to interact with Swi6HP1 in the presence of an H3K9me3 peptide. (D) Western blots of in vitro binding assays between recombinant Epe1 and Swi6HP1 in the presence of differentially methylated histone H3 tail peptides (1–15 or 1–21 amino acids). Increasing amounts of wild-type MBP-Epe1 are added while maintaining a fixed amount of 3XFLAG-Swi6HP1 on beads. Experiments were performed in the presence of an unmethylated H3 peptide (H3K9me0), an H3K9 tri-methylated peptide (H3K9me3), or an H3K4 tri-methylated peptide (H3K4me3). Epe1 exhibits a significant increase in its ability to interact with Swi6HP1 in the presence of an H3K9me3 but not an H3K4me3 peptide. (E) Western blots of in vitro binding assays between recombinant Epe1 and Swi6HP1 W104A in the presence of histone H3 tail peptides (1–15 or 1–21 amino acids). Increasing amounts of wild-type MBP-Epe1 are added while maintaining a fixed amount of 3XFLAG- Swi6HP1 W104A on beads. Experiments were performed in the presence of an unmethylated H3 peptide (H3K9me0) or an H3K9 tri-methylated peptide (H3K9me3). Epe1 exhibits an increase in its ability to interact with Swi6HP1 W104A in the presence of an H3K9me3 peptide.
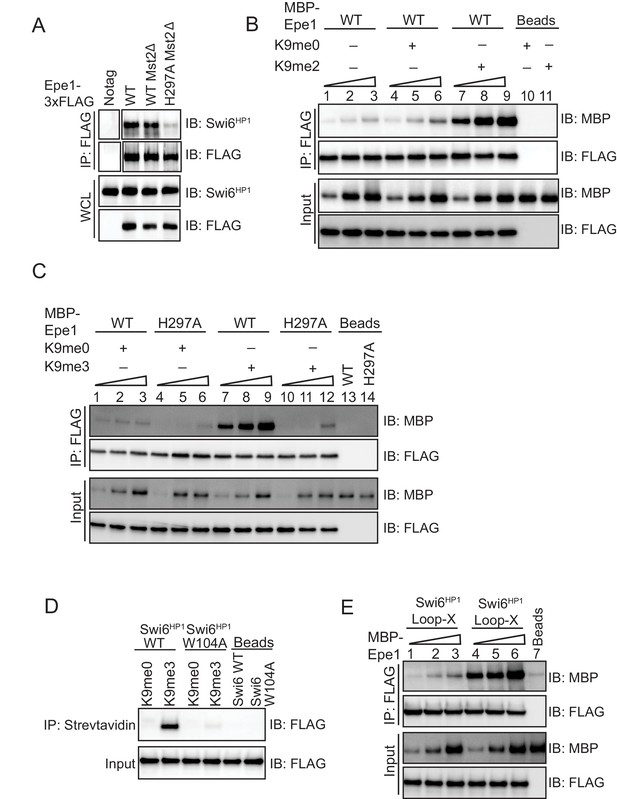
An Epe1 mutant fails to exhibit H3K9me dependent stimulation in its interaction with Swi6HP1.
(A) Deleting Mst2 does not alter the interaction between Epe1-3X FLAG or Epe1 H297A−3X FLAG and Swi6HP1. Western blots of a co-immunoprecipitation assay between Epe1 and Swi6HP1 in an mst2Δ background. (B) Western blots of in vitro binding assays between recombinant Epe1 and Swi6HP1 in the presence of histone H3 tail peptides (1–15 or 1–21 amino acids). Increasing amounts of wild-type MBP-Epe1 are added while maintaining a fixed amount of 3XFLAG-Swi6HP1 on beads. Experiments were performed in the presence of an unmethylated H3 peptide (H3K9me0) or an H3K9 dimethylated peptide (H3K9me2). Epe1 exhibits an increase in its ability to interact with Swi6HP1 in the presence of an H3K9me2 peptide similar to the type of stimulation we observed upon the addition of an H3K9me3 peptide. (C) Western blots of in vitro binding assays between recombinant Epe1 WT and Epe1 H297A with Swi6HP1 in the presence of an H3K9me3 tail peptide. Increasing amounts of wild-type MBP-Epe1 and MBP-Epe1 H297A are added while maintaining a fixed amount of 3XFLAG- Swi6HP1 on beads in the presence of an H3K9me3 peptide or an unmodified H3K9me0 peptide. We observed a stimulation in the interaction between Epe1 WT and Swi6HP1 (lanes 7–9) but not in the case of Epe1 H297A and Swi6HP1 (lanes 10–12). (D) Peptide binding assays measuring Swi6HP1 and Swi6HP1 W104A binding to an H3K9me3 peptide. Biotinylated H3K9me3 or H3K9me0 peptides immobilized on streptavidin beads were used to pull-down Swi6HP1 or Swi6HP1 W104A. Swi6HP1 W104A exhibits a substantial defect in its interaction with an H3K9me3 peptide. (E) Western blots of in vitro binding assays between recombinant Epe1 and Swi6HP1 Loop-X mutant in the presence of histone H3 tail peptides (1–15 or 1–21 amino acids). Increasing amounts of wild-type MBP-Epe1 are added while maintaining a fixed amount of 3XFLAG-Swi6HP1 Loop-X mutant on beads. Experiments were performed in the presence of an unmethylated H3 peptide (H3K9me0) or an H3K9 tri-methylated peptide (H3K9me3). Epe1 exhibits an increase in its ability to interact with Swi6HP1 Loop-X mutant in the presence of an H3K9me3 peptide.
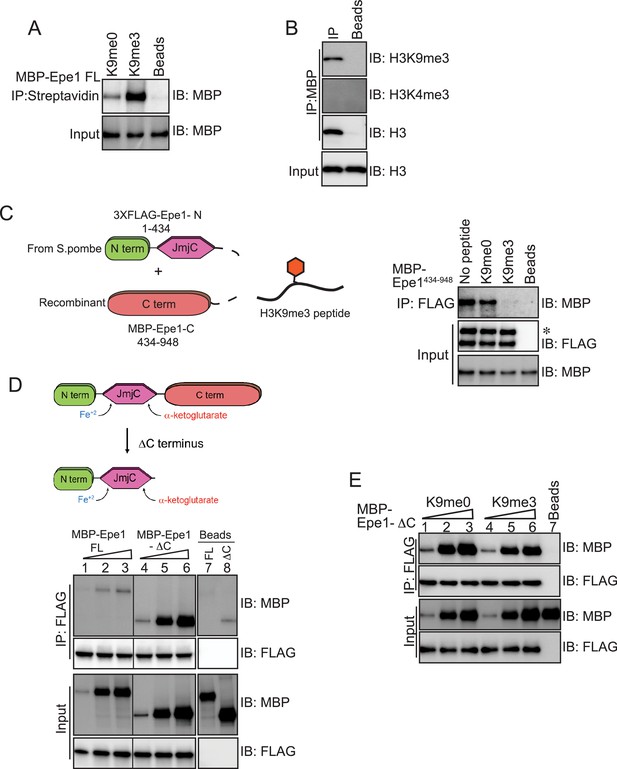
An Epe1 C-terminal truncation mutant exhibits enhanced binding to Swi6 and is insensitive to H3K9 methylation.
(A) Biotinylated peptides (H3K9me3 or H3K9me0) are immobilized on streptavidin beads and incubated with MBP-Epe1. MBP-Epe1 preferentially associates with an H3K9me3 peptide compared to an H3K9me0 peptide. (B) Epe1 specifically interacts with H3K9me3 histones. Calf-thymus histones containing an unspecified mixture of differentially modified histones were incubated with MBP-Epe1 immobilized on an amylose resin. Epe1 preferentially interacts with H3K9 methylated histones as opposed to H3K4 methylated histones. (C) Western blots of an in vitro binding assay to determine whether an H3K9me3 peptide disrupts a trans interaction between the N- and C-terminal halves of Epe1. The N-terminus of Epe1, 3X FLAG-Epe1-N (1-434) was purified using a FLAG antibody from fission yeast cells. The purified protein was incubated with recombinant MBP-Epe1434-948 protein in the presence of an H3K9me0 peptide and an H3K9me3 peptide. The H3K9me3 peptide specifically disrupts a direct trans interaction between the N- and C-terminal halves of Epe1. (D) Western blots of in vitro binding assays comparing the interactions between recombinant Epe1-ΔC and Swi6HP1. Increasing amounts of wild-type MBP-Epe1 or MBP-Epe1-ΔC are added while maintaining a fixed amount of 3XFLAG- Swi6HP1 on beads. Epe1-ΔC (lanes 4–6) clearly surpasses the full-length protein (lanes 1–3) in terms of its ability to interact with Swi6HP1. (E) Western blots of in vitro binding assays between recombinant Epe1-ΔC and Swi6 HP1 in the presence of histone H3 tail peptides (1–15 amino acids). Increasing amounts of MBP-Epe1-ΔC are added while maintaining a fixed amount of 3XFLAG-Swi6HP1 on beads. Experiments were performed in the presence of an unmethylated H3 peptide (H3K9me0) (lanes 1–3) or an H3K9 tri-methylated peptide (H3K9me3) (lanes 4–6). Epe1-ΔC exhibits no change in its interaction with Swi6HP1 in the presence of an H3K9me3 peptide.
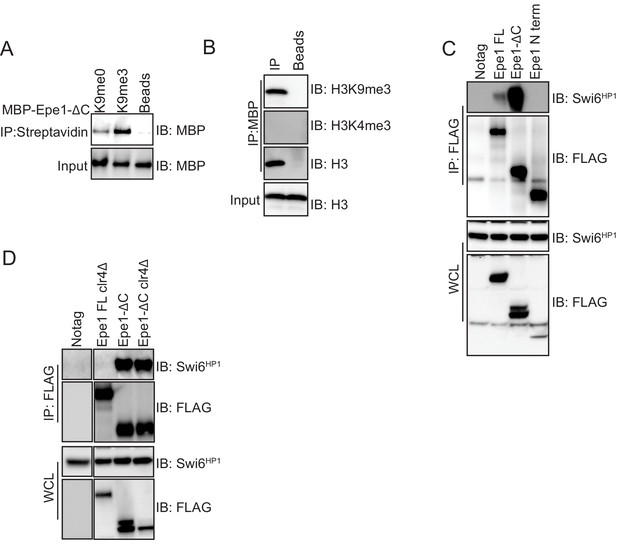
An Epe1 C-terminal truncation mutant interacts with Swi6HP1 in the absence of H3K9 methylation in cells.
(A) Biotinylated peptides (H3K9me3 or H3K9me0) were immobilized on streptavidin beads and incubated with MBP-Epe1-ΔC. MBP- Epe1-ΔC preferentially associates with an H3K9me3 peptide. (B) Calf-thymus histones containing an unspecified mixture of differentially modified histones were incubated with MBP- Epe1-ΔC immobilized on an amylose resin. Epe1-ΔC preferentially interacts with H3K9 methylated histones as opposed to H3K4 methylated histones. Therefore, the N-terminus of Epe1 harbors H3K9 methylation binding activity. (C) Western blots of co-immunoprecipitation measurements to test the interaction between Epe1 truncations and Swi6HP1. The full-length Epe1 protein and Epe1 truncations are detected using a FLAG antibody and Swi6HP1 is detected using a primary antibody. The deletion of the Epe1 C-terminus (Epe1-ΔC) leads to a substantial increase in its interaction with Swi6HP1 relative to the full-length protein. Epe1-N (1-434) fails to interact with Swi6HP1. (D) Western blots of co-immunoprecipitation measurements to test the interaction between Epe1-ΔC and Swi6HP1 in clr4Δ cells. Epe1-ΔC is detected using a FLAG antibody and Swi6HP1 is detected using a primary antibody. The interaction between Epe1-ΔC, and Swi6HP1 remains unaltered upon loss of H3K9 methylation, unlike its full-length counterpart, which is highly sensitive to heterochromatin disruption.
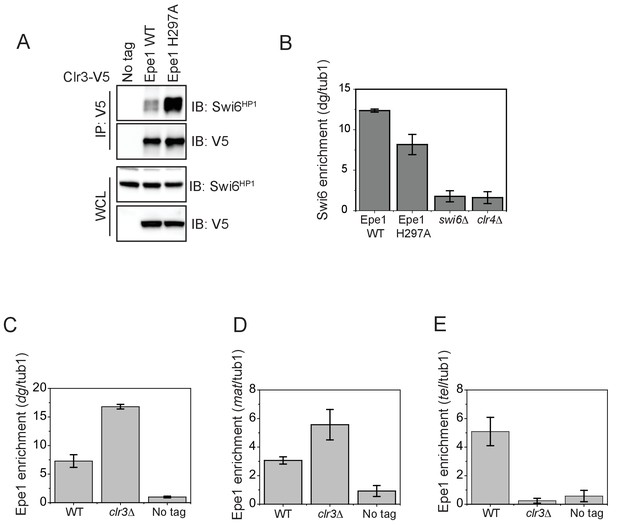
The interaction between Epe1 and Swi6 displaces Clr3 from localizing to sites of heterochromatin formation.
(A) Epe1 displaces Clr3 from sites of heterochromatin formation. Western blots of a co-immunoprecipitation experiment reveal that Clr3-3X V5 interacts weakly with Swi6HP1 in a wild-type Epe1 background. This interaction increases significantly in an Epe1 H297A background. (B) ChIP-qPCR measurements of Swi6HP1 occupancy at sites of constitutive heterochromatin in wild-type and Epe1 mutant cells. The increase in interaction between Swi6HP1 and Clr3 is independent of Swi6HP1 levels at constitutive heterochromatin. (C) ChIP-qPCR measurements of Epe1 occupancy at dg pericentromeric repeats in clr3Δ cells (N = 2). Error bars represent standard deviations. Epe1 enrichment is increased two to three-fold in cells at dg in clr3Δ strains. (D) ChIP-qPCR measurements of Epe1 occupancy at the mating-type locus (mat) in clr3Δ cells (N = 2). Error bars represent standard deviations. Epe1 enrichment is increased two to three-fold at the mat locus in clr3Δ strains. (E) ChIP-qPCR measurements of Epe1 occupancy at telomeres (tlh1) in clr3Δ cells (N = 2). Error bars represent standard deviations. Epe1 localization is completely eliminated at the telomeres in clr3Δ strains.
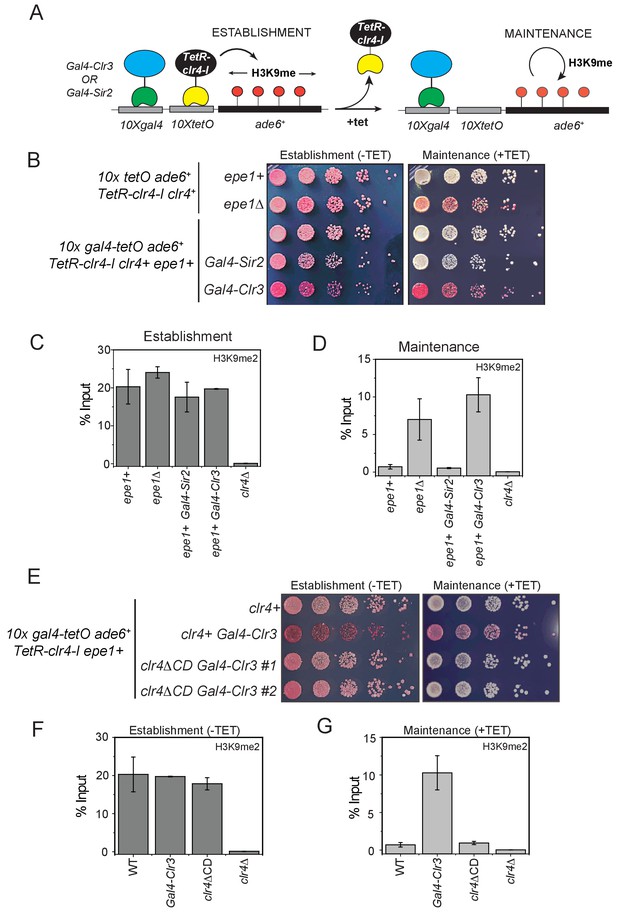
H3K9 methylation regulates the non-enzymatic functions of a putative histone demethylase Epe1, which interacts weakly with Swi6HP1.
(A) A reporter system to detect epigenetic inheritance of H3K9 methylation in the presence of orthogonal chromatin effectors. Heterochromatin initiation depends on TetR-Clr4-I binding (-tetracycline). Orthogonal chromatin effectors can be recruited to the 10xgal4 DNA binding site via a Gal4 DNA binding domain (Gal4 DBD). The addition of tetracycline promotes TetR-Clr4-I dissociation to measure epigenetic inheritance in the presence of an orthogonal chromatin effector. (B) A color-based assay to detect the establishment and maintenance of epigenetic states. The establishment of epigenetic silencing (-tetracycline) results in red colonies. Tethering Gal4-Clr3 but not Gal4-Sir2 at an ectopic site promotes epigenetic inheritance of H3K9 methylation resulting red or sectored colonies in +tetracycline containing medium in epe1+ (C) ChIP-qPCR measurements of H3K9me2 levels at the ectopic site (10X tetO ade6+) during establishment (-tetracycline). (N=2). Error bars represent standard deviations. (D) ChIP-qPCR measurements of H3K9me2 levels at the ectopic site (10X tetO ade6+) during maintenance (+tetracycline) (N=2). Error bars represent standard deviations. (E) The deletion of the Clr4 chromodomain (clr4ΔCD) inactivates read-write functions which affects epigenetic inheritance. Cells that are initially red during heterochromatin establishment (-tetracycline) turn white during maintenance (+tetracycline) in a clr4ΔCD background. (F) ChIP-qPCR measurements of H3K9me2 levels at the ectopic site (10X tetO ade6+) during establishment (-tetracycline) in clr4ΔCD mutant cells (N=2). Error bars represent standard deviations. ChIP-qPCR values from Figure 7C are plotted as a reference. (G) ChIP-qPCR measurements of H3K9me2 levels at the ectopic site (10X tetO ade6+) during maintenance (+tetracycline) in clr4ΔCD mutant cells (N=2). Error bars represent standard deviations. Error bars represent standard deviations. ChIP-qPCR values from Figure 7D are plotted as a reference.
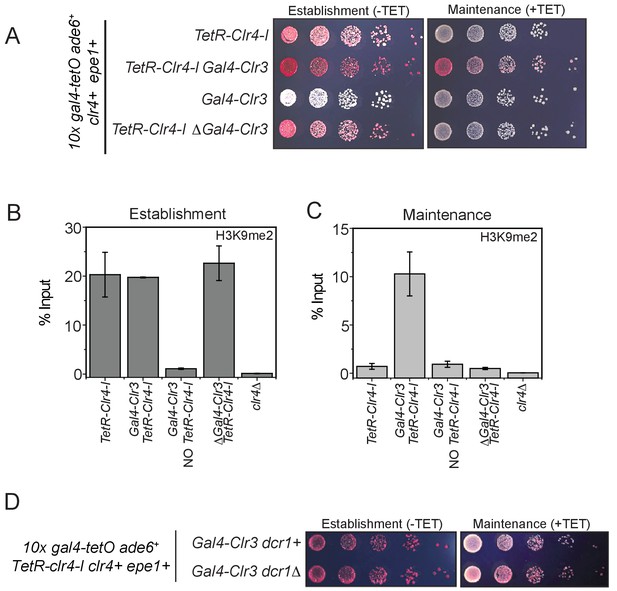
Clr3 tethering in cis and the read-write activity of Clr4 is required for epigenetic inheritance.
(A) Color-based assays to measure epigenetic inheritance in the presence of Gal4-Clr3 variants. Our results reveal that Gal4-Clr3 fusions cannot initiate heterochromatin de novo in the absence of TetR-Clr4-I (line 3). In the absence of sequence-dependent tethering of Clr3, cells establish but do not maintain epigenetic silencing (line 4). (B) ChIP-qPCR measurements of H3K9me2 levels at the ectopic site (10X tetO ade6+) during establishment (-tetracycline) in the presence of Gal4-Clr3 variants. ChIP-qPCR values from Figure 7C are plotted as a reference. (C) ChIP-qPCR measurements of H3K9me2 levels at the ectopic site (10X tetO ade6+) during maintenance (+tetracycline) in the presence of Gal4-Clr3 variants (N=2). Error bars represent standard deviations. ChIP-qPCR values from Figure 7D are plotted as a reference. (D) Epigenetic inheritance of H3K9 in the presence of Gal4-Clr3 is independent of the RNAi pathway.
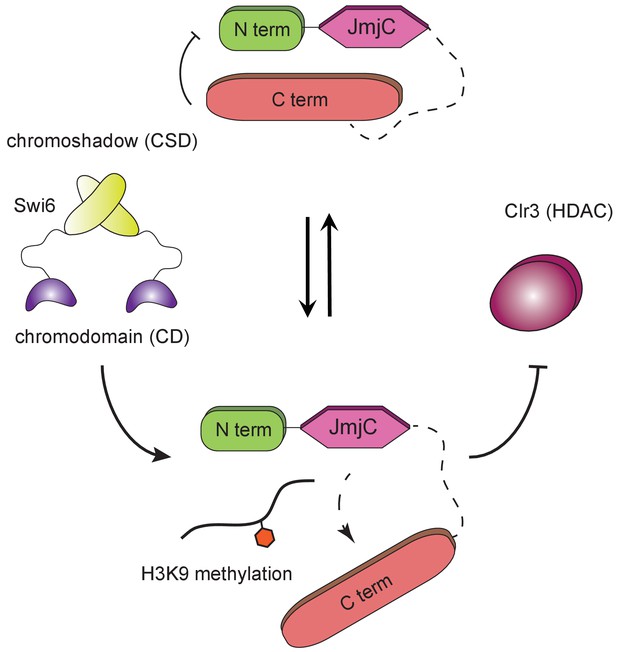
H3K9 methylation regulates the non-enzymatic functions of a putative histone demethylase.
Epe1 interacts weakly with Swi6HP1. Point mutations within the catalytic JmjC domain of Epe1 disrupts its interaction with Swi6HP1. H3K9 methylation binding stimulates the interaction between Epe1 and Swi6HP1 which stabilizes complex formation at sites of heterochromatin formation. The Epe1 C-terminus enforces an H3K9 methylation dependent mode of interaction between Epe1 and Swi6HP1 which displaces the histone deacetylase, Clr3 from sites of heterochromatin formation. Our model reveals how H3K9 methylation stabilizes an inhibitory complex consisting of Epe1 and Swi6HP1 and suggests how a non-enzymatic function associated with Epe1 C-terminus regulates epigenetic inheritance.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information | |
|---|---|---|---|---|---|
| Chemical compound, drug | Trizma base | Sigma aldrich | Cat# T1503-5KG | ||
| Chemical compound, drug | Boric acid | Sigma Aldrich | Cat# B6768-5KG | ||
| Chemical compound, drug | EDTA | Sigma Aldrich | Cat# EDS-500G | ||
| Chemical compound, drug | DTT | Thermo Fisher Scientific | Cat# BP17225 | ||
| Chemical compound, drug | HEPES | Sigma Aldrich | Cat# H4034-100G | ||
| Chemical compound, drug | Proteinase K | Thermo Fisher Scientific | Cat# FEREO0491 | ||
| Chemical compound, drug | Magnesium chloride hexahydrate | Sigma Aldrich | Cat# M2670-500G | ||
| Chemical compound, drug | Potassium chloride | Sigma Aldrich | Cat# P9541-500G | ||
| Chemical compound, drug | Sodium chloride | Thermo Fisher Scientific | Cat# S271-3 | ||
| Chemical compound, drug | Sodium acetate | Sigma Aldrich | Cat# S2889-250G | ||
| Chemical compound, drug | Tryptone | Thermo Fisher Scientific | Cat# BP1421-500 | ||
| Chemical compound, drug | SDS micropellets | Thermo Fisher Scientific | Cat# BP8200-500 | ||
| Chemical compound, drug | Agarose | Thermo Fisher Scientific | Cat# BP1356-500 | ||
| Chemical compound, drug | PMSF | Calbiochem | Cat# 7110–5 GM | ||
| Chemical compound, drug | Triton X-100 | Sigma Aldrich | Cat# T8787-250ML | ||
| Chemical compound, drug | Lithium chloride | Sigma Aldrich | Cat# L4408-500G | ||
| Chemical compound, drug | Lithium acetate dihydrate | Sigma Aldrich | Cat# CAS6108-17-4 | ||
| Chemical compound, drug | Tween 20 | Thermo Fisher Scientific | Cat# CAS9005-64-5 | ||
| Chemical compound, drug | Peptone | RPI Research Products | Cat# P20240-1000.0 | ||
| Chemical compound, drug | Leupeptin | RPI Research Products | Cat# L22035-0.025 | ||
| Chemical compound, drug | Aprotinin | RPI Research Products | Cat# A20550-0.05 | ||
| Chemical compound, drug | Pepstatin | RPI Research Products | Cat# P30100-0.025 | ||
| Chemical compound, drug | N-Oxalylglycine | Sigma Aldrich | Cat# O9390-10MG | ||
| Chemical compound, drug | α-Ketoglutaric acid disodium salt dihydrate | Sigma Aldrich | Cat# 75892–25G | ||
| Chemical compound, drug | SuperSignal West Pico PLUS Chemiluminescent Substrate | Thermo Scientific | Cat# B2162617 | ||
| Chemical compound, drug | Trichloroacetic acid | Sigma Aldrich | Cat# T0699-100ML | ||
| Chemical compound, drug | Phenol:chloroform: isoamyl alcohol | Sigma Aldrich | Cat# P3803-100ML | ||
| Chemical compound, drug | L-ascorbic acid | Fisher Chemical | Cat# C6H8O6 | ||
| Chemical compound, drug | Coblat(II) chloride hexahydrate | Sigma Aldrich | Cat# 255599–100G | ||
| Chemical compound, drug | Ammonium iron (II) sulfate hexahydrate | ACROS ORGANICS | Cat# 423721000 | ||
| Chemical compound, drug | Formaldehyde | Sigma Aldrich | Cat# 252549–500 ML | ||
| Chemical compound, drug | Acetonitrile | OmniSolv | Cat# AX0156-1 | ||
| Chemical compound, drug | Trifluoroacetic acid | Fisher Chemical | Cat# A116−10 × 1 AMP | ||
| Chemical compound, drug | Dimethyl pimelimidate | Thermo Scientific | Cat# 21667 | ||
| Chemical compound, drug | Ammonium hydroxide solution | Sigma Aldrich | Cat# 221-228-100ML-A | ||
| Chemical compound, drug | Ethanolamine | Sigma Aldrich | Cat# 411000–100 ML | ||
| Chemical compound, drug | Ethyleneglycol bis succinimidylsuccinate | Thermo Scientific | Cat# 21565 | ||
| Chemical compound, drug | Glycogen | Sigma Aldrich | Cat# 10901393001 | ||
| Chemical compound, drug | GelGreen Nucleic Acid Stain | BioTium | Cat# 41004 | ||
| Chemical compound, drug | 30% Acrylamide/ Bis solution, 37.5:1 | Bio-Rad | Cat# 1610158 | ||
| Chemical compound, drug | Ammonium persulfate | Bio-Rad | Cat# A3678-100G | ||
| Chemical compound, drug | TEMED | Sigma Aldrich | Cat# T9281-50ml | ||
| Chemical compound, drug | Hydrochloric acid | Thermo Fisher Scientific | Cat# A14-500 | ||
| Chemical compound, drug | Agar | Sigma Aldrich | Cat# A1296-1KG | ||
| Chemical compound, drug | IPTG | Thermo Fisher Scientific | Cat# BP1755-10 | ||
| Antibody | Anti-H3K9me2 (Mouse monoclonal) | Abcam | Cat# ab1220 RRID:AB_449854 | IF (1:1000), WB (1:2500) | |
| Antibody | Anti-H3K9me3 (Rabbit polyclonal) | Abcam | Cat# ab8898 RRID:AB_306848 | IF (1:1000), WB (1:5000) | |
| Antibody | Anti-H3 (Rabbit polyclonal) | Abcam | Cat# ab1791 RRID:AB_302613 | IF (1:1000), WB (1:5000) | |
| Antibody | Anti-H3K4me3 (Rabbit polyclonal) | Abcam | Cat# ab8580 RRID:AB_2827504 | IF (1:1000), WB (1:1000) | |
| Antibody | Monoclonal ANTI- FLAG M2 Antibody | Abcam | Cat# F1804-5MG RRID:AB_262044 | IF (1:5000), WB (1:5000) | |
| Antibody | THE V5 Tag Antibody | GenScript | Cat# A01724-100 RRID:AB_2827501 | IF (1:1000), WB (1:5000) | |
| Antibody | Anti-MBP (Mouse monoclonal) | New England Biolabs | Cat# E8032S RRID:AB_2827502 | WB (1:5000) | |
| Antibody | Anti-Swi6 (Rabbit polyclonal) | Custom | WB (1:2500) | ||
| Peptide, recombinant protein | H3K9me3 (1603 Da) | New England Peptide | Custom | H2N-ARTKQTAR(K9me3) STGGKA-amide | |
| Peptide, recombinant protein | H3K9me0 (1560 Da) | New England Peptide | Custom | ARTKQTKARKSTGGKA-amide | |
| Peptide, recombinant protein | H3K9(me2) (2751 Da) | Anaspec peptide | Cat# AS-64359 | H-ARTKQTARK(ME2)STGGKAPPKQLAGGK(biotin)-OH | |
| Peptide, recombinant protein | H3K9(me3) (2766 Da) | Anaspec peptide | Cat# AS-64360 | H-ARTKQTARK(ME3)STGGKAPPKQLAGGK(biotin)-OH | |
| Peptide, recombinant protein | H3 (2722 Da) | Anaspec peptide | Cat# AS-61702 | H-ARTKQTARKSTGGKAP PKQLAGGK(biotin)-OH | |
| Other | Dynabeads Protein G | Thermo Fisher Scientific | LOT# 00448217 | ||
| Other | Dynabeads M-280 Streptavidin | Thermo Fisher Scientific | LOT# 00448388 | ||
| Other | Dynabeads Protein A | Thermo Fisher Scientific | LOT# 00689576 | ||
| Other | ANTI-FLAG M2 Affinity Gel | Sigma Aldrich | LOT# A2220-5ML | ||
| Other | Amylose Resin | New England Biolabs | LOT# E8021L | ||
| Other | Pierce Gutathione Agarose, 100 ml | Thermo Scientific | LOT# R1241698 | ||
Additional files
-
Supplementary file 1
Description of genotypes of strains used in this study.
- https://cdn.elifesciences.org/articles/53155/elife-53155-supp1-v2.docx
-
Supplementary file 2
Description of plasmids used in this study.
- https://cdn.elifesciences.org/articles/53155/elife-53155-supp2-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/53155/elife-53155-transrepform-v2.docx





