Previously undetected super-spreading of Mycobacterium tuberculosis revealed by deep sequencing
Figures
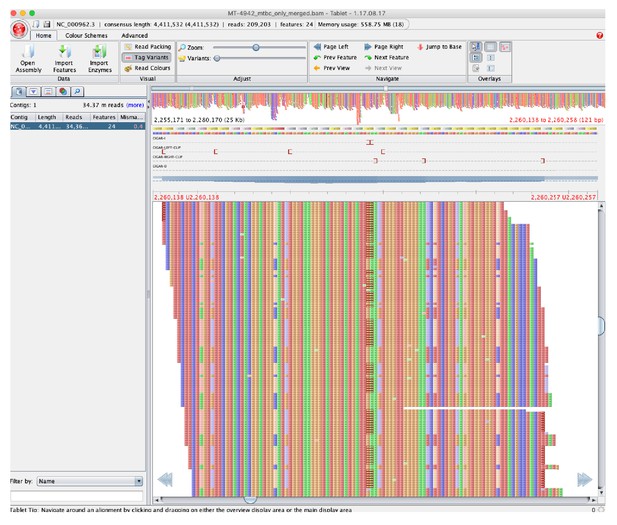
Pileup of reads showing hSNPs suspected to be due to alignment error as listed in Source data 3, with MT-4942 used as an example and zoomed on position 2,255,171 to 2,280,170 in H37Rv (National Center for Biotechnology Information RefSeq Database Accession NC_000962.3).
Binary Alignment Map (BAM) file were loaded into Tablet (v.1.17.08.17, Milne et al., 2013) to visualize the pileup compared to H37Rv.
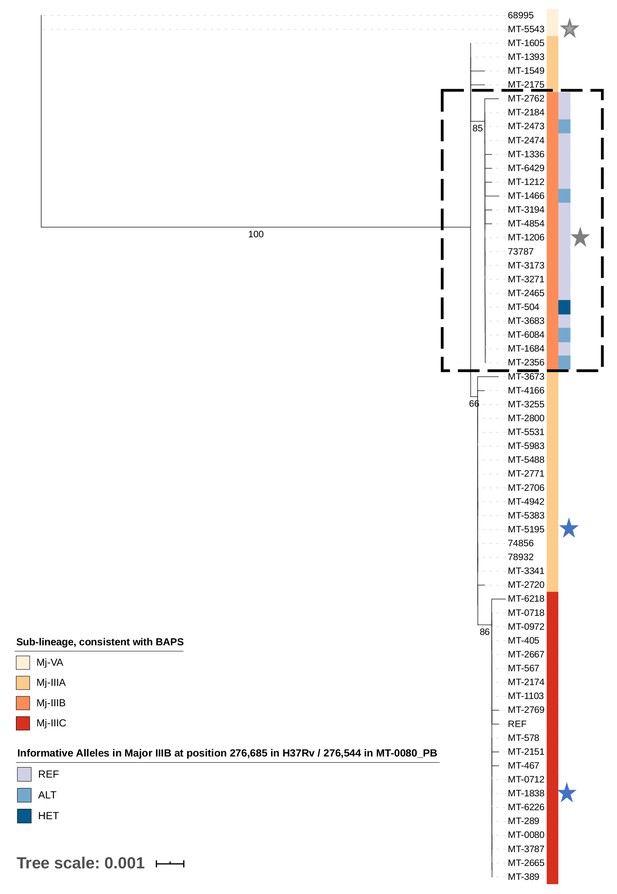
Transmission of M. tuberculosis in village K.
Maximum likelihood tree of 62/65 cases diagnosed between 2007–2012 in village K based on consensus single nucleotide polymorphisms (cSNPs). After aligning to a local reference, MT-0080_PB, cSNPs were identified based on a minimum threshold of ≥95% of reads supporting the alternative allele. A core cSNP alignment was then produced with 90 positions.and IQ-Tree (v.1.6.8 Nguyen et al., 2015) was used to generate the tree using a KP3 model with correction for ascertainment bias. Model selection was based on the lowest Bayesian Information Criterion. 1000 bootstrap replicates were done; only p values > 60% are shown. Clusters were identified using hierarchical Bayesian Analysis of Population Structure (Cheng et al., 2013). These clusters were consistent with the sub-lineages previously identified in Lee et al. (2015a); Lee et al. (2015b), thus only sub-lineage names are indicated (Major sub-lineages [Mj]-IIIA, B, C, and Mj-VA). Only Mj-IIIA/B/C were present in 2011–2012; Mj-IIIA was first seen in village K in 2007, IIIB was first seen in 2009, and IIIC was first seen in 2012. Alleles informative for transmission in Mj-IIIB, identified using deep sequencing, are indicated. Between 2007–2012, there were two individuals who had a second episode of TB; stars are used to highlight these samples, with a different colour for each patient. MT-0080 is included in the alignment as the deep sequencing data from a sweep of all colonies identified a cSNP compared to the MT-0080_PB reference, which itself was generated from a single colony pick.
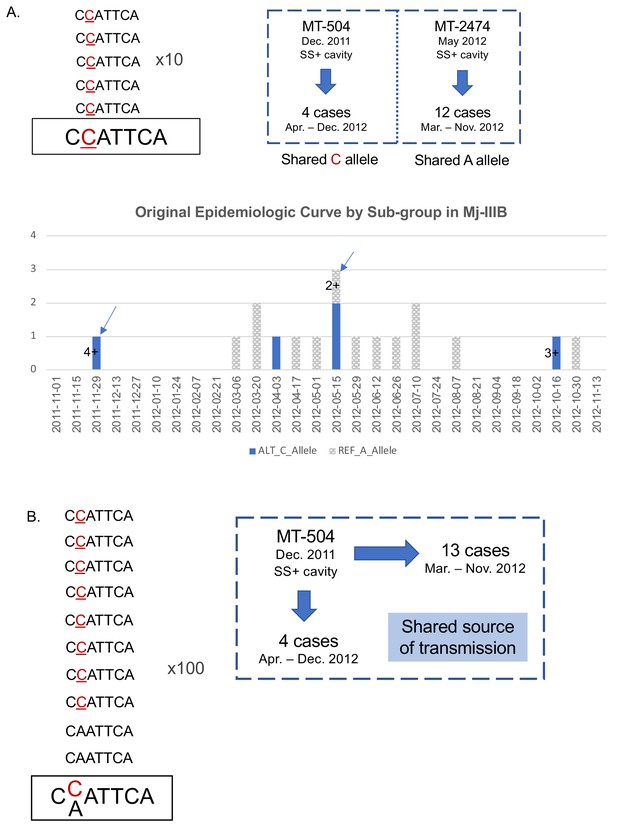
Comparison of epidemiologic inferences using 'routine' versus 'deep' sequencing.
(A) Routine sequencing (to ~40–50x) had indicated there were three major clusters present in village K in 2011–2012, Mj-IIIA, B and C. Further analysis of sequencing and epidemiological data indicated that the 2011–2012 cases in Mj-IIIB could be divided into two subgroups of transmission – comprised of five and 13 patients, respectively (Lee et al., 2015b). MT-504 was the suspected source case for the subgroup of five, which all shared a ‘C’ allele at position 276,685 in H37Rv (position 276,544 in MT-0080_PB). In contrast, all members of the subgroup of 13 shared an ‘A’ at this position. Previously, MT-2474 was the suspected source case for this subgroup; this case was the first person with smear-positive (SS+) cavitary disease diagnosed in this subgroup. The epidemiologic curve for the 2011–2012 Mj-IIIB cases is shown, stratified by sub-group based on the alleles detected with routine sequencing. MT-504 and MT-2474, the putative sources within each sub-group are indicated. Sputum smear grade is given where cases were positive on microscopy. Dates of diagnosis are classified into biweekly intervals. (B) In contrast to standard sequencing, deep sequencing data revealed that, in fact, MT-504 – the presumed source for the subgroup of five cases and the first highly contagious case diagnosed in Mj-IIIB during the outbreak year – had both ‘C’ and ‘A’ alleles at this position (563:133 of reads, respectively), suggesting this was in fact the most probable source for both subgroups.
Tables
Comparison of alignments to H37Rv and MT-0080_PB.
Based on these filters: Phred < 50, Root Mean Square Mapping Quality (RMS-MQ) ≤ 30, depth (DP) < 20, Fisher Strand Bias (FS) ≥ 60 and read position strand bias (ReadPos) < −8 and an allelic fraction of ≥ 95% for cSNPs, with hSNPs classified when 5% < ALT < 95%. Quality metrics for the individual cSNPs/hSNPs identified in each sample are given in Source data 2.
| H37Rv (4,411,532 bp) | MT-0080_PB (4,426,525 bp) | P value | |
|---|---|---|---|
| Number of positions according to reference genome | |||
| Invariant reference across all samples, n (%) | 4,018,786 (91·10%) | 4,084,195 (92·27%) | <0·00005 a |
| Position was missing/low quality in at least one sample, n (%) | 391,761 (8·88%) | 342,179 (7·73%) | <0·00005 a |
| Position was an c/hSNP in at least one sample, n (%) | 985 (0·22%) | 152 (0·00%) | <0·00005 a |
| Shared cSNPs across all samples, n (%) | 764 (0·02%) | 1 (0·00%) | <0·00005 a |
| Shared hSNPs across all samples, n (%) | 42 (0·00%) | 0 (0%) | <0·00005 a |
| Core pairwise distances | |||
| Core cSNPs vs. reference, median (range) | 791 (790–792) | 3 (1–65) | <0·00005 b |
| Core cSNPs between samples, median (range) | 3 (0–64) | 3 (0–66) | <0·00005 b |
-
a Two sample test for difference in proportions.
b Wilcoxin Signed Rank test.
Additional files
-
Source data 1
Percent of kmers classified as Mycobacterium tuberculosis Complex (MTBC).
hSNP frequency is shown using the alignment to MT-0080_PB after removing non-MTBC reads, and filtering with the following thresholds: Phred score < 50, Root Mean Square Mapping Quality [RMS-MQ] ≤ 30, depth [DP] < 20, Read Position Rank Sum [ReadPosRankSum] < −8, Fisher Strand Bias [FS] ≥ 60
- https://cdn.elifesciences.org/articles/53245/elife-53245-data1-v2.docx
-
Source data 2
Comparison of consensus single-nucleotide polymorphisms (cSNPs) and heterogeneous alleles (hSNPs) in all samples aligned to H37Rv versus MT-0080_PB, after initial filtering with Allelic Fraction for cSNPs ≥ 0·95 and 0·05 < hSNP < 0·95.
Initial filtering thresholds used were: Phred score < 50, Root Mean Square Mapping Quality [RMS-MQ] ≤ 30, depth [DP] < 20, Read Position Rank Sum [ReadPosRankSum] < −8, Fisher Strand Bias [FS] ≥ 60. As a consequence of the depth of coverage, where allelic fraction was 0·05 < alternative allele [ALT] < 0·95, all hSNPs had at least 2 REF and ALT alleles by default. This includes all hSNPs and cSNPs identified across all samples, except variants in PE_PGRS and PPE genes, as well as those in mobile elements; some of these variants will be in positions that are excluded from the core alignment, as they failed quality control or are missing in at least one sample in the dataset. Read Position Rank Sum can only be calculated when both reference and alternative alleles are present at a position, therefore the number of cSNPs included in the summary statistics for this variable are 14720 for the H37Rv alignment and 153 for the alignment to MT-0080. *As samples were downsampled to this threshold, this is truncated at 1500.
- https://cdn.elifesciences.org/articles/53245/elife-53245-data2-v2.docx
-
Source data 3
Positions with shared heterogeneous alleles (hSNPs) in all 62 samples versus H37Rv.
- https://cdn.elifesciences.org/articles/53245/elife-53245-data3-v2.xlsx
-
Source data 4
Quality metrics for each heterogeneous allele (hSNPs) that was shared across all 62 samples versus H37Rv.
- https://cdn.elifesciences.org/articles/53245/elife-53245-data4-v2.xlsx
-
Source data 5
Assembly metrics for Single Molecule Real-Time sequencing of MT-0080 (‘MT-0080_PB’), aligned to NC_000962.3 (H37Rv).
Quast (v.5.0.2 Gurevich et al., 2013) was used to tabulate the above statistics, with the exception of the number of CDS and RNA, where annotation was done using RASTtk (v.2.0 Brettin et al., 2015).
- https://cdn.elifesciences.org/articles/53245/elife-53245-data5-v2.docx
-
Source data 6
Core cSNPs and hSNPs between samples, where cSNPs >= 0.95 and 0.05 < hSNP < 0.95 and Phred < 50, RMS-MQ <= 30, DP < 20, FS >= 60, ReadPos < −8, with a minimum of 2 ALT and 2 REF alleles to call hSNPs.
This alignment was also filtered to remove variants in PE_PGRS and PE genes, as well as transposons, phage, and integrase as annotated using RASTtk (v.2.0). The original clusters and subgroups identified in Lee et al. (2015b) are indicated using different colours. cSNPs and hSNPs are indicated in grey, while cells with alleles that are the same as the reference are filled with white. Due to the minimum DP and allele frequency, a minimum of 2 ALT and 2 REF alleles were needed to call hSNPs by default.
- https://cdn.elifesciences.org/articles/53245/elife-53245-data6-v2.xlsx
-
Source data 7
Core cSNPs and hSNPs between samples, where cSNPs >= 0.95 and 0.05 < hSNP < 0.95 and Phred < 596, RMS-MQ <= 39, DP < 20, FS >= 46, ReadPos < −6, with a minimum of 2 ALT and 2 REF alleles to call hSNPs.
This alignment was also filtered to remove variants in PE_PGRS and PE genes, as well as transposons, phage, and integrase as annotated using RASTtk (v.2.0). The original clusters and subgroups identified in Lee et al. (2015b) are indicated using different colours. cSNPs and hSNPs are indicated in grey, while cells with alleles that are the same as the reference are filled with white. Due to the minimum DP and allele frequency, a minimum of 2 ALT and 2 REF alleles were needed to call hSNPs by default.
- https://cdn.elifesciences.org/articles/53245/elife-53245-data7-v2.xlsx
-
Source data 8
Core cSNPs and hSNPs between samples, where cSNPs >= 0.99 and 0.01 < hSNP < 0.99 and Phred < 50, RMS-MQ <= 30, DP < 20, FS >= 60, ReadPos < −8, minimum of 2 ALT and 2 REF alleles.
This alignment was also filtered to remove variants in PE_PGRS and PE genes, as well as transposons, phage, and integrase as annotated using RASTtk (v.2.0). The original clusters and subgroups identified in Lee et al. (2015b) are indicated using different colours. cSNPs and hSNPs are indicated in grey, while cells with alleles that are the same as the reference are filled with white.
- https://cdn.elifesciences.org/articles/53245/elife-53245-data8-v2.xlsx
-
Source data 9
Comparison of consensus single-nucleotide polymorphisms (cSNPs) and heterogeneous alleles (hSNPs) in all samples aligned to H37Rv versus MT-0080_PB, after initial filtering with Allelic Fraction for cSNPs ≥ 0·99 and 0·01 < hSNP < 0·99.
Initial filtering thresholds used were: Phred score < 50, Root Mean Square Mapping Quality [RMS-MQ] ≤ 30, depth [DP] < 20, Read Position Rank Sum [ReadPosRankSum] < −8, Fisher Strand Bias [FS] ≥ 60. Where 0·01 < alternative allele [ALT] < 0·99, a minimum of 2 REF/ALT alleles were required for all hSNPs to reduce risk of including sequencing error; those that failed to meet these criteria were excluded. This includes all hSNPs and cSNPs identified across all samples, except variants in PE_PGRS and PPE genes, as well as those in mobile elements; some of these variants will be in positions that are excluded from the core alignment, as they failed quality control or are missing in at least one sample in the dataset. Read Position Rank Sum can only be calculated when both reference and alternative alleles are present at a position, therefore the number of cSNPs included in the summary statistics for this variable are 13255 for the H37Rv alignment and 148 for the alignment to MT-0080 in the cSNPs ≥ 0·99 and 0·01 < ALT < 0·99 analysis. *As samples were downsampled to this threshold, this is truncated at 1500. P values were calculated using on the Wilcoxon-Mann-Whitney test.
- https://cdn.elifesciences.org/articles/53245/elife-53245-data9-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/53245/elife-53245-transrepform-v2.docx





