Dual histone methyl reader ZCWPW1 facilitates repair of meiotic double strand breaks in male mice
Figures
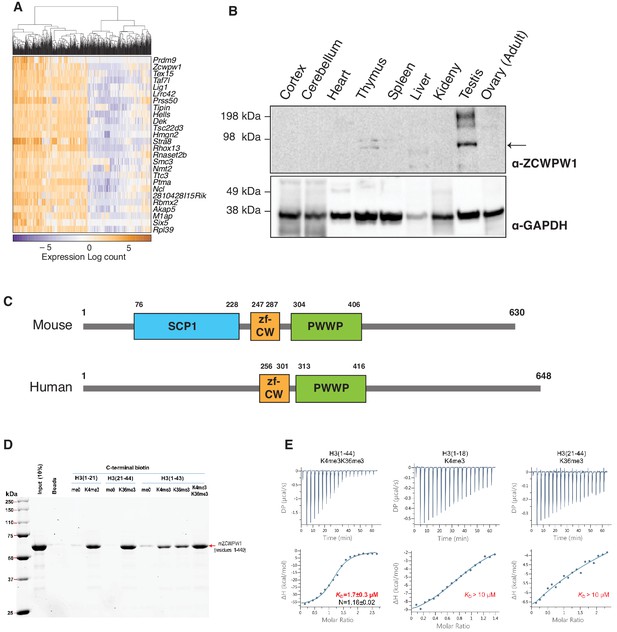
ZCWPW1 tissue expression and methyl histone binding activity in vitro.
(A) Heatmap of the top 25 genes co-expressed with PRDM9 during meiosis in single cells (Chen et al., 2018). Rows (genes) are ordered based on PRDM9 correlation coefficient (rho), from Figure 1—source data 1. Columns (single cells) were clustered by hierarchical clustering using euclidean distance and complete linkage. (B) Western blot for endogenous ZCWPW1 protein expression in different tissues from WT B6 mice. The arrow indicates ZCWPW1 band. (C) Schematic representation of mouse and human ZCWPW1 proteins with structural domains shown as annotated from NCBI Conserved Domain Database. (D) Recombinant ZCWPW1 was mixed with indicated biotinylated histone peptides and bound proteins were identified by Coomassie staining. (E) Isothermal titration calorimetry measurements were performed with recombinant ZCWPW1 and indicated histone peptides. The top panel is the raw data showing the heat released and measured by the sensitive calorimeter during gradual titration of the peptide into the sample cell containing ZCWPW1 until the binding reaction has reached equilibrium. The bottom panel shows each peak in the raw data is integrated and plotted versus the molar ratio of peptide to protein. The resulting isotherm can be fitted to a binding model from which the affinity (KD) is derived. The Y-axis measures enthalpy change (ΔH) using the relationship ΔG=ΔH-TΔS where ΔG is the Gibbs free energy, ΔS is the entropy change and T is the absolute temperature. N refers to the reaction stoichiometry.
-
Figure 1—source data 1
Correlation of expression between cellular genes and Prdm9 in single cell RNA-seq analysis.
- https://cdn.elifesciences.org/articles/53360/elife-53360-fig1-data1-v2.xlsx
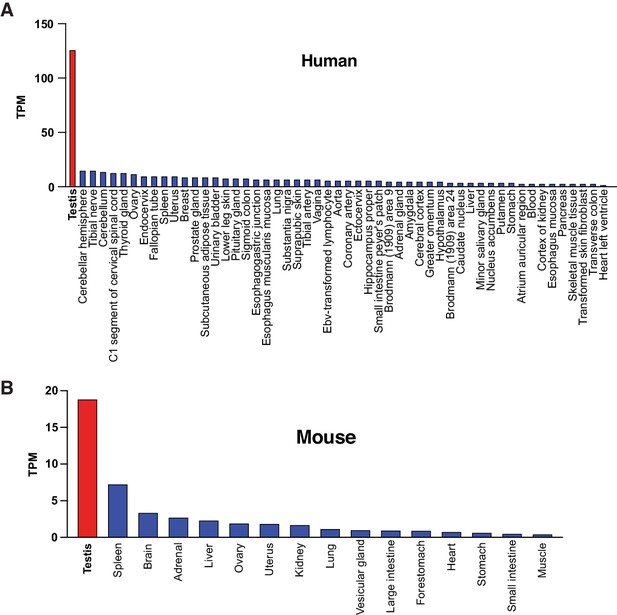
Zcwpw1 transcript expression in human and mouse tissues.
ZCWPW1 transcript expression from different tissues in human (GTEx) (A), and mouse (Li et al., 2017) (B).
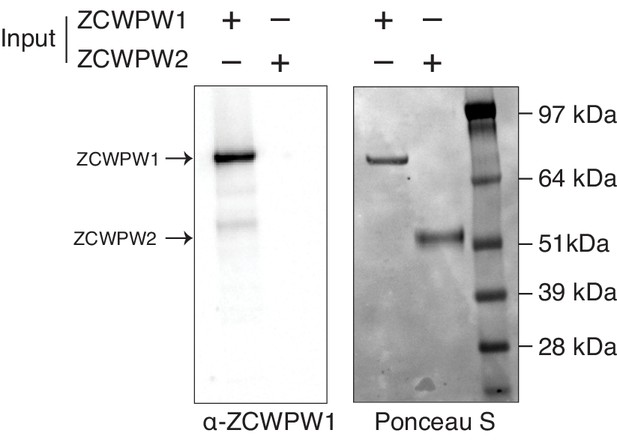
ZCWPW1 antibody specificity.
Testing ZCWPW1 antibody specificity for ZCWPW1 and potential cross-reactivity with ZCWPW2.
His-tagged recombinant ZCWPW1 (~70 kDa) and ZCWPW2 (~49 kDa) were loaded in SDS-PAGE gel and transferred to PVDF membrane. Membranes were then subjected for either Ponceau S staining (right) or immunoblotting with anti-ZCWPW1 rabbit polyclonal antibody (left).
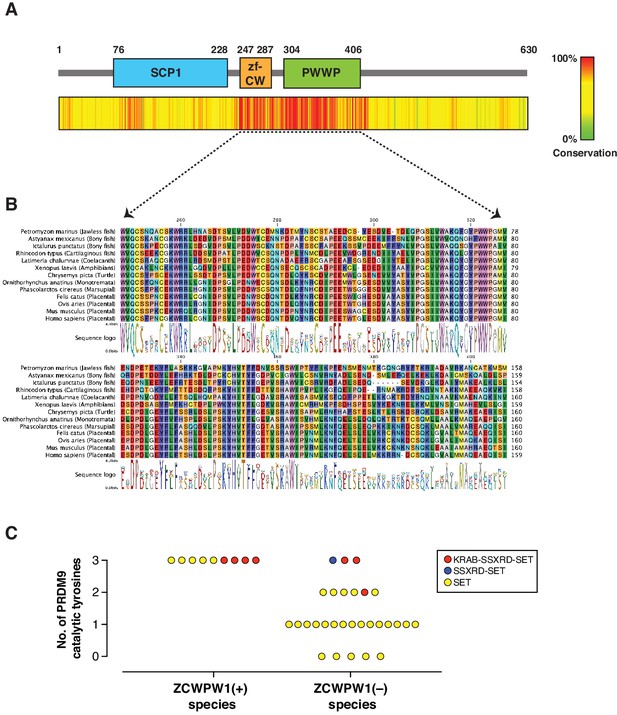
Evolution of Zcwpw1 in vertebrates.
(A) ZCWPW1 protein sequences available from public databases (174 species) were aligned by CLC Genomics Workbench using MUSCLE algorithm (Edgar, 2004). Alignment conservation score is plotted as heat map across different regions of mouse ZCWPW1 shown in the schematic figure above the heatmap. Alignment segments corresponding to gaps in the reference sequence (mouse ZCWPW1) were removed. (B) Multiple sequence alignment from (A) showing regions corresponding to zf-CW and PWWP domains in 13 species representing different vertebrate clades. Consensus sequence logo is shown below the alignment. (C) Plot showing number of conserved catalytic tyrosines (n = 3) in the SET domain of PRDM9 in bony fish species that have a PRDM9 ortholog. PRDM9 orthologs are divided into two groups depending on whether the corresponding species of that ortholog has ZCWPW1 ortholog (with ZCWPW1 group) or lacks ZCWPW1 ortholog (without ZCWPW1 group). Different colors represent different domain architecture of PRDM9 ortholog in the particular species. PRDM9 information reproduced from Baker et al., 2017.
-
Figure 2—source data 1
ZCWPW1 orthologs used in evolution analysis.
- https://cdn.elifesciences.org/articles/53360/elife-53360-fig2-data1-v2.xlsx
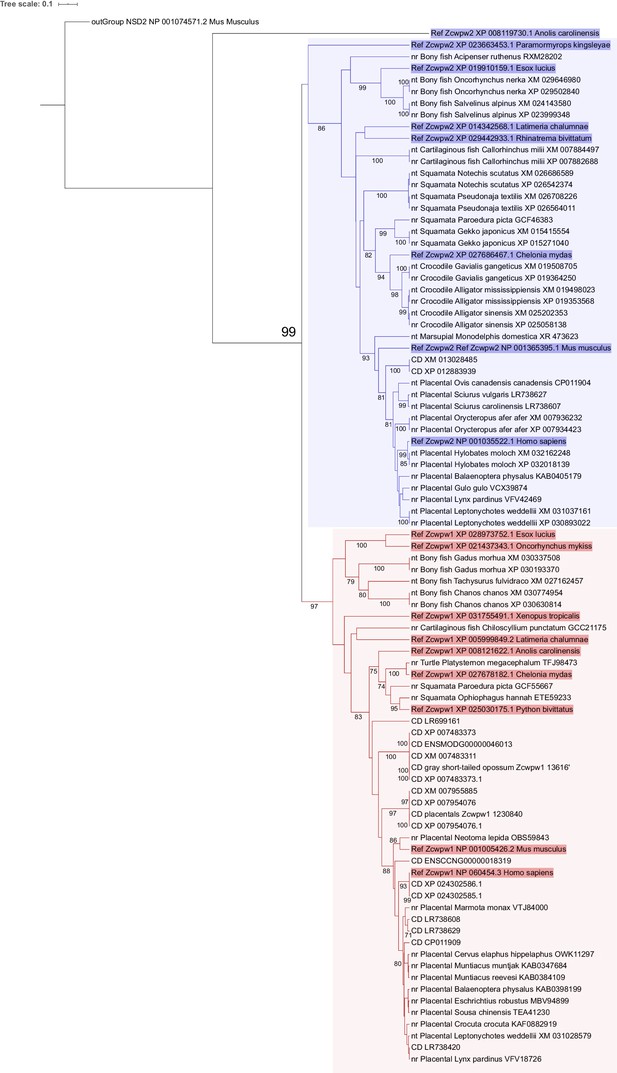
Phylogenetic analysis of potential ZCWPW1 orthologs discovered by BLAST.
Maximum likelihood phylogenetic tree was made to differentiate ZCWPW1 orthologs from ZCWPW2 orthologs among tentative sequences discovered by BLAST (see methods). Using CLC Genomics Workbench (progressive alignment algorithm), tentative sequences were aligned with known ZCWPW1 and ZCWPW2 reference sequences (text highlighted with blue and red respectively for ZCWPW1 and ZCWPW2 reference sequences). Mouse NSD2 was used as outgroup sequence. 1000 bootstrap replicates were performed, with bootstrap values ≥ 70% are shown in the tree. ZCWPW1 and ZCWPW2 sequences clustered into distinct branches (blue and red box respectively) with node support of 99%. ‘Ref’: known reference sequence for ZCWPW1 or ZCWPW2 ortholog, ‘nr’: sequence discovered using BLAST blastp, ‘nt’: sequence discovered using BLAST tblastn, ‘CD’: evidence for presence of zf-CW and PWWP domain was through NCBI conserved domain database search (evidence for other sequences is by ScanProsite).
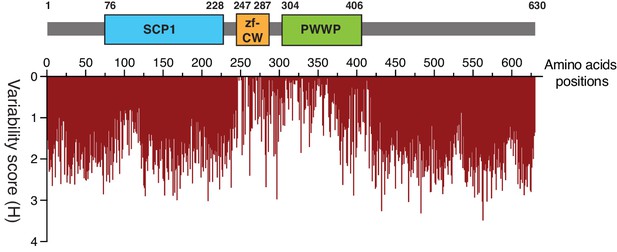
Amino acid variability in ZCWPW1 orthologs.
Plot for variability of amino acid residues in aligned sequences of ZCWPW1 orthologs in Figure 2A. Amino acid positions in mouse ZCWPW1 were used as reference for plotting. Shannon entropy analysis was used to calculate variability scores (Shannon, 1948. see methods). X axis represents amino acid positions in mouse ZCWPW1 (schematic representation of it shown above the plot). Y axis represents Shannon entropy (H) for every position in X. Small H score means low variability of amino acid reside in the corresponding position, and therefore greater conservation.
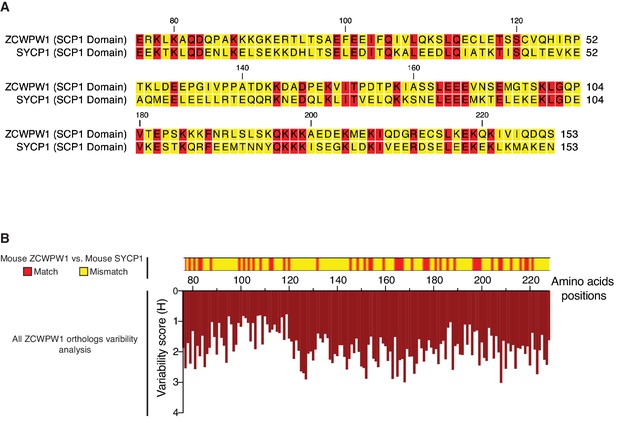
Alignment of SCP1 domains from mouse ZCWPW1 and SYCP1.
(A) Alignment of SCP1 domain from mouse ZCWPW1 (76–228 aa) with SCP1 domain from mouse SYCP1. Alignment is done by CLC Genomics Workbench using MUSCLE algorithm (Edgar, 2004). Alignment segments corresponding to gaps in ZCWPW1 were removed. (B) Upper: Heatmap representation for the alignment in (A). Lower: Plot for variability of amino acid residues in aligned sequences of ZCWPW1 orthologs. It is partial subplot from the plot in Figure 2—figure supplement 2), showing only the SCP1 domain region (76–228 aa).
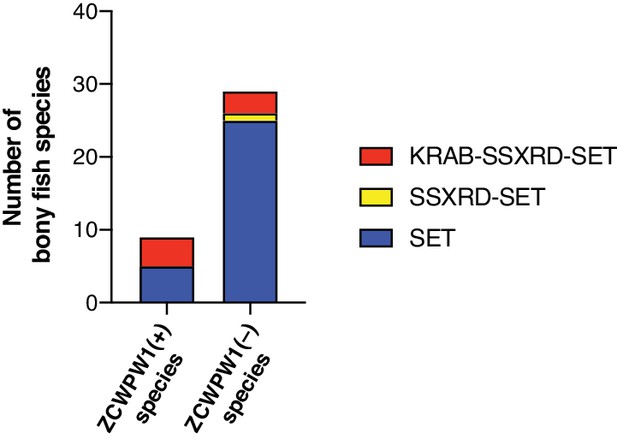
PRDM9 domain structure in bony fishes.
Comparison of PRDM9 domain structure in bony fish species with or without ZCWPW1 ortholog. PRDM9 information reproduced from Baker et al., 2017.
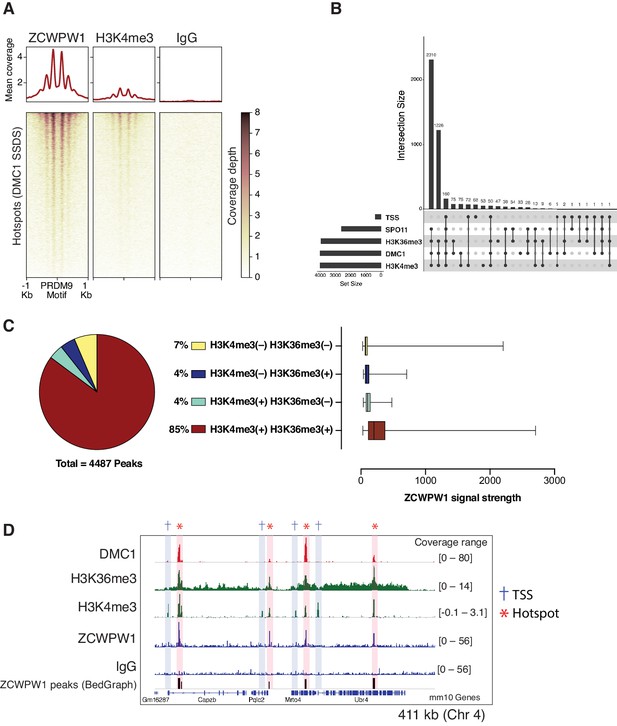
Mapping of ZCWPW1 chromatin biding in vivo using CUT&RUN in spermatocytes from B6/B6 mice.
(A) Heatmaps representing ZCWPW1, H3K4me3 and IgG (anti-GFP) CUT&RUN read coverage in B6/B6 mouse spermatocytes at hotspots determined by DMC1 SSDS (GSE99921). Signals are centered around PRDM9 motifs. Hotspots with multiple motifs were excluded from plotting (11,350 DSB hotspots were used in total). Y-axis of line plots in the top represents mean coverage (trimmed fragments/50 bp) (B) Upset plot showing intersections of ZCWPW1 peaks (n = 4,487) with SPO11 oligos (GSE84689), DMC1 SDDS (GSE99921), prophase H3K4me3 and H3K36me3 peaks (GSE121760) and transcription starting sites (TSS). Y-axis represents number of ZCWPW1 peaks intersecting with the specified regions. (C) Left: pie chart showing tri-methylation states of lysine 4 and 36 of histone H3 at ZCWPW1 peaks in B6/B6 spermatocytes. Right: ZCWPW1 signal strength at different ZCWPW1 peak subsets shown in the pie chart on the left. X-axis represents the strength of ZCWPW1 signal (read coverage) per peak. (D) Read coverage plots for DMC1 SSDS hotspots (GSE35498/red), H3K4me3/H3K36me3 (GSE121760/Green) and ZCWPW1/IgG (CUT&RUN/blue) across a region on chromosome 4. Hotspots and TSSs are highlighted. Y-axis represents coverage (fragments/bin). The track in the bottom represents the bedGraph of ZCWPW1 called peaks using MACS2.
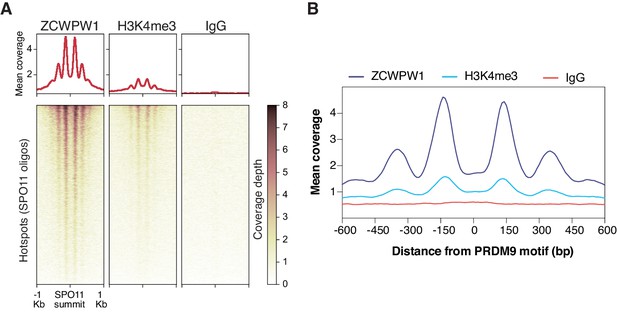
Mapping of ZCWPW1 chromatin biding in vivo using CUT&RUN in spermatocytes from B6/B6 mice.
(A) Heatmaps representing ZCWPW1, H3K4me3 and IgG (anti-GFP) CUT&RUN read coverage in B6/B6 mice spermatocytes at SPO11 oligo summits (GSE84689). Y-axis of line plots in the top represents mean coverage (trimmed fragments/50 bp) (B) Representation of mean coverage plot from Figure 3A to emphasize the signal pattern around PRDM9 motifs. Y-axis represents mean coverage (trimmed fragments/50 bp). The central regions lack ZCWPW1 and H3K4me3 signal. These H3K4me3/ZCWPW1 depleted regions are surrounded by symmetrically distributed peaks of H3K4me3/ZCWPW1 indicative of nucleosome positioning around the hotspots’ center.
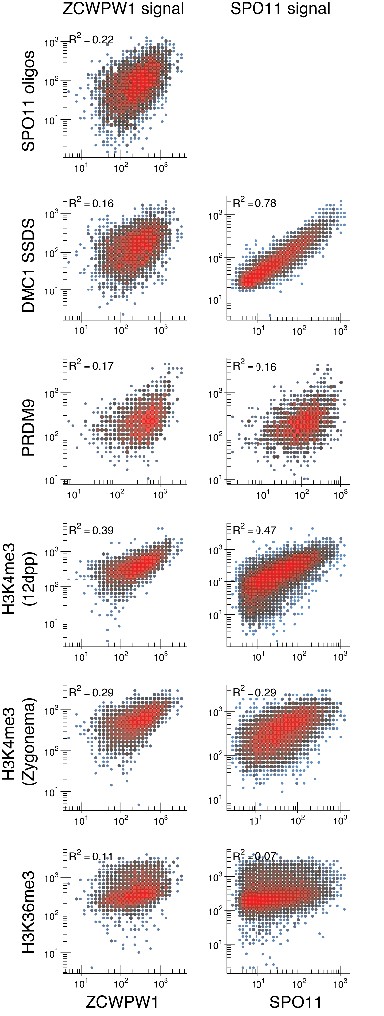
ZCWPW1 correlation with other metrics of meiotic recombination at DSB hotspots.
Correlation of ZCWPW1 CUT&RUN signal strength (left) and SPO11 oligo intensity (GSE84689) (right) with other metrics of recombination at DSB hotspots: DMC1 SSDS (GSE35498), H3K4me3 12dpp (GSE35498), PRDM9 (GSE93955), H3K36me3 (GSE93955) and H3K4me3 Zygonema (GSE121760). ZCWPW1 CUT&RUN data is from B6 mice. For all panels, only autosomal hotspots that coincide with a ZCWPW1 peak were used. ZCWPW1 strength is calculated as the sum of in-peak sequencing CUT&RUN reads at each hotspot. SPO11 strength is represented as SPO11-oligo density (Lange et al., 2016). DMC1 strength is represented as DMC1 SSDS signal at hotspots (Khil et al., 2012). For remaining metrics, the strength is calculated as the sum of all reads at the SSDS-defined hotspot.
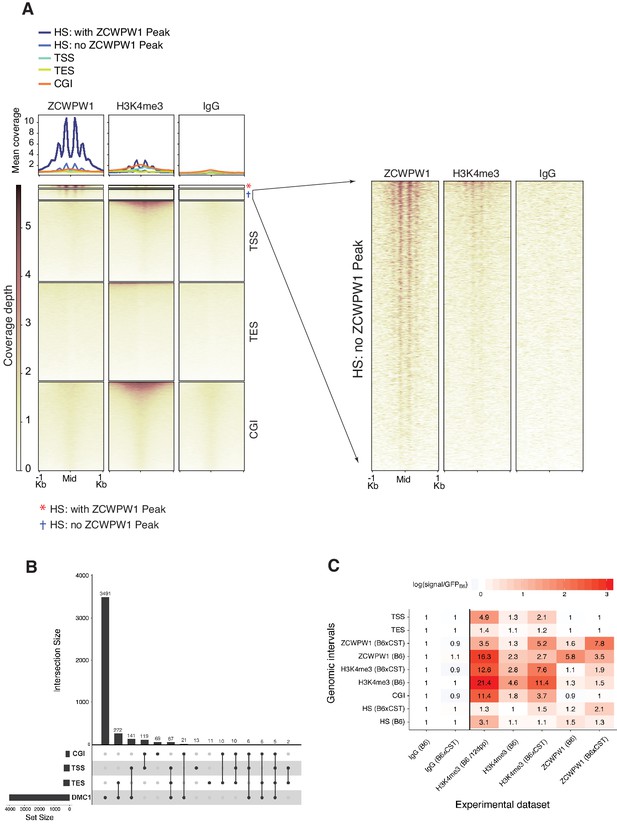
ZCWPW1 binding at functional genomic sites.
(A) Heatmaps representing ZCWPW1, H3K4me3 and IgG (anti-GFP) CUT&RUN read coverage in B6/B6 mouse spermatocytes at indicated regions. Y-axis of line plots in the top represents mean coverage (trimmed fragments/50 bp). The purpose is to compare ZCWPW1 occupancy at DSB hotspots with occupancy at functional sites in the genome: Transcription Start Sites (TSS), Transcription End Site (TES) and CpG islands (CGI). Hotspots regions are based on DMC1 SSDS (GSE99921), and they are categorized into two groups based on whether ZCWPW1 peak was called in the individual hotspot (n = 2,446) or not called (n = 8,904). Heatmap for ZCWPW1 signal around hotspots without called ZCWPW1 peak is zoomed in at the right panel. For hotspots, signals are centered around PRDM9 motifs, and hotspots with multiple motifs were excluded from plotting. TSS and TES were retrieved from GENCODE release M20. After mm10 blacklisted regions in TSS and TES were filtered out, 67,579 TSS and 82,380 TES sites were used for plotting. (B) Upset plot showing intersection of ZCWPW1 peaks (n = 4,487) with DMC1 SSDS hotspots (GSE99921), TSSs, TESs and CGIs. Y-axis represents number of ZCWPW1 peaks intersecting with the specified regions. (C) Enrichment of reads from different experiments at indicated genomic regions. All experiments are from CUT&RUN with exception of 12dpp H3K4me3 reads which are from ChIP-seq (GSE35498). Genomic regions represent (i) peaks called from ZCWPW1 and H3K4me3 CUT&RUN from both B6 and B6XCAST mice (ii) hotspots (HS) regions from DMC1 SSDS (B6: GSE99921), (B6/CAST: GSE73833) and (iii) functional genomic sites: Transcription Start Sites (TSS), Transcription End Site (TES) and CpG islands (CGI). Sequencing reads (q > 30) at each set of genomic intervals were counted for our four CUT&RUN experiments as well as for H3K4me3 ChIP-Seq data in 12dpp testis. Thereafter, percentage of total reads in each set of genomic intervals was determined. For all experiments, the enrichment of percentage reads relative to IgG control (anti-GFP) CUT&RUN in B6 mice is shown. Color legend is scaled by the log of enrichment values.
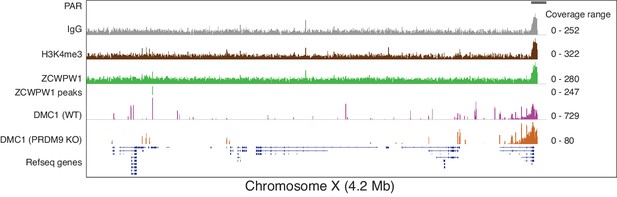
ZCWPW1 binding at chromosome X pseudoautosomal region.
Read coverage plots for ZCWPW1, H3K4me3 and IgG (anti-GFP) CUT&RUN, and DMC1 SSDS hotspots from WT and PRDM9 KO (GSE35498) in chromosome X pseudoautosomal region (PAR) or at PAR-adjacent PRDM9-independent hotspots. Y-axis represents coverage (fragments/bin).
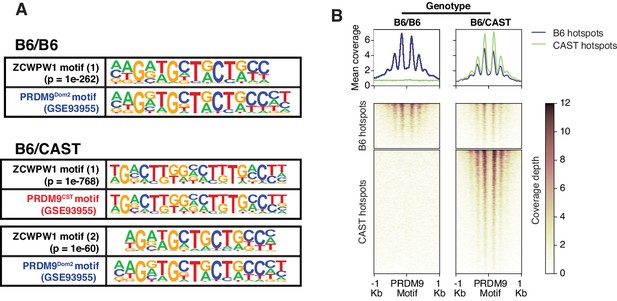
Mapping of ZCWPW1 chromatin biding in vivo using CUT&RUN in spermatocytes from F1 B6/CAST hybrid mice.
(A) Comparisons of de novo discovered motifs for PRDM9Dom2, PRDM9Cast and ZCWPW1. For PRDM9Dom2 and PRDM9Cast, PRDM9 ChIP-seq peaks (GSE93955) for either allele were queried by HOMER to identify the consensus DNA motif for their binding sites. For ZCWPW1, CUT&RUN peaks from either B6/B6 or B6/CAST F1 hybrid were used to identify ZCWPW1 binding motifs in pure and mixed genomic backgrounds, respectively. The first motif hit is shown for binding in B6/B6 and the first and second hits are shown for B6/CAST. (B) Heatmaps comparing ZCWPW1 read coverage for CUT&RUN performed in spermatocytes from either B6/B6 or B6/CAST F1 hybrid. ZCWPW1 signal is plotted around F1 hybrid hotspots defined by DMC1 SSDS (GSE73833) and regions categorized based on PRDM9 allele motif in the center of the hotspot (Prdm9Dom2 and Prdm9Cast for B6 and CAST strains respectively). Signals are centered around PRDM9 motifs, and hotspots with multiple motifs were excluded from plotting (10,466 DSB hotspots from B6xCAST F1 were used in total). Y-axis of line plots in the top represents mean coverage (trimmed fragments/50 bp).
-
Figure 4—source data 1
Enrichment of known motifs at ZCWPW1 binding sites in B6/B6 mice.
- https://cdn.elifesciences.org/articles/53360/elife-53360-fig4-data1-v2.xlsx
-
Figure 4—source data 2
De novo motifs discovery at ZCWPW1 binding sites in B6/B6 and F1 B6/CAST hybrid mice.
- https://cdn.elifesciences.org/articles/53360/elife-53360-fig4-data2-v2.xlsx
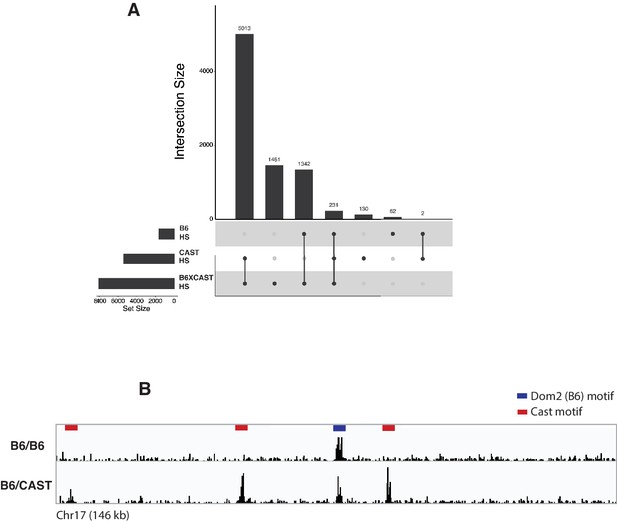
Mapping of ZCWPW1 chromatin biding in vivo using CUT&RUN in spermatocytes from F1 B6/CAST hybrid mice.
(A) Upset plot showing intersections of ZCWPW1 CUT&RUN peaks (from B6/CAST F1 hybrid spermatocytes, n = 8,976 peaks) with DMC1 SSDS hotspots (GSE75419) from (i) B6/B6, (ii) CAST/CAST and (iii) B6/CAST F1 hybrid spermatocytes. Y-axis represents number of ZCWPW1 peaks intersecting with the specified regions. (B) Read coverage plots for ZCWPW1 occupancy in B6/B6 and B6/CAST spermatocytes. Y-axis represents coverage (fragments/bin).
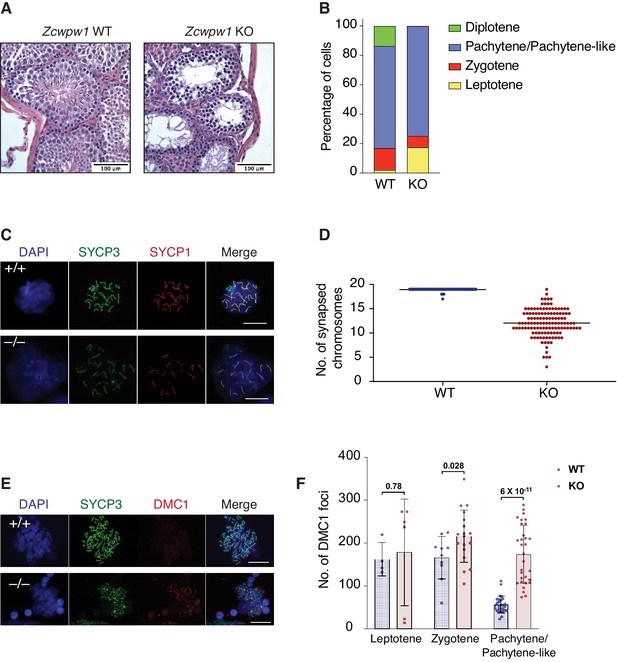
Zcwpw1 KO mice are azoospermic and display hallmarks of defective synapsis and DSB repair.
(A) Hematoxylin and Eosin staining of paraffin embedded tissue sections from testes of Zcwpw1 WT or Zcwpw1 KO. Scale bar is 100 µm. (B) Percentage distribution for meiotic prophase I stages in spermatocyte spreads from Zcwpw1 WT (n = 183) or Zcwpw1 KO (n = 194). Staging is based on double immunofluorescence staining of SYCP3 and SYCP1. (C) Indirect immunofluorescence staining with SYCP3 and SYCP1 in spermatocyte spreads from Zcwpw1 WT or Zcwpw1 KO. Scale bar is 10 µm (D) Counts for number of fully synapsed homologous chromosomes (autosomes) per spermatocyte. Counting was performed on spermatocyte spreads with double staining of SYCP3 and SYCP1. Only spermatocytes in pachytene/pachytene-like stage are considered for counting in Zcwpw1 WT or Zcwpw1 KO (n = 127 for each genotype). (E) Indirect immunofluorescence staining with SYCP3 and DMC1 in spermatocyte spreads from Zcwpw1 WT or Zcwpw1 KO. Scale bar is 10 µm (F) DMC1 foci count per cell in spermatocyte spreads double stained with SYCP3 and DMC1 in Zcwpw1 WT (Leptotene = 4, Zygotene = 10, Pachytene = 27) or Zcwpw1 KO (Leptotene = 6, Zygotene = 18, Pachytene-like = 31) spermatocytes. p-value shown is calculated with Welch’s t-test.
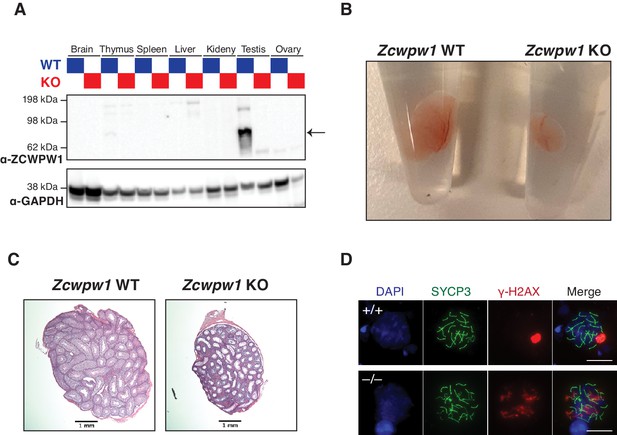
Phenotyping of Zcwpw1 KO spermatocytes.
(A) Western blot with ZCWPW1 specific polyclonal antibody in tissues from WT (blue lanes) or KO male mouse (red lanes) tissues. The arrow indicates ZCWPW1 band (B) Photo comparing size of testes from Zcwpw1 WT (left) and Zcwpw1 KO mice (right) (C) Hematoxylin and Eosin staining of testes sections from Zcwpw1 WT or Zcwpw1 KO mice. Scale bar is 1 mm. (D) Indirect immunofluorescence staining with SYCP3 and γ-H2AX in spermatocyte spreads from Zcwpw1 WT or Zcwpw1 KO. Scale bar is 10 µm.
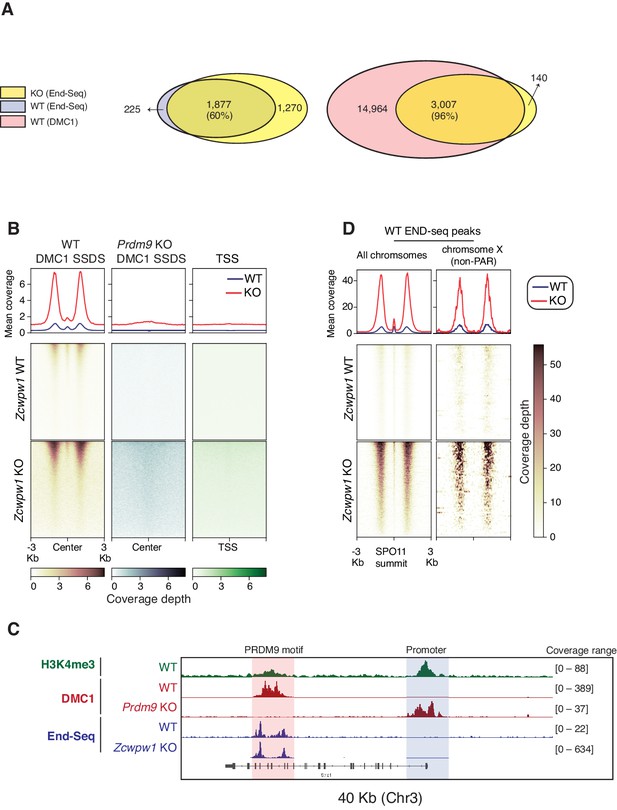
Double Strand Break mapping by END-seq in spermatocytes from adult Zcwpw1 WT and Zcwpw1 KO mice.
(A) Venn diagram showing END-seq peak overlap between Zcwpw1 KO and Zcwpw1 WT (left) or between Zcwpw1 KO END-seq peaks and DMC1 SSDS hotspots (GSE35498) (right). (B) END-seq read coverage heatmaps comparing Zcwpw1 WT and Zcwpw1 KO DSBs at different indicated regions: DMC1 SSDS hotspots (WT and PRDM9 KO/GSE35498) and transcription starting sites (TSS). All signals are normalized with spike-in controls. Y-axis of line plots in the top represents spike normalized Reads Per Million Reads (RPM) (C) Read coverage plots for H3K4me3 (CUT&RUN/green), DMC1 SSDS hotspots from WT and PRDM9 KO (GSE35498/red) and END-seq (blue). (D) Read coverage heatmaps for END-seq reads from Zcwpw1 WT and Zcwpw1 KO spermatocytes. Signal is plotted around WT END-seq called peaks (total peaks on the left, and non-PAR of chromosome X peaks on the right). Signals are centered at the overlapping SPO11 oligo summits (GSE84689). Signal is normalized with spike-in controls. Y-axis of line plots in the top represents spike normalized Reads Per Million Reads (RPM).
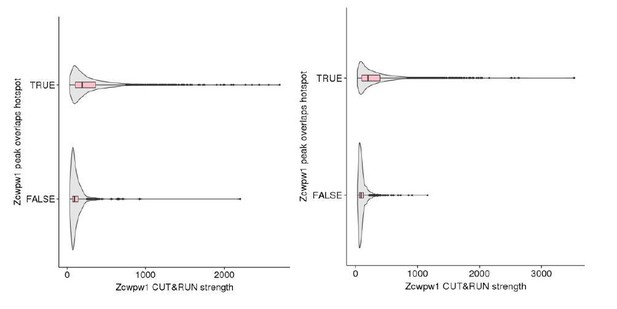
ZCWPW1 peaks overlapping hotspots are stronger than non-hotspots.
Left panel: B6, Right panel B6xCAST.
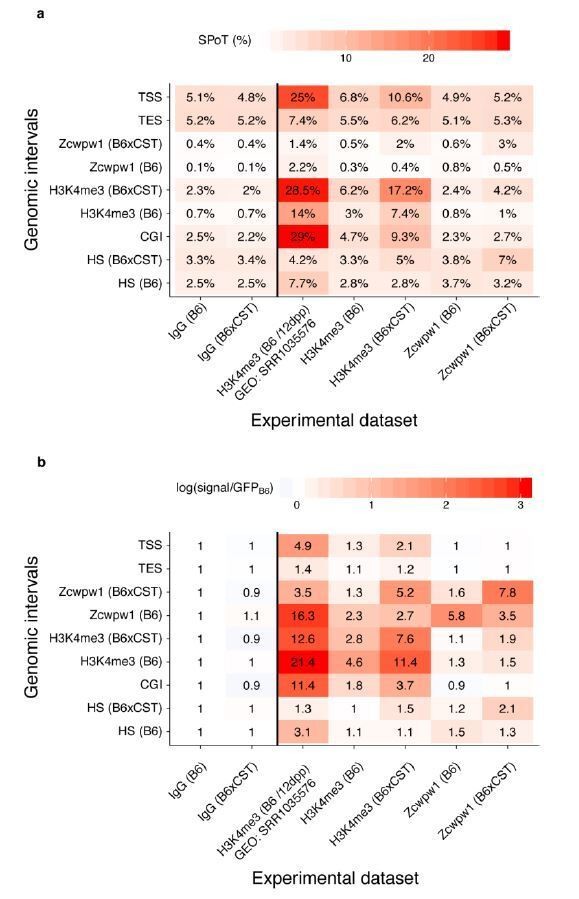
ZCWPW1 CUT&RUN shows very little enrichment at functional sites in the genome.
Sequencing reads (q > 30) at each set of genomic intervals were counted for our four CUT&RUN experiments as well as for H3K4me3 ChIP-Seq data in 12dpp testis. The genomic intervals used are TSS and TES from GENCODE vM20 (gencodeTSS, gencodeTES), peaks from our CUT&RUN experiments (ZCWPW1 (B6xCST), ZCWPW1 (B6), H3K4me3 (B6), H3K4me3 (B6xCST)), CpG islands (CGI), DSB hotspots (HS (B6), HS (B6XCST)). (a) The percentage of total reads in each set of genomic intervals is shown. (b) The enrichment relative to GFP CUT&RUN in B6 mice is shown. Color scale in b is scaled by the log of enrichment values.
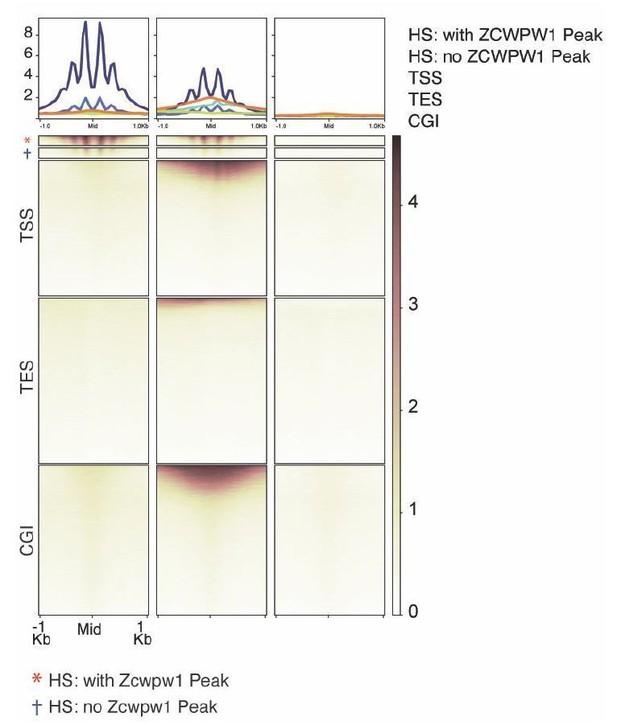
ZCWPW1 CUT&RUN shows very little enrichment at functional sites in the genome.
Only sequencing reads with a mapping quality of >30 were used. The top and second-to-top panels represent DSB hotspots with / without a called ZCWPW1 peak, respectively. Data here are from B6 x CAST mice.
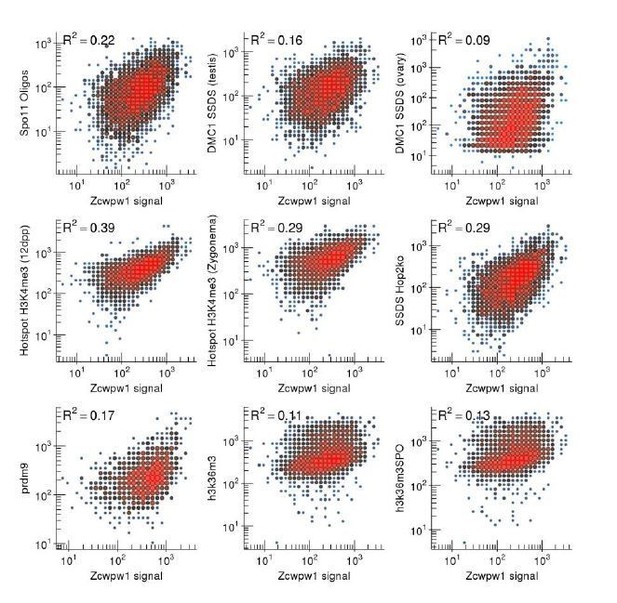
ZCWPW1 CUT&RUN signal correlates with other metrics of recombination at DSB hotspots.
Data are shown for ZCWPW1 CUT&RUN in B6 mice. For all panels, only autosomal hotspots that coincide with a ZCWPW1 peak were used. Strength for SSDS and H3K4me3 12dpp are taken from Brick et al., Nature 2018. CUT&RUN strength for H3K4me3 and ZCWPW1 is calculated as the sum of in-peak sequencing reads at each hotspot. The strength of H3K36me3 is calculated as the sum of all reads at the SSDS-defined hotspot. ZCWPW1 signal correlates best with H3K4m3 ChIP-Seq data from 12dpp mice. Notably however, it correlates less well with H3K4me3 from isolated Zygotene spermatocytes. This variation is likely a reflection of variation in ChIP / experimental enrichment and highlights the difficulty of attributing any of these measures as being “best” correlated with ZCWPW1 binding. ZCWPW1 appears to correlate equally well with Spo11-oligo and DMC1-SSDS measures of DSB frequency. Interestingly however, the correlation with DMC1-SSDS is notably improved in mice that lack the ability to repair DSBs (Hop2ko). The correlation with H3K36me3 is relatively lower, however the H3K36me3 signal at hotspots is notably weaker than that of the other metrics. Thus, the low correlation may be strongly influenced by background noise.
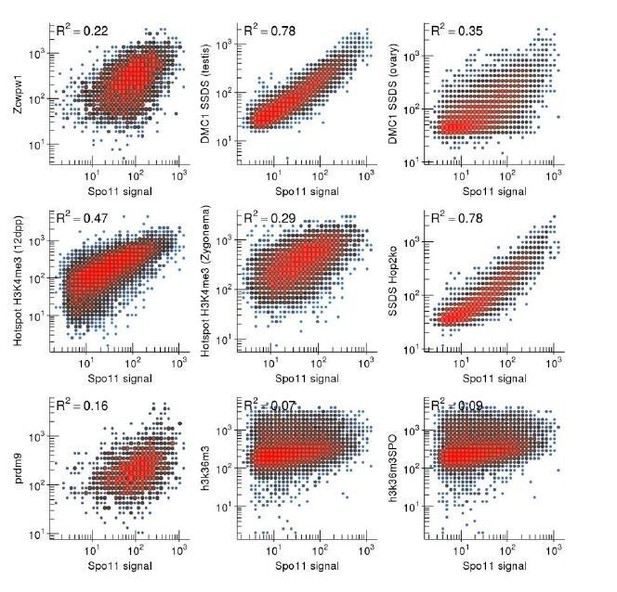
This figure is simply to provide a comparison point to Author response image 3.
Spo11-oligo signal correlates with other metrics of recombination. For all panels, only autosomal hotspots that coincide with a ZCWPW1 peak were used. Spo11 signal correlates best with DMC1-SSDS. Notably however, it correlates less well with DMC1-SSDS from female mice. As with ZCWPW1, the correlation with H3K36me3 is poor, since the H3K36me3 signal at hotspots is notably weaker than that of the other metrics.





