EDEM2 stably disulfide-bonded to TXNDC11 catalyzes the first mannose trimming step in mammalian glycoprotein ERAD
Figures
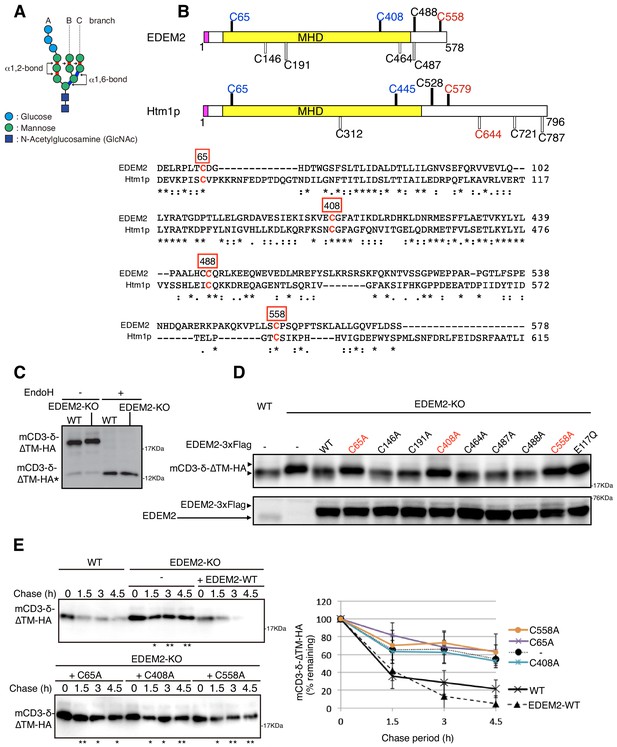
Effect of mutation of various cysteine residues in EDEM2 on gpERAD.
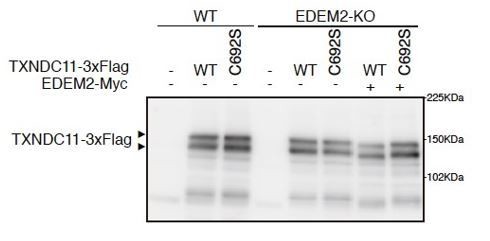
(A) Structure of Glc3Man9GlcNAc2 is schematically shown. Mannose residues are α1,2-bonded or α1,6-bonded as indicated. (B) Structures of human EDEM2 and yeast Htm1p are schematically shown with cysteine residues (C) highlighted together with their positions (black bars underneath C indicate conserved cysteine residues, whereas white bars over C indicate non-conserved cysteine residues). The purple and yellow boxes denote the signal sequence and mannosidase homology domain (MHD), respectively. Sequence comparison around the four cysteine residues conserved between EDEM2 and Htm1p (marked in red color) is shown below (asterisk and colon indicate identical and similar amino acids, respectively). (C) Cell lysates were prepared from WT and EDEM2-KO cells expressing mCD3-δ-ΔTM-HA by transfection, treated with (+) or without (-) EndoH, and analyzed by immunoblotting using anti-HA antibody. mCD3-δ-ΔTM-HA* denotes deglycosylated mCD3-δ-ΔTM-HA. (D) Cell lysates were prepared from WT and EDEM2-KO cells expressing WT or one of various cysteine mutants of 3x Flag-tagged EDEM2 together with mCD3-δ-ΔTM-HA by transfection, and analyzed by immunoblotting using anti-HA and anti-EDEM2 antibodies. E117Q is an enzymatically inactive mutant of EDEM2. (E) Cycloheximide chase was conducted to determine the degradation rate of mCD3-δ-ΔTM-HA in WT and EDEM2-KO cells expressing WT or one of the three cysteine mutants of 3x Flag-tagged EDEM2 by transfection, and analyzed by immunoblotting using anti-HA antibody (n = 3). Quantified data are shown on the right.
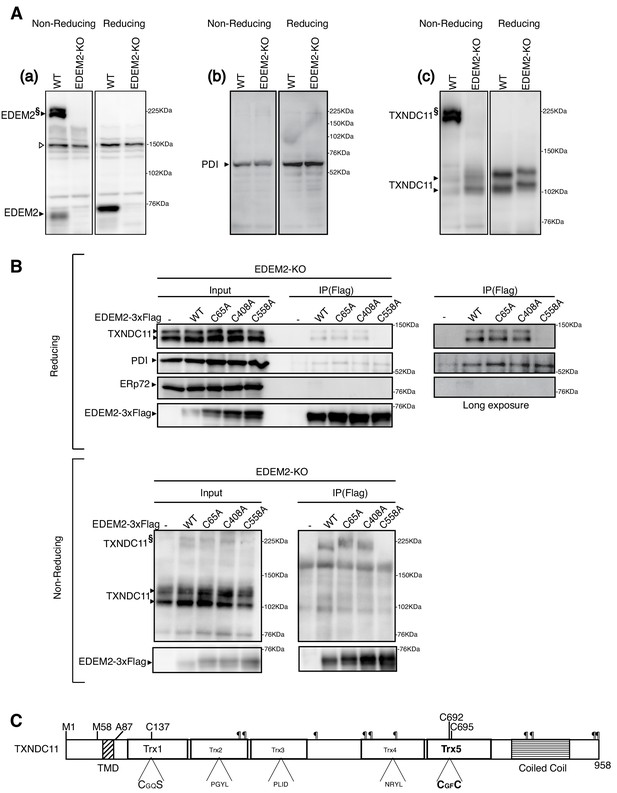
Disulfide bond formation between EDEM2 and TXNDC11.
(A) Cell lysates were prepared from WT and EDEM2-KO cells, subjected to SDS-PAGE under reducing and non-reducing conditions, and analyzed by immunoblotting using anti-EDEM2 (a), anti-PDI (b) and anti-TXNDC11 (c) antibodies. § denotes high molecular weight forms of EDEM2 and TXNDC11. Open triangle indicates a non-specific band. (B) Cell lysates were prepared from EDEM2-KO cells expressing WT or one of the three cysteine mutants of 3x Flag-tagged EDEM2 by transfection, and subjected to immunoprecipitation using anti-Flag antibody. An aliquot of cell lysates (Input) and immunoprecipitates {IP(Flag)} were subjected to SDS-PAGE under reducing and non-reducing conditions, and analyzed by immunoblotting using anti-TXNDC11, anti-PDI, anti-ERp72, and anti-Flag antibodies. (C) Structure of human TXNDC11 containing the TMD, five Trx domains, and coiled coil domain is schematically shown. ¶ denote potential N-glycosylation sites. The positions of two initiation methionines are also shown.
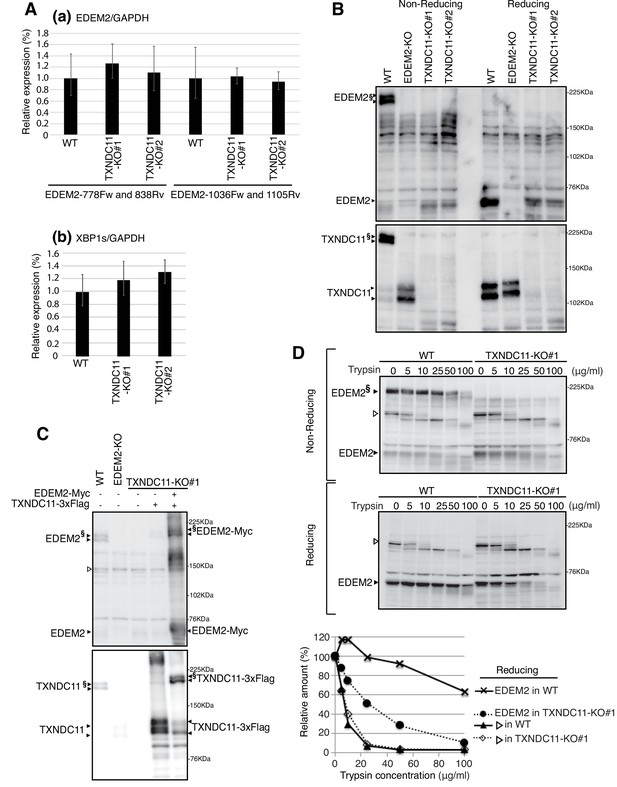
Effect of TXNDC11 knockout on EDEM2.
(A) Quantitative RT-PCR was conducted to determine the levels of endogenous EDEM2 mRNA (a) using the two primer sets indicated as well as spliced XBP1 mRNA (b) relative to the level of GAPDH mRNA in WT and two TXNDC11-KO cells (n = 3). (B) Cell lysates were prepared from WT, EDEM2-KO, and two TXNDC11-KO cells, subjected to SDS-PAGE under reducing and non-reducing conditions, and analyzed by immunoblotting using anti-EDEM2 and anti-TXNDC11 antibodies. (C) Cell lysates were prepared from WT, EDEM2-KO, and TXNDC11-KO#1 cells expressing 3x Flag-tagged TXNDC11 or both 3x Flag-tagged TXNDC11 and Myc-tagged EDEM2 by transfection, subjected to SDS-PAGE under non-reducing conditions, and analyzed by immunoblotting using anti-EDEM2 and anti-TXNDC11 antibodies. (D) Cell lysates were prepared from WT and TXNDC11-KO#1 cells, treated with the indicated amount of trypsin at 4°C for 15 min, subjected to SDS-PAGE under reducing and non-reducing conditions, and analyzed by immunoblotting using anti-EDEM2 antibody. The band with open triangle denotes a non-specific protein which serves as a control for trypsin digestion. Quantified data are shown at the bottom.
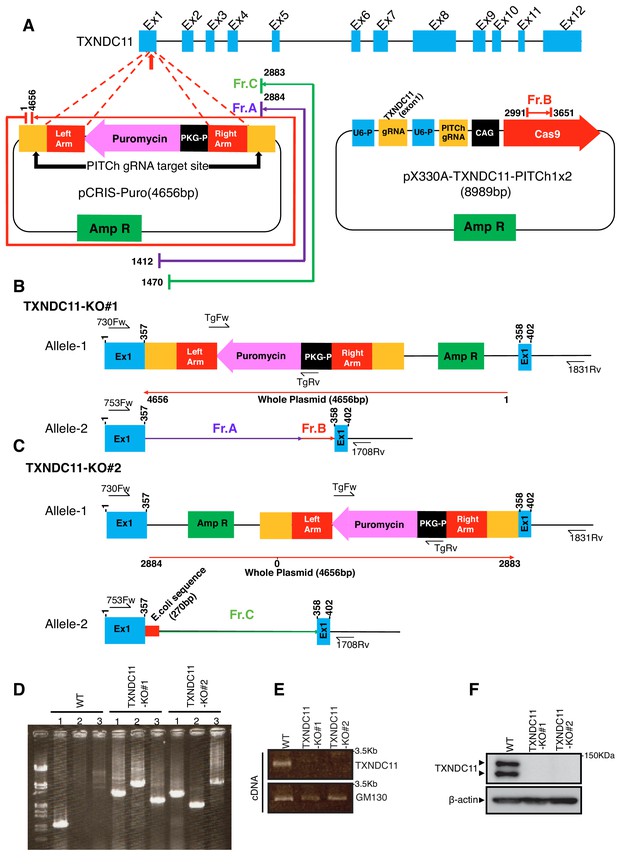
Construction of TXNDC11-KO cells.
(A) Strategy of the CRISPR/Cas9-based PITCh method to target exon 1 of the TXNDC11 gene is shown. (B) Structures of the TXNDC11 locus in TXNDC11-KO#1 cells are schematically shown. (C) Structures of the TXNDC11 locus in TXNDC11-KO#2 cells are schematically shown. (D) Genomic PCR was carried out to confirm recombination. Primers for lanes 1, 2 and 3 are 753Fw and 1708Rv, TgFw and 1831Rv, and 730Fw and TgRv, respectively. (E) Total RNA was prepared from WT and two TXNDC11-KO cells, and subjected to RT-PCR to amplify cDNA corresponding to full length TXNDC11 and GM130. (F) Cell lysates were prepared from WT and two TXNDC11-KO cells, and analyzed by immunoblotting using anti-TXNDC11 and β-actin antibodies.
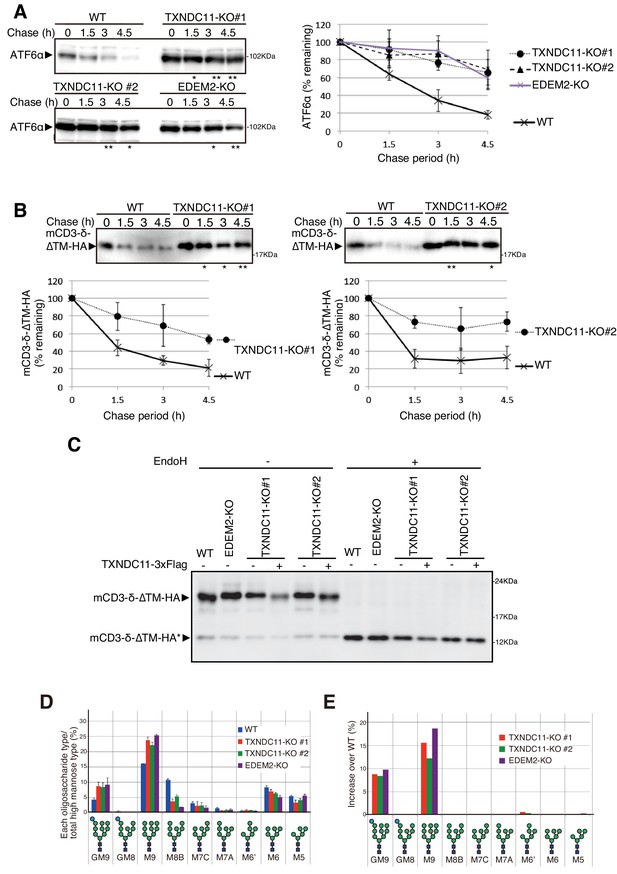
Effect of TXNDC11 knockout on gpERAD.
(A) Cycloheximide chase was conducted to determine the degradation rate of endogenous ATF6α in WT, EDEM2-KO and two TXNDC11-KO cells, and cell lysates were analyzed by immunoblotting using anti-ATF6α antibody (n = 3). Quantified data are shown on the right. (B) Cycloheximide chase was conducted to determine the degradation rate of mCD3-δ-ΔTM-HA in transfected WT and two TXNDC11-KO cells, and cell lysates were analyzed by immunoblotting using anti-HA antibody (n = 3). Quantified data are shown below. (C) WT and EDEM2-KO cells were transfected with the plasmid to express mCD3-δ-ΔTM-HA. The two TXNDC11-KO cells were also transfected with the plasmid to express mCD3-δ-ΔTM-HA together with (+) or without (-) the plasmid to express 3x Flag-tagged TXNDC11(WT). Cell lysates were then prepared, treated with (+) or without (-) EndoH, and analyzed by immunoblotting using anti-HA antibody. (D) Isomer composition of N-glycans prepared from total cellular glycoproteins of WT, TXNDC11-KO#1, TXNDC11-KO#2 and EDEM2-KO cells is shown. This experiment was completed once. (E) Oligosaccharides obtained in (D) whose contents in TXNDC11-KO#1, TXNDC11-KO#2 and EDEM2-KO cells exceeded those in WT cells are displayed with increase over WT (%).
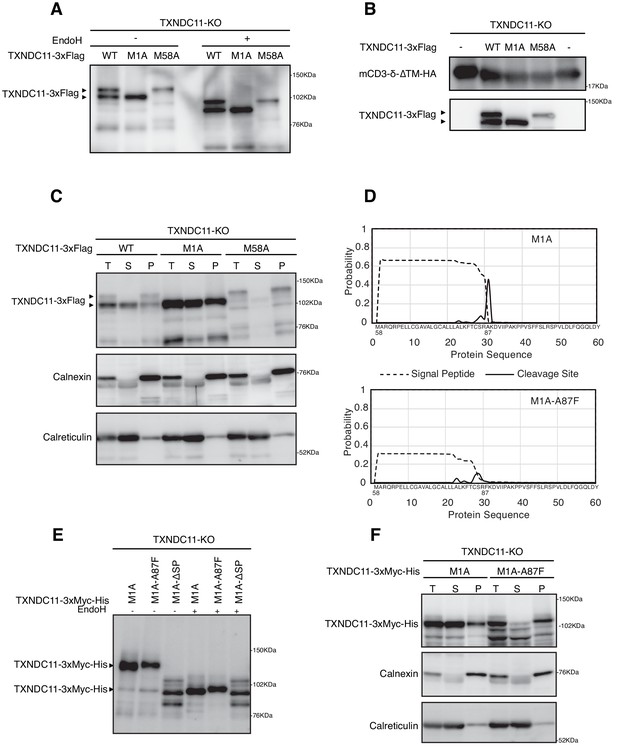
Effect of alternative translation on solubility of TXNDC11.
(A) Cell lysates were prepared from TXNDC11-KO cells expressing WT, M1A or M58A mutant of 3x Flag-tagged TXNDC11 by transfection, treated with (+) or without (-) EndoH, and analyzed by immunoblotting using anti-Flag antibody. (B) Cell lysates were prepared from TXNDC11-KO cells expressing mCD3-δ-ΔTM-HA together with WT, M1A, or M58A mutant of 3x Flag-tagged TXNDC11 by transfection, and analyzed by immunoblotting using anti-TXNDC11 and anti-HA antibodies. (C) TXNDC11-KO cells expressing WT, M1A or M58A mutant of 3x Flag-tagged TXNDC11 by transfection were subjected to repeated freezing and thawing, and then centrifuged as described in Materials and methods. An aliquot of total membrane fraction (T) and resulting supernatant (S) and pellet (P) were analyzed by immunoblotting using anti-TXNDC11, anti-calnexin and anti-calreticulin antibodies. (D) SignalP-5.0-mediated prediction of the probability for functionality of the TMD as a signal peptide (broken line) and the probability for its cleavage by signal peptidase (solid line) in M1A (upper) and M1A-A87F (lower). (E) Cell lysates were prepared from TXNDC11-KO cells expressing M1A, M1A-A87F, or M1A-ΔSP mutant of 3x Myc-His-tagged TXNDC11 by transfection, treated with (+) or without (-) EndoH, and analyzed by immunoblotting using anti-TXNDC11 antibody. (F) TXNDC11-KO cells expressing M1A or M1A-A87F mutant of 3x Myc-His-tagged TXNDC11 by transfection were analyzed as in (C).
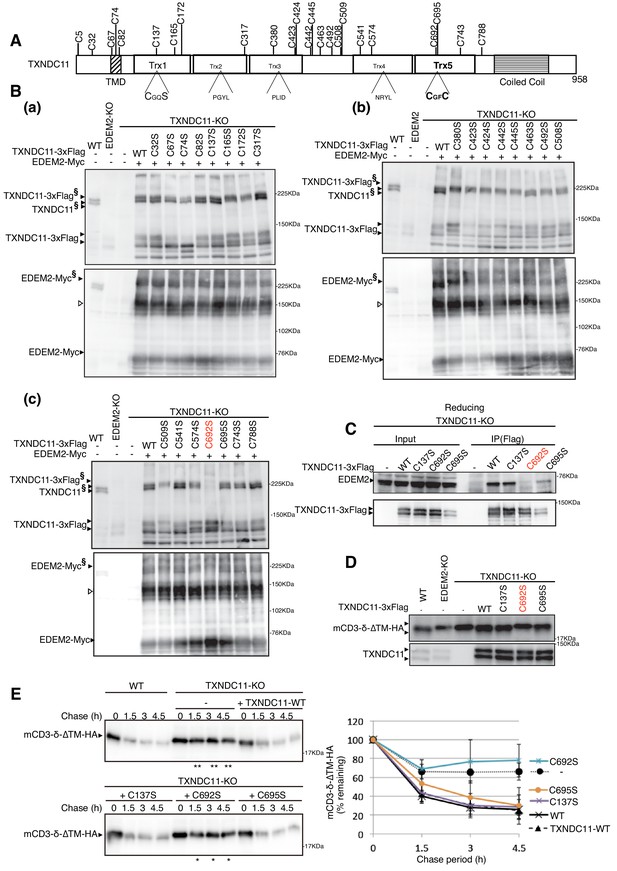
Effect of mutation of various cysteine residues of TXNDC11 on its disulfide-bonding to EDEM2 and on gpERAD.
(A) Structure of human TXNDC11 is schematically shown with cysteine residues (C) highlighted together with their positions. (B) Cell lysates were prepared from WT, EDEM2-KO, and TXNDC11-KO cells expressing WT or one of various cysteine mutants of 3x Flag-tagged TXNDC11 together with Myc-tagged EDEM2 by transfection, subjected to SDS-PAGE under non-reducing conditions, and analyzed by immunoblotting using anti-TXNDC11 and anti-EDEM2 antibodies. (C) Cell lysates were prepared from TXNDC11-KO cells expressing WT or one of the three cysteine mutants of 3x Flag-tagged TXNDC11, and subjected to immunoprecipitation using anti-Flag antibody. An aliquot of cell lysates (Input) and the immunoprecipitates {IP(Flag)} were subjected to SDS-PAGE under reducing conditions, and analyzed by immunoblotting using anti-EDEM2 and anti-TXNDC11 antibodies. (D) Cell lysates were prepared from WT, EDEM2-KO, and TXNDC11-KO cells expressing WT or one of the three cysteine mutants of 3x Flag-tagged TXNDC11 together with mCD3-δ-ΔTM-HA by transfection, and analyzed by immunoblotting using anti-HA and anti-TXNDC11 antibodies. (E) Cycloheximide chase was conducted to determine the degradation rate of mCD3-δ-ΔTM-HA in WT and TXNDC11-KO cells expressing WT or one of the three cysteine mutants of 3x Flag-tagged TXNDC11 by transfection, and cell lysates were analyzed by immunoblotting using anti-HA antibody (n = 3). Quantified data are shown on the right.
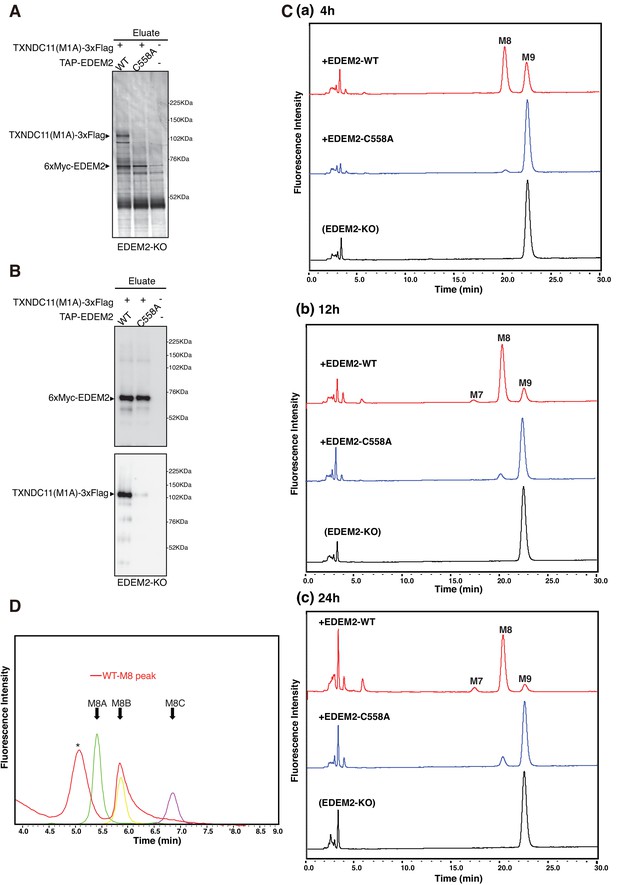
Mannosidase activity of the EDEM2-TXNDC11 complex.
(A) EDEM2-KO cells were untransfected (-) or transfected (+) with the indicated plasmids, and subjected to purification using IgG Sepharose beads. Samples eluted from the beads by digestion with TEV protease were subjected to reducing SDS-PAGE and silver-staining. (B) The amounts of 6xMyc-EDEM2 and TXNDC11(M1A)-3xFlag in samples for in vitro assay were checked by immunoblotting using anti-EDEM2 and anti-TXNDC11 antibodies. (C) PA-M9 was incubated with samples in (B) for 4 hr (a), 12 hr (b) and 24 hr (c) as indicated, and then analyzed by HPLC (amide column) for mannose contents. This experiment was completed once. (D) The M8 peak obtained in (C) after incubation with WT EDEM2 was analyzed by HPLC (ODS column) for isomer identification. Green, yellow, and purple peak indicate the elution position of M8A, M8B, and M8C, respectively. The asterisk observed during analysis of the M8 peak denotes a fluorescent peak unrelated to oligosaccharides. This experiment was completed once.
Tables
| Reagent type or resources | Designation | Source or reference | Identifier | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | colorectal carcinoma | ATCC | HCT116 | Parental HCT116 cell line have been authenticated and all cell lines have been tested negative for mycoplasma. |
| Recombinant DNA reagent | p3xFlag-CMV-14 | Sigma-Aldrich | ||
| Recombinant DNA reagent | pcDNA3.1-MycHis | ThermoFisher | ||
| Antibody | anti-TXNDC11 (Rabbit monoclonal) | Abcam | Cat#: ab188329 | WB (1:500) |
| Antibody | anti-EDEM2 (Rabbit polyclonal) | Novusbio | Cat#: NBP2-37921 | WB (1:500) |
| Antibody | anti-HA (Rabbit polyclonal) | Recenttec | Cat#: R4-TP1411100 | WB (1:1000) |
| Antibody | anti-calnexin (Rabbit polyclonal) | Enzo Life Sciences | Cat#: ADI-SPA-865 | WB (1:1000) |
| Antibody | anti-PDI (Rabbit polyclonal) | Enzo Life Sciences | Cat#: ADI-SPA-890 | WB (1:1000) |
| Antibody | anti-ERp72 (Rabbit polyclonal) | Enzo Life Sciences | Cat#: ADI-SPA-720 | WB (1:1000) |
| Antibody | anti-calreticulin (Rabbit polyclonal) | Enzo Life Sciences | Cat#: ADI-SPA-600 | WB (1:1000) |
| Antibody | anti-Flag (Mouse monoclonal) | Sigma | Cat#: F3165 | WB (1:1000) IP (2.5 μl) |
| Antibody | anti-β-actin (Mouse monoclonal) | Wako | Cat#: 017–24573 | WB (1:2000) |
| Antibody | Anti-human ATF6α (Rabbit polyclonal) | Haze et al., 1999 | WB (1:1000) |
Additional files
-
Supplementary file 1
Table of primers.
- https://cdn.elifesciences.org/articles/53455/elife-53455-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/53455/elife-53455-transrepform-v2.pdf