Conservation and divergence of related neuronal lineages in the Drosophila central brain
Figures
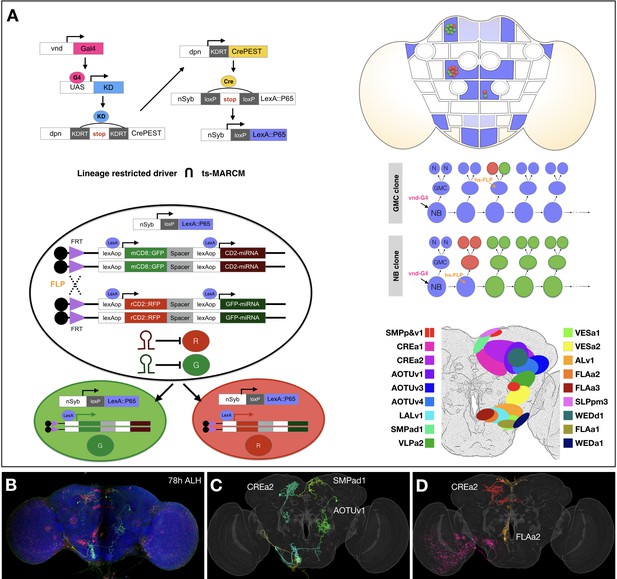
Concurrent mapping of Vnd neuronal lineages with twin-spot MARCM.
(A) Schematic illustration of twin-spot MARCM with vnd-specific lineage-restricted driver. Top left: LexA driver is restricted to Vnd+ lineages via a multi-step cascade. vnd-GAL4 (pink) reconstitutes dpn-CrePEST (yellow), which in turn reconstitutes nSyp-LexA::P65 (blue). LexA::P65 is only expressed in neurons produced by Vnd+ NBs. Bottom left: mitotic recombination leads to differential labeling of paired sister cells by twin-spot MARCM. Top right: Twin-spot MARCM clones (red/green cells) are stochastically introduced only in LexA::P65 expressing lineages (blue intensity indicates frequency of nSyb-LexA::P65 reconstitution). Middle right: mitotic recombination (hs-FLP) in a GMC elicits paired single-cell clones, whereas recombination in a NB leads to GMC offspring (red) paired with the remainder of the lineage (green). Bottom right: approximate distribution of Vnd neuronal cell bodies in a standard fly brain template. (B–D) Example mapping of multiple twin-spot MARCM clones. A representative nc82-counterstained (blue) adult fly brain (B) carrying multiple twin-spot clones (green/red), induced at 78 hr ALH. The green neurons (C) and red neurons (D) were segmented out and then warped onto a standard adult fly brain. Lineage origin is annotated for neurons with cell bodies in the central brain.
-
Figure 1—source data 1
Deconstructing 18 Vnd lineages into serially derived neuron types.
Serially derived distinguishable twin-spot clones present in the same fly brain template (grey) for each of the 18 mapped Vnd lineages (shown in alphabetical order from ALv1 [A] to WEDd1 [R]). Note presence of either only one viable neuron or two sister neurons in GMC-derived twin-spot clones. Non: green; Noff: magenta. Within each hemilineage, we cluster neuron types (see text for neuron type nomenclature) into morphological groups named after main common neuropil targets (indicated on bottom right corner). The immediately following hemilineage-based heatmaps show NBLAST scores of pairwise neuron-type comparison, sorted based on the birth order. Other accompanying hemilineage-based heatmaps show orderly arrangement of neuron types or morphological groups on the Y axis based on their sequential recovery following clone induction at serial time points along the X axis (see Materials and methods for birth order analysis). For the type-level heatmaps, blue to red color represents the actual single-cell clone numbers (max = 10) recovered from induction at given time points. For the group-level heatmaps, the sample distribution was normalized to one for each separate production window.
- https://cdn.elifesciences.org/articles/53518/elife-53518-fig1-data1-v2.pdf
-
Figure 1—source data 2
Reconstituting full-size NB clones with identified neuron types.
(A–R) Representative twin-spot NB clones (GMC side: green, NB side: red) induced at the beginning of larval neurogenesis, shown in the standard fly brain template (grey). (A’–R’) For each of the 18 mapped Vnd lineages, we merged all morphologically distinguishable neuron types (green) for close comparison with the corresponding twin-spot NB clone. The twin spots of each twin-spot NB clone were pseudo-colored the same to derive the whole full-size pattern (red). To account for the entire pattern (red) with a minimal number of single neurons (green), we merged one neuron per type or one neuron per subtype for types with obvious variations. Note extensive overlap between the green and red patterns, except in [L’] where an unrelated SEZ single-cell clone existed coincidently with the VESa2 twin-spot NB clone.
- https://cdn.elifesciences.org/articles/53518/elife-53518-fig1-data2-v2.pdf
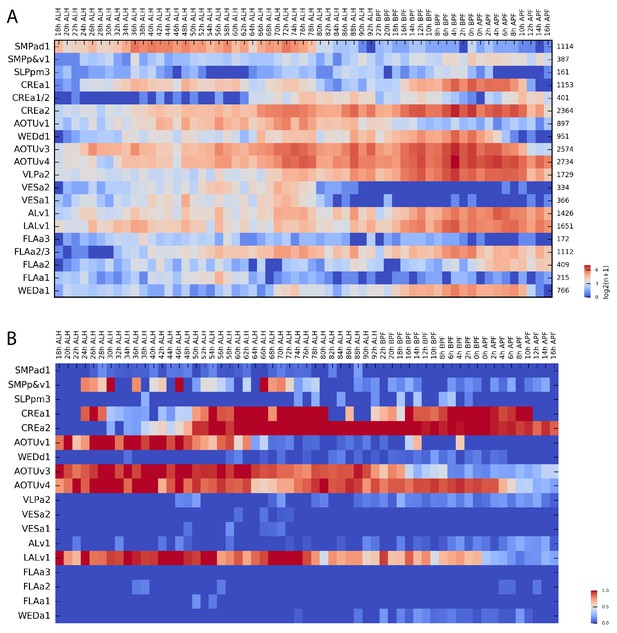
Distribution of ~21 k twin-spot clones among 18 Vnd lineages across 59 two-hour windows.
(A) A log2-scale heatmap to show the twin-spot clone numbers for each of the 18 Vnd lineages (Y axis) across 59 time points (X axis). The total clone number for each lineage is shown to the right. ALH: after larval hatching; BPF: before pupa formation; APF: after pupa formation. (B) Heatmap of the percentage of twin-spot clones, derived from specific Vnd lineages (Y axis) at specific time points (X axis), that exist as one green and one red neuron or as two viable neurons accompanied by a multi-cellular NB clone. The co-existent two neurons in such twin-spot clones were mostly made by a common GMC, although could possibly arise from separate precursors due to independent mitotic recombination events.
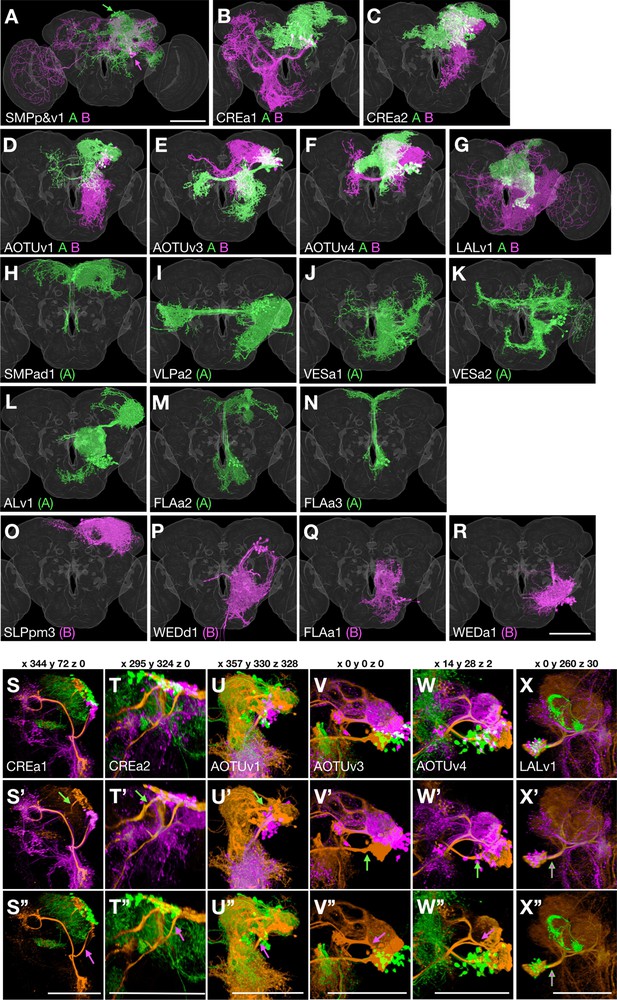
Sister hemilineage distinctions.
(A–R) Hemilineage morphology is revealed by pseudo-coloring Non/A neurons green and Noff/B neurons magenta based on the Notch state (judged from notch RNAi phenotypes). For the seven Vnd lineages composed of dual hemilineages (A–G), representative single-cell clones of A or B neurons were assembled to create synthetic NB clones with sister hemilineages in distinct colors. For the 11 unpaired Vnd hemlineages (H–R), the full pattern was shown by merging the first larval-born neuron with its accompanying NB clone and then pseudo-coloring the merged clone according to A/B fate. (S–X’’) Full-size NB clones (orange) overlaid with both A (green) and B (magenta) hemilineage masks (S–X) or either B (S’–X’) or A (S’’–X’’) hemilineage mask, to examine the hemilineage-structure correspondence in dual-hemilineage NB clones (except SMPp&v1 with widely separate A/B cell body clusters). Composite confocal images viewed from various angles (x, y, and z coordinates indicated above) demonstrate distinct hemilineage-specific neurite fascicles (indicated with green/magenta arrows), extending out of fully or partially separate A and B cell body clusters, in six of the seven Vnd lineages (S–W’’). Only the LALv1 lineage has a mixed cell body region that extends a single neurite bundle projecting posteriorly before dividing into multiple fascicles (X–X’’).

Notch-dependent sister hemilineage projections in seven Vnd lineages.
(A) Schematic illustration showing how a conditional GAL4 driver can be irreversibly activated in random type I NBs (through a cascade of pattered recombinase activities) at the timing of interest (controlled by heat shock). The reconstituted ubiquitous GAL4 driver can then activate notch RNAi in addition to the reporter continuously in a lineage-restricted manner. (B–H) Representation of distinct sister hemilineages (Non: green, Noff: red) reconstituted through merging the identified neuron types of distinct Notch states separately in the standard fly brain template (grey). The Notch states were determined based on phenotypes shown in [B’] to [H’]. (B’–H’) Representative notch-RNAi NB clones (green) in nc82-counterstained adult fly brains (blue). Note persistence of only a half pattern (corresponding to the red Noff hemilineage shown in [B] to [H]) in the labeled notch-RNAi NB clones. Multiple notch-RNAi NB clones could coexist; and some bilaterally symmetrical neurons with midline crossing existed as background clones in almost all samples.
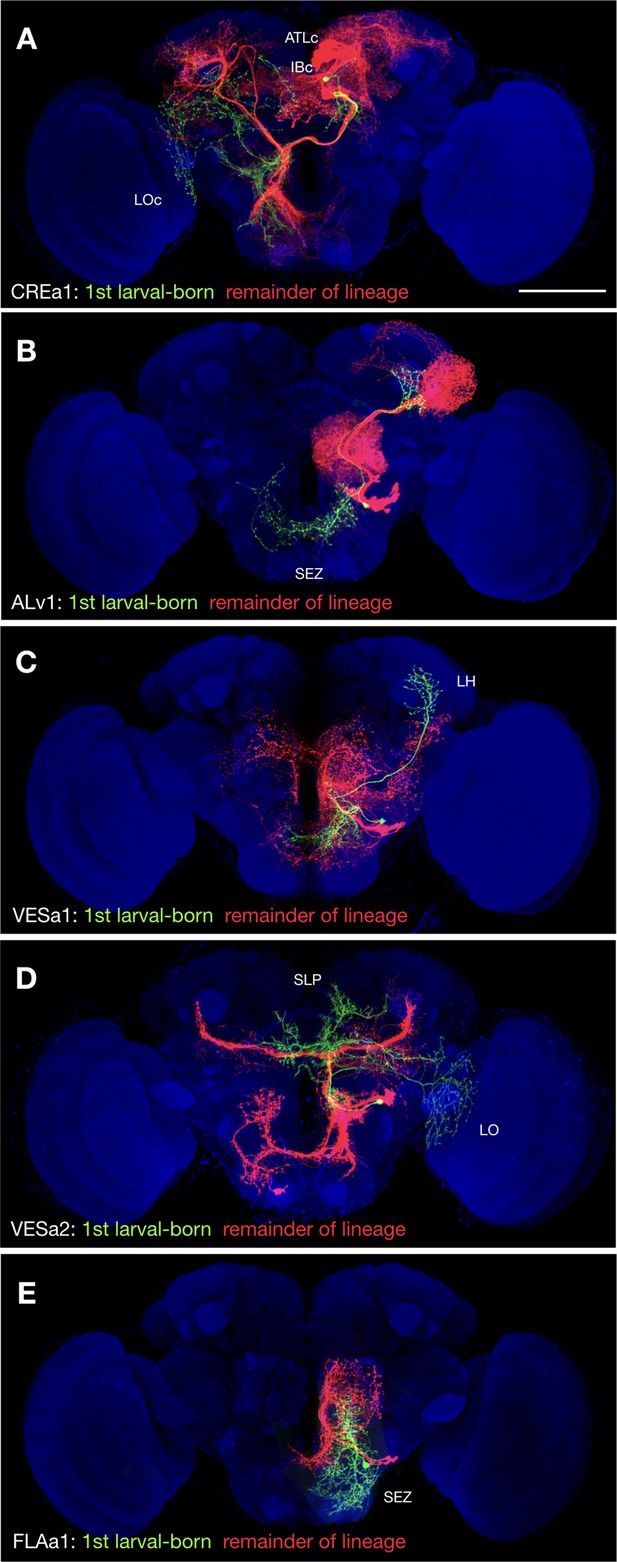
Uniquely exuberant first larval-born neurons.
(A-E) Representative twin-spot NB clones in nc82-counterstained (blue) adult fly brains. Each twin-spot clone consists of one to two (only CREa1 [A] has two) first larval-born neurons (green) paired with all subsequently born neurons (red) of the same lineage. Neuropils uniquely innervated by the shown first larval-born neurons are indicated.
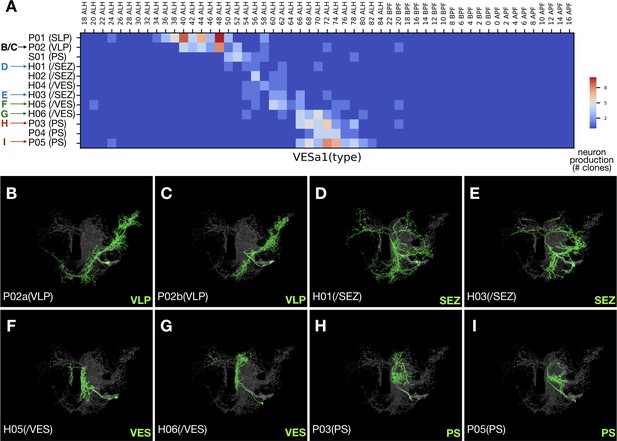
Late-born neurons show simplified morphology.
(A) Heatmap of sample distribution of the annotated VESa1 neurons types (Y-axis) vs. timing of clone induction (X-axis). Blue to red color represents the actual single-cell clone numbers (max = 10) recovered from induction at given time points. Neuron types were manually sorted to reflect their ordered production. Colored arrows indicate distinct morphological groups shown in [B-I]. See text for neuron type nomenclature. (B–I) Serially derived single neuron types (green) shown in the context of all recovered VESa1 neuron types (grey). Note reduction in neurite elaboration along birth order.
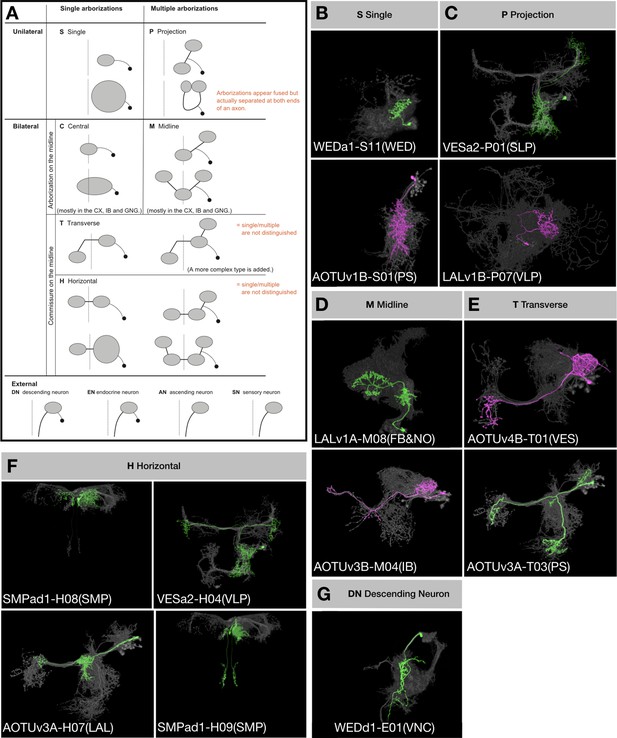
Topological classification of single neurons.
(A) Schematic illustration of neuron morphology classification—multiple examples are given for each class within the brain. Dashed lines indicate the brain midline. Black dots represent cell bodies and grey ovals represent neurite elaborations. (B–G) Representative single neurons of each morphological classification (green: A/Non, magenta: B/Noff) shown in the context of all recovered neurons (grey) of the same hemilineages (indicated in the beginning of neuron type names). Multiple neuron types corresponding to each illustrated class are shown, with the exception of Central and External neurons. In the 18 Vnd lineages, no C-topology neurons were found and only the DN type of external neurons was found.
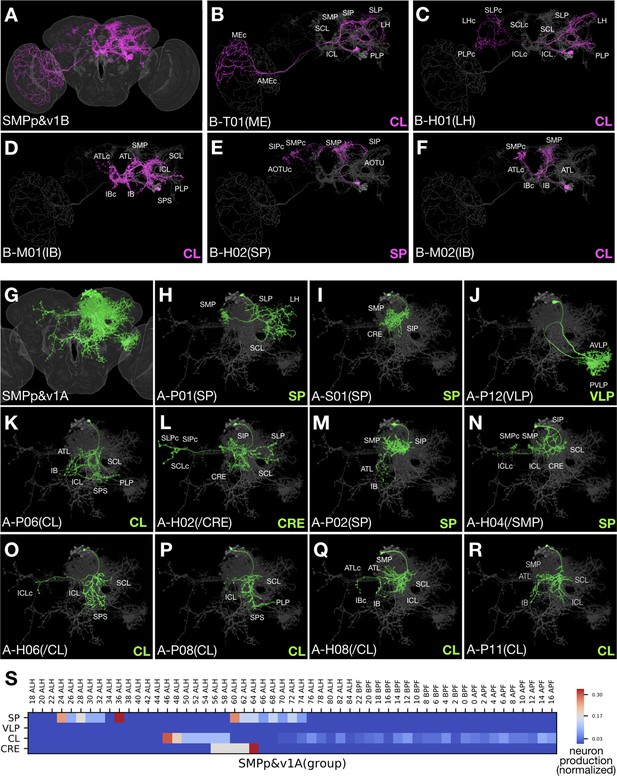
Clonally related neurons show diverse topology.
(A) All identified SMPp&v1B (Noff) neuron types merged onto the standard fly brain template. (B–F) Serially derived single neuron types (magenta) shown in the context of all identified SMPp&v1B neuron types merged together (grey). Note presence of T-, M-, H-topolgy neurons with comparable ipsilateral elaborations. (G) All identified SMPp&v1A (Non) neuron types merged onto the standard fly brain template. (H–R) Serially derived single neuron types (green) shown in the context of all identified SMPp&v1A neuron types merged together (grey). Note presence of P-, S-, H-topology neurons in the same hemilineage, and similarities among certain S-, P-, and H-topology neurons (I,M,N). (S) Heatmap of sample distribution of the morphological groups of SMPp&v1A neurons (Y-axis) over time of clone induction (X-axis). The sample distribution was normalized to one for each separate production window. Note presence of two recovery windows for both SP and CL groups, and no detectable recovery window (yielding six or more samples from an uninterrupted interval spanning at least two time points) for the minor VLP group.
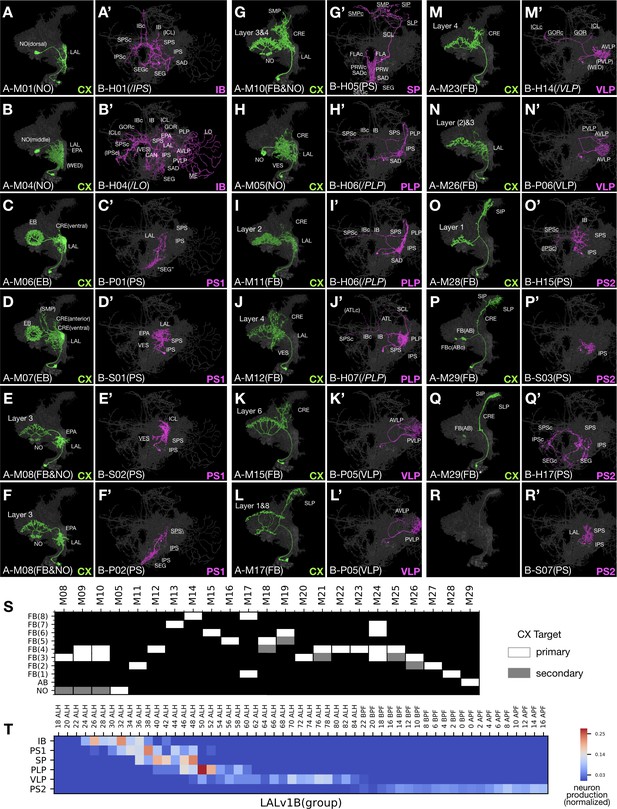
Uniform CX neurons pair with diverse groups of non-CX neurons in LALv1.
(A–R’) Representative pairs of LALv1A (Non green) and LALv1B (Noff magenta) neuron types arranged in birth-order, shown in the context of all identified LALv1A neuron types merged together (grey). In the LALv1A hemilineage, there is serial innervation of various CX neuropils in the sequence of NO, EB, FB/NO, NO, then FB (with AB-targeting neurons at the end). Note the recurrent targeting of FB layers ([J] and [M]). The LALv1B hemilineage produces six morphological groups in oder, targeting IB, PS, SP, PLP, VLP, and PS again. Note the presence of P-topology neurons in the PS1 group (C’,F’) versus H-topology neurons in the PS2 group (O’,Q’), as well as similar S-topology neurons in both PS1 and PS2 groups (D’,R’). (S) Graphical representation of LALv1A hemilineage CX targets throughput the FB targeting window. Neuron types are arranged in birth-order. (T) Birth-order heatmap of LALv1B morphological groups, demonstrating sequential production of distinct morphological groups.
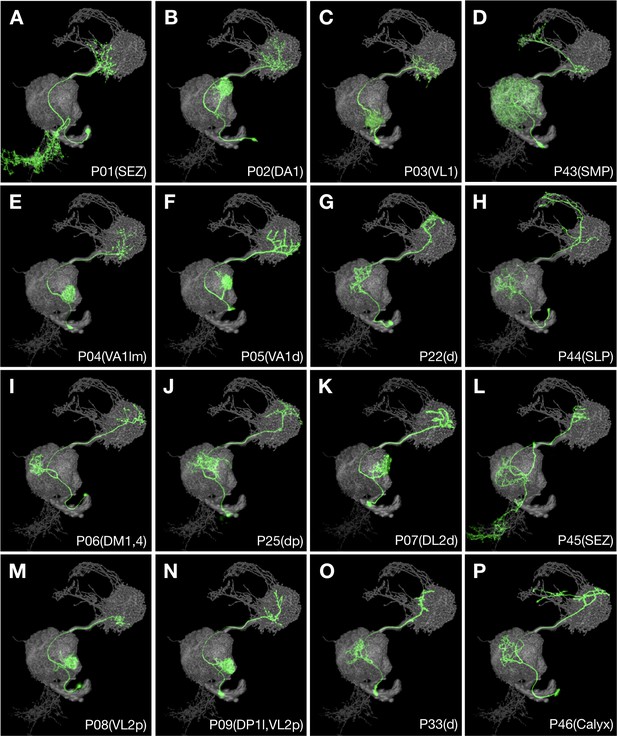
Multiple recurrent features in the ALv1 hemilineage.
Representative ALv1 neuron types (Non green) arranged in birth-order, shown in the context of all identified ALv1 neuron types merged together (grey). Note several recurrent features, including mono-glomerular AL innervation (B, C, E, F, I, K, M and N), extension beyond LH (D, H and P), and SEZ targeting (A, L).
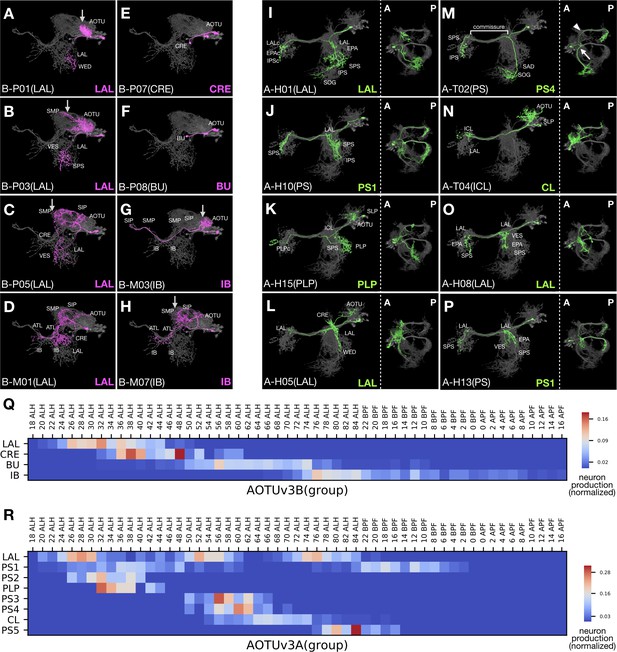
Progressive innervation in AOTUv3B and recurrent innervation in AOTUv3A.
(A–H) Representative AOTUv3B neuron types (Noff magenta) arranged in birth-order, shown in the context of all identified AOTUv3B neuron types merged together (grey). Note one M-topology neuron type (D) produced much earlier than many other M-topology types (e.g. [G] and [H]). Both LAL and IB groups show progressive AOTU-to-SP proximal elaborations (arrows). (I–P) Representative AOTUv3A neuron types (Non green) arranged in birth-order, shown in the context of all identified AOTUv3A neuron types merged together (grey). Note multiple recovery windows for LAL (I,L,O) and PS1 (J,P) groups. Tilted views at a lower magnification are showed to the right; anterior (A) to the left, posterior (P) to the right. Note variable length and direction of extensions at the entry and exit of the shared commissure, and recurrence of similar extension patterns (e.g. [J] and [P]). The commissure entry (arrow) and exit (arrowhead) points are indicated in [M]. (Q–R) Birth-order heatmaps of AOTUv3B (Q) and AOTUv3A (R) morphological groups. Note that the LAL group of neurons appear in a single window in the AOTUv3B hemilineage, but multiple windows in the AOTUv3A hemilineage.
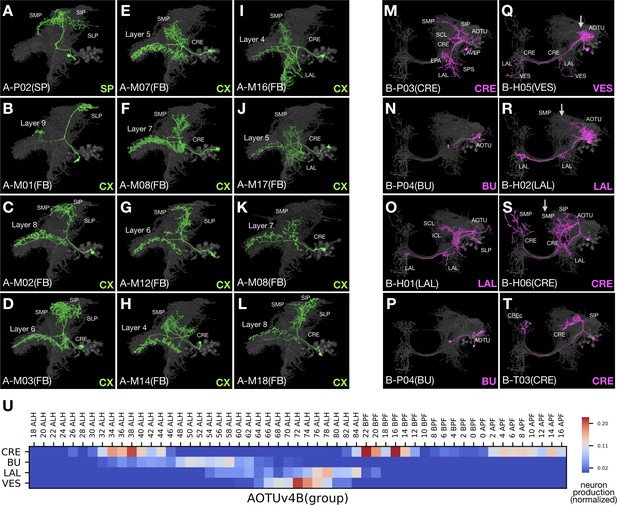
AOTUv4A resembles LALv1A and AOTUv4B resembles AOTUv3B.
(A–L) Representative AOTUv4A neuron types (Non green) arranged in birth-order, shown in the context of all identified AOTUv4A neuron types merged together (grey). Note the early transition from non-CX neurons (A) to FB neurons (B–L). Note also that the first FB neuron type innervates layer 9 (B), and FB layers 4–8 are recurrently innervated (C–L). (M–T) Representative AOTUv4B neuron types (Noff magenta) arranged in birth-order, shown in the context of all identified AOTUv4B neuron types merged together (grey). Note one H-topology LAL-group neuron type (O) made in the middle of P-topology BU-group types (N,P) and much earlier than many other H-topology types (Q–S). Both the beginning P-topology CRE-group neurons (see Figure 1—source data 1D, magenta in [1] to [4]) and the later multi-group H-topology neurons show progressive AOTU-to-SP proximal elaborations (Q–S), as in the LAL and IB groups of AOTUv3B (arrows). (U) Birth-order heatmap of AOTUv4B morphological groups.
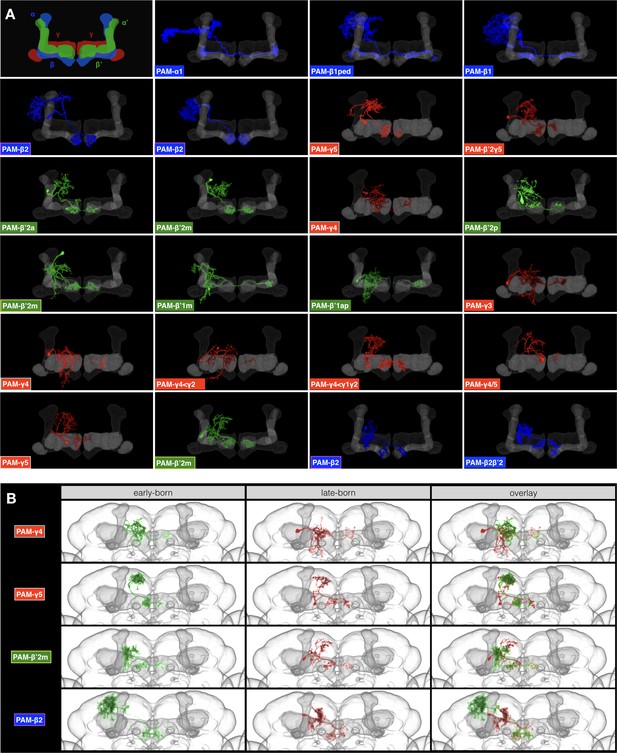
Patterned MB innervation by serially derived CREa1A/CREa2A PAM neurons.
(A) PAM neuronal elaborations (pseudo-colored based on lobe identity) within the MB medial lobes (grey) showing progressive innervation of neighboring MB lobe zones. Note recurrence of four targeting patterns: beta2, gamma5, beta’2m, gamma4. (B) Representative PAM neurons (early-born: green and late-born: red) with the same target zone merged onto an adult brain template. Note differential proximal elaborations in early- vs. late-born neurons targeting the same zone of MB medial lobes.
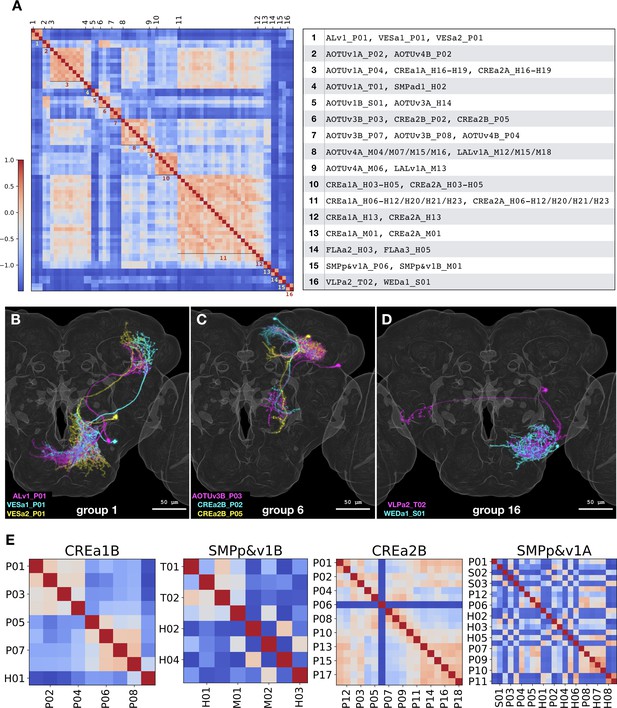
NBLAST analysis of inter- vs intra-hemilineage relatedness.
(A) Sixteen groups of morphologically related neuron types were recovered across independent hemilineages, based on the adjacency matrix derived from NBLAST scores larger than 0.3. Shown side by side are the heatmap of NBLAST scores and the list of Vnd neuron types for the 16 non-overlapping similarity groups. Note that the extensive CREa1A/CREa2A and AOTUv4A/LALv1A inter-hemilineage analogies account for all four (#3/8/10/11) large groups with six or more members. (B–D) Representative neurons of the related neuron types for the similarity group #1 (B), #6 (C), and #16 (D). Note extensive overlap in both proximal and distal (B and C) or just proximal neurite elaborations (D), despite distinct hemilineage-characteristic primary projections. (E) Heatmaps of intra-hemilineage NBLAST scores, sorted based on the birth-order of neuron types (indicated with alternating x-tick and y-tick labels). Note distinct temporal patterns of relatedness in diverse hemilineages. The CREa1B hemilineage displays progressive changes, and the SMPp&v1B hemilineage exhibits both progressive and cyclic changes. By contrast, the CREa2B and SMPp&v1A hemilineages yield more neuron types that appear in various cyclic manners. Please find the complete set of hemilineage NBLAST heatmaps in Figure 1—source data 1.
-
Figure 12—source data 1
All-to-all NBLAST score matrix of 464 annotated Vnd neuron types.
- https://cdn.elifesciences.org/articles/53518/elife-53518-fig12-data1-v2.csv
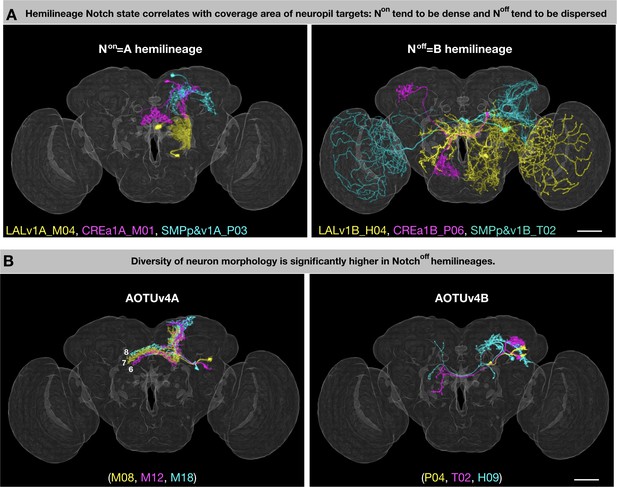
General patterns of lineage-guided morphological diversification.
(A) Notch state correlates with coverage area of neuropil targets. Representative pairs of Non/A (i) and Noff/B (ii) neurons from the LALv1 (yellow), CREa1 (magenta), and SMPp&v1 (cyan) lineages. (B) Relatively uniform Non/A neurons pair with grossly diverse Noff/B neurons. Representative pairs of Non/A (i) and Noff/B (ii) neurons, color-coded based on birth order (yellow, magenta, cyan) in the AOTUv4 lineage.
-
Figure 13—source data 1
Neuron skeleton lengths of 464 annotated Vnd neuron types.
- https://cdn.elifesciences.org/articles/53518/elife-53518-fig13-data1-v2.xlsx
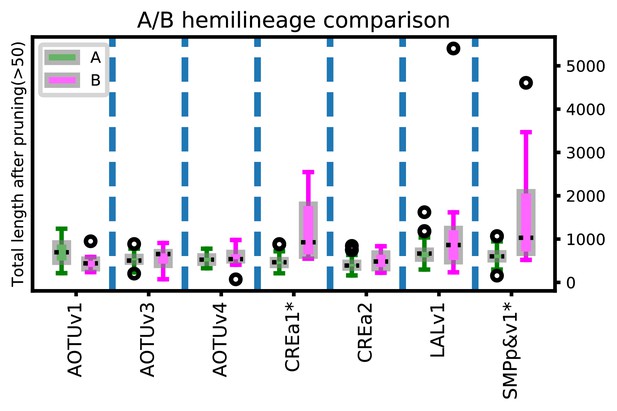
Total length of main trajectory in Non/A vs Noff/B neurons.
Distribution of the main trajectory lengths of the A versus B neuron types identified from the seven paired Vnd hemilineages, shown in standard boxplot with interquartile-range boxes, minimum and maximum whiskers, and circles for individual outliners. * indicates p-value<0.01 for two-tailed T test.
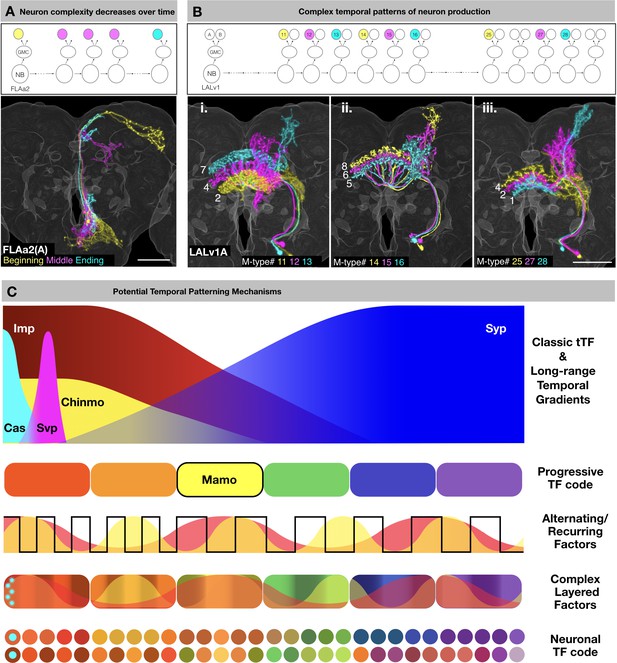
Common themes of neuronal lineage temporal patterning.
(A) Neuron complexity decreases over time. Top: Schematic of FLAa2 postembryonic lineage development showing beginning neuron (yellow), middle neurons (magenta) and ending neuron (cyan). Bottom: Neurons of the FLAa2(A) lineage show gradual restriction in neurite elaboration. (B) Complex temporal patterns of neuron production. Top: Schematic of LALv1 lineage development. Select M-type FB innervating neurons are color-coded based on birth order (yellow, magenta, cyan) in three series. Bottom: Serially derived LALv1A neurons with progressive changes in neurite elaboration patterns. Neurons in each panel are color-coded based on birth-order in the same sequence of yellow, magenta and cyan. Note that the ventral-to-dorsal innervation of specific FB layers (i) is followed by reverse dorsal-to-ventral innervation (ii and iii). Also note progressive changes in non-FB elaborations in each round of serial layer-specific targeting. (C) Schematic illustration of potential temporal patterning mechanisms, including long-range temporal patterning orchestrated by graded Imp/Syp expressions and asynchronous cyclic changes elicited by unknown alternating/recurring factors. Cas and Svp, as classic temporal transcription factors (tTF), are expressed at the beginning of post-embryonic neurogenesis. Svp triggers the opposite Imp/Syp gradients that in turn shape the descending Chinmo protein gradient. The hierarchical Imp/Syp and Chinmo gradients (plus other undiscovered gradients) could subdivide a protracted neuronal lineage into multiple temporal windows with a progressive TF code (e.g. Mamo). Cas expression (cyan), in the beginning of the lineage may cause the initial exuberant neuron types. In parallel, there could exist various alternating/recurring factors which diversify each temporal window further in a combinatorial manner. Together the tTFs, progressive TF code and alternating/recurring factors lead to a complex layering of factors which are interpreted differentially by sister neurons (depending on Notch state) to reveal the neuronal TF code.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | anti-GFP (Rabbit polyclonal) | Thermo Fisher Scientific | Cat # A-11122; RRID:AB_221569 | (1:1000) |
| Antibody | anti-mCD8 (Rat monoclonal) | Thermo Fisher Scientific | Cat # MCD0800; RRID:AB_10392843 | (1:100) |
| Antibody | anti-RFP (Rabbit polyclonal) | Clontech | Cat # 632496 | (1:1000) |
| Antibody | anti-nc82 (Mouse monoclonal) | Developmental Studies Hybridoma Bank | nc82; Registry ID:AB_2314866 | (1:100) |
| Antibody | anti-rabbit, Alexa488 (Goat) | Thermo Fisher Scientific | Cat # A-11034; RRID:AB_2576217 | (1:500) |
| Antibody | anti-Rat, Alexa488 (Goat) | Thermo Fisher Scientific | Cat # A-11006; RRID:AB_2534074 | (1:500) |
| Antibody | anti-rabbit, Alexa568 (Goat) | Thermo Fisher Scientific | Cat # A-11036; RRID:AB_10563566 | (1:500) |
| Antibody | anti-mouse, Alexa647 (Donkey) | Jackson ImmunoResearch lab, Inc | Cat # 715-605-151 | (1:500) |
| Chemical compound drug | Paraformadehyde 20% Solution, EM Grade | Electron Microscopy Sciences | Cat # 15713 | |
| Chemical compound drug | Phosphate Buffered Saline 10X,Molecular Biology Grade | Thermo Fisher Scientific | Cat # 46–013 CM | |
| Chemical compound drug | Triton X-100 | Sigma-Aldrich | Cat # 329830772 | |
| Chemical compound drug | SlowFadeTM Gold antifade Mountant | Thermo Fisher Scientific | Cat # S36936 | |
| Chemical compound drug | Ethyl alcohol, pure | Sigma-Aldrich | Cat # 459844 | |
| Chemical compound drug | Xylenes | Thermo Fisher Scientific | Cat # X5-500 | |
| Chemical compound drug | DPX mountant | Electron Microscopy Sciences | Cat # 13512 | |
| Genetic reagent (D. melanogaster) | UAS-Notch-RNAi | BloomingtonDrosophilastock center | BDSC:33611; FBti0140084; RRID:BDSC_33611 | FlyBase symbol:P{TRiP.HMS00001}attP2 |
| Genetic reagent (D. melanogaster) | UAS-mCD8-GFP | Lee and Luo (1999) | ||
| Genetic reagent (D. melanogaster) | hs-ATG > KOT > FLP | Ren et al., 2018 | ||
| Genetic reagent (D. melanogaster) | dpn > FRT-STOP-FRT>Cre::PEST | Awasaki et al. (2014) | ||
| Genetic reagent (D. melanogaster) | lexAop2-rCD2::RFP-insulator-lexAop2-GFP-RNAi, FRT40A | Awasaki et al. (2014) | ||
| Genetic reagent (D. melanogaster) | hs-FLP,dpn > KDRT-stop-KDRT>Cre PEST; lexAop2-mCD8::GFP-insulator-lexAop2-rCD2-RNAi, FRT40A; nSyb > loxP-stop-loxP>LexA::P65,UAS-KD1 | Awasaki et al. (2014) | ||
| Genetic reagent (D. melanogaster) | vnd-T2A-Gal4 | This paper: Materials and methods | Lee T, Janelina Research Campus, HHMI | |
| Genetic reagent (D. melanogaster) | ase-KD1 | This paper: Materials and methods | Lee T, Janelina Research Campus, HHMI | |
| Genetic reagent (D. melanogaster) | act > loxP-STOP-loxP>Gal4 | This paper: Materials and methods | Lee T, Janelina Research Campus, HHMI | |
| Software and Algorithms | Fiji | NIH; Schindelin et al., 2012 | https://fiji.sc | |
| Software and Algorithms | Adobe Photoshop | Adobe Systems, San Jose, CA | https://www.adobe.com | |
| Software and Algorithms | Adobe Illustrator | Adobe Systems, San Jose, CA | https://www.adobe.com | |
| Software and Algorithms | Python | Python Software Foundation | https://www.python.org | |
| Software and Algorithms | Flybase 2.0 | FlyBase Consortium et al., 2019 | http://flybase.org |
Additional files
-
Supplementary file 1
Lists of annotated neuron types/groups and their sample numbers recovered from serial 2-hr windows for each of the 25 Vnd hemilineages.
- https://cdn.elifesciences.org/articles/53518/elife-53518-supp1-v2.xlsx
-
Supplementary file 2
Lists of annotated neuron types and their neuropil innervation patterns for each of the 25 Vnd hemilineages.
- https://cdn.elifesciences.org/articles/53518/elife-53518-supp2-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/53518/elife-53518-transrepform-v2.docx





