HOPS recognizes each SNARE, assembling ternary trans-complexes for rapid fusion upon engagement with the 4th SNARE
Figures
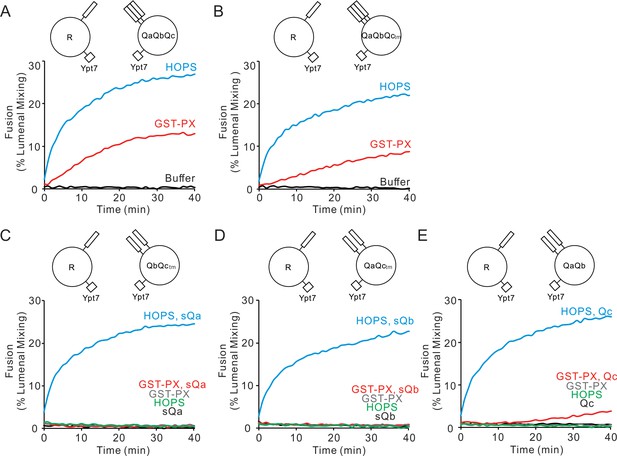
HOPS recruits each Q-SNARE, whereas a simple tether (GST-PX) does not.
(A–E) Fusion reactions had proteoliposomes bearing either R- or Q-SNARE combinations as indicated at 1:16000 SNARE:lipid molar ratio. Fusion was assayed between R and (A) QaQbQc, (B) QaQbQctm, (C) QbQctm, (D) QaQctm, or (E) QaQb proteoliposomes as described in Materials and methods. Fusion reactions had 500 nM GST-PX or 50 nM HOPS as indicated. (A, B) Mixed proteoliposomes were incubated with HOPS (blue), GST-PX (red), or buffer (black). (C–E) HOPS or GST-PX and 4 μM soluble Q-SNAREs (sQ) were added: HOPS and sQ (blue), GST-PX and sQ (red), GST-PX alone (gray), HOPS alone (green), sQ alone (black). All proteoliposomes had Ypt7-tm at a 1:8000 protein:lipid molar ratio. Kinetic curves of content mixing assays in this figure are representative of n ≥ 3 experiments; average and standard deviations of fusion from three independent experiments are in Figure 1—figure supplement 1.
-
Figure 1—source data 1
Source data file (Excel) for Figure 1A,B,C,D and E.
- https://cdn.elifesciences.org/articles/53559/elife-53559-fig1-data1-v2.xlsx
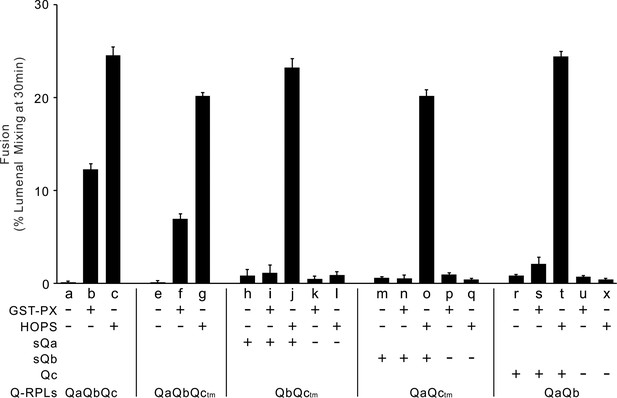
HOPS, but not GST-PX, supports fusion of R- and 2Q-SNARE proteoliposomes mixed with the third soluble Q-SNARE.
Fusion assays were conducted as described in Figure 1, with R and QaQbQc, QaQbQctm, QbQctm, QaQctm or QaQb proteoliposomes. All proteoliposomes had SNAREs at a 1:16000 and Ypt7-tm at a 1:8000 protein: lipid molar ratio. Average and standard deviations of fusion 30 min after addition of HOPS, for triplicate assays relative to total mixed contents.
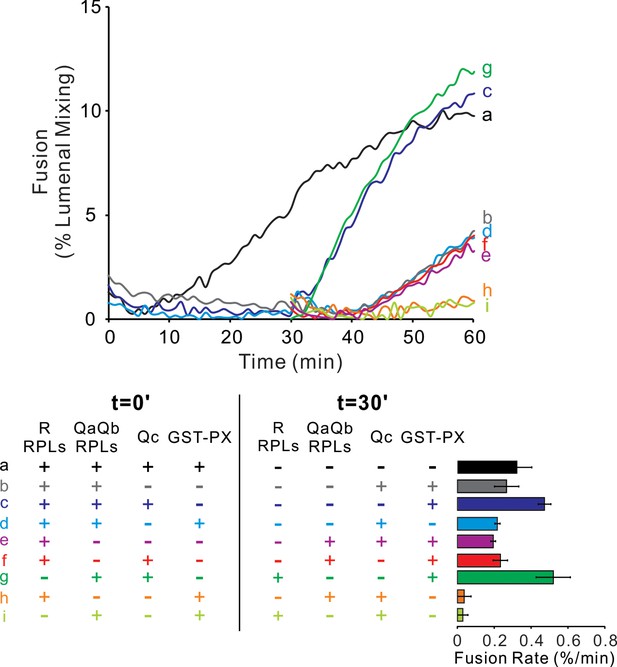
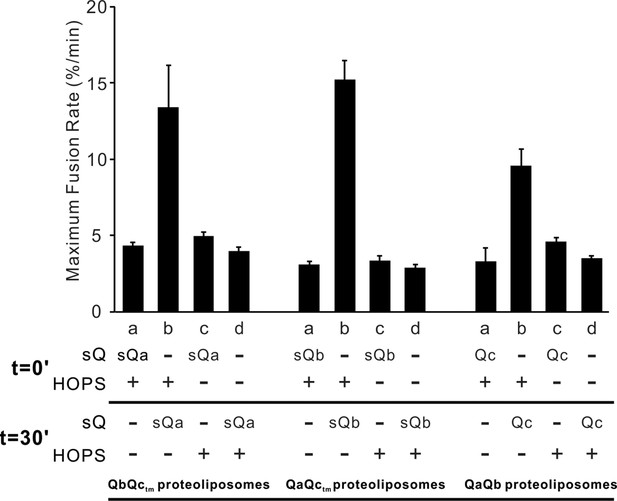
The Qc-SNARE can slowly and stably assemble spontaneously with QaQb-proteoliposomes.
Fusion reactions had proteoliposomes bearing Ypt7 and either the R- or QaQb-SNAREs at 1:16000 SNARE:lipid molar ratio. R and QaQb proteoliposomes were mixed without preincubation (a, b, c and d) or after 30 min preincubation at 27°C (e, f, g, h and i). Fusion incubations received 500 nM GST-PX and/or 4 μM Qc where indicated. The bar graph quantifies the maximal rate of content mixing from three independent experiments. The rate of fusion was determined as the slope of the content mixing reaction after fusion initiation. Kinetic curves of contents mixing assays in this figure are representative of n ≥ 3 experiments.
Fusion inhibition by sR.
Fusion reactions had proteoliposomes bearing either the R or (A) QbQctm, (B) QaQctm, or (C) QaQb SNAREs and Ypt7-tm at 1:16000 SNARE:lipid and 1:8000 Ypt7:lipid molar ratios. Fusion incubations received 50 nM HOPS at t = 30 with 4 μM of the soluble form of the third Q-SNARE at t = 0 (a, b, e) or t = 30 (c, d, f) as indicated. Soluble Nyv1 (sR) was added to 4 μM at t = 0 (a, c) or t = 30 (b, d). Content mixing assays in this figure are representative of n ≥ 3 experiments; means and standard deviations for each experiment are presented.
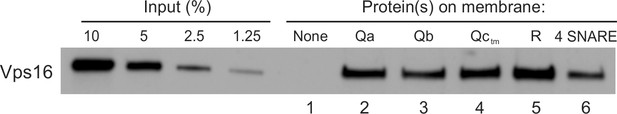
HOPS binds directly to each vacuolar SNARE.
PC liposomes with no integral SNAREs, with each individual integrally-bound SNARE, or with all four wild-type SNAREs were incubated with HOPS at a twofold molar excess to SNAREs and floated. Flotation assays were conducted as described (Orr et al., 2017) with modifications. Liposomes (7.5 µl) were incubated for 1 hr at 30°C in 30 µl reactions (0.5 mM lipid, 500 nM HOPS, 0.2% defatted bovine serum albumin (BSA; Sigma-Aldrich), and 1 mM MgCl2 in RB150). Reactions were gently vortexed with 90 µl of 54% (wt/vol) Histodenz (Sigma-Aldrich) in iso-osmolar RB150/Mg2+ (containing a reduced level (2%) of glycerol) and 80 µl were transferred to 7 × 20 mm polycarbonate tubes (Beckman Coulter, Brea CA), overlaid with 80 µl of 35%, then 80 µl of 30% Histodenz in iso-osmolar RB150+Mg2+ and finally 50 µl of RB150+Mg2+. The remaining portions of the starting incubations were solubilized with 1 µl of 5% (vol/vol) Thesit for determination of lipid recovery. Reactions were centrifuged in a Beckman TLS-55 rotor, 4°C, 55,000 rpm, 30 min. Samples were harvested by pipetting 80 µl from the top of the tube and solubilized with 2 µl of 5% Thesit. Lipid recovery was assayed as described (Orr et al., 2015), measuring either rhodamine fluorescence (excitation, 560 nm; emission, 580 nm; cutoff 570) or NBD fluorescence (excitation, 460 nm; emission, 538 nm; cutoff 515), depending on the composition of the liposomes. Bound protein determination was performed as described (Orr et al., 2015) by immunoblot of its Vps16 subunit with a standard curve of the input. Quantification and statistical analysis of HOPS binding from three independent experiments is in Figure 2—figure supplement 1.
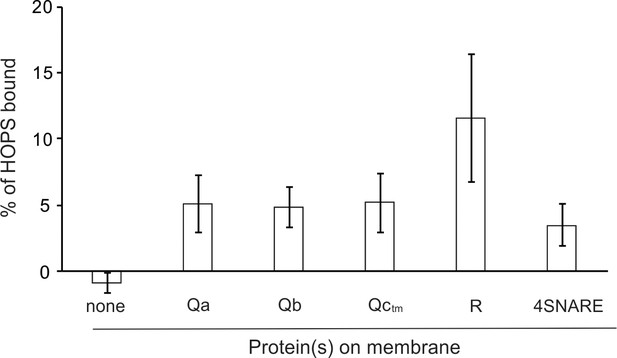
HOPS binds to each SNARE.
Western blots of 3 independent experiments were analyzed with UN-SCAN-IT software (Silk Scientific, Orem UT).
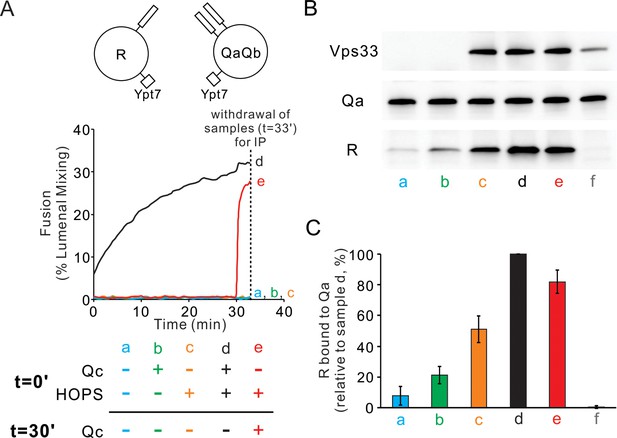
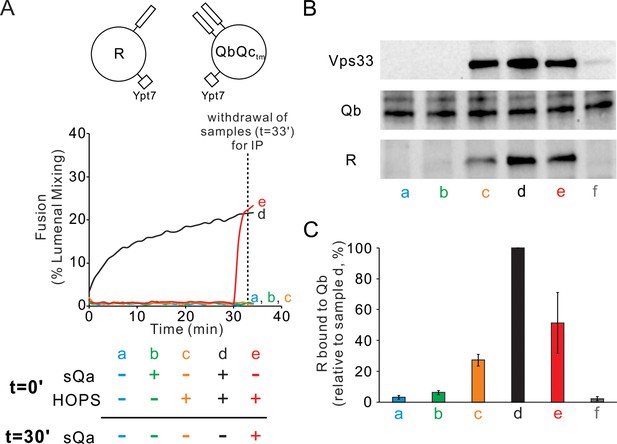
HOPS induces formation of a rapid-fusion intermediate which includes the R- and Qa-SNAREs in trans association with each other and/or the same HOPS molecule.
Proteoliposomes with Ypt7 and R (1:8000 and 1:16,000 molar ratio to lipids, respectively) were mixed with proteoliposomes with Ypt7 and Qa and Qb SNAREs and with 50 nM HOPS and 4 μM Qc where indicated, added either at the start of incubation or after 30 min. (A) Fusion was assayed as lumenal FRET. After 30 min, Qc was added to one sample (e, red). (B) To measure complex formation, the amount of R SNARE that was immunoprecipitated from a detergent extract with anti-Qa antibody was determined. After incubation for 33 min, samples were placed on ice and mixed with five volumes of ice-chilled modified RIPA buffer [20 mM Hepes·NaOH, pH 7.4, 150 mM NaCl, 0.2% (wt/vol) BSA, 1% (vol/vol) Triton X-100,1% (wt/vol) sodium cholate, 0.1% (wt/vol) SDS] containing RIPA buffer-washed protein A magnetic beads (ThermoFisher), 5 μM GST-R and 5 μg anti-Qa antibody. After the mix was nutated at 4°C for 2 hr, beads were washed three times with 1 mL of RIPA buffer. Proteins were eluted with 100 μL of SDS sample buffer at 95°C for 5 min. Eluates were assayed by immunoblot with antibodies to R, Qa and Vps33. For sample f, the separate proteoliposomes, Qc, and HOPS were each mixed with RIPA buffer, then combined. (C) Immunoblots for the R-SNARE were scanned from five experiments, the band intensity of sample d (HOPS and Qc added at t = 0 min) was set to 100%, and the means and standard deviations are shown.
-
Figure 3—source data 1
Source data file (Excel) for Figure 3A and C.
- https://cdn.elifesciences.org/articles/53559/elife-53559-fig3-data1-v2.xlsx
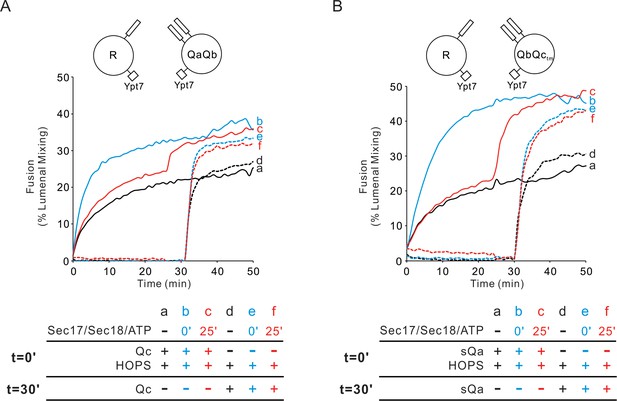
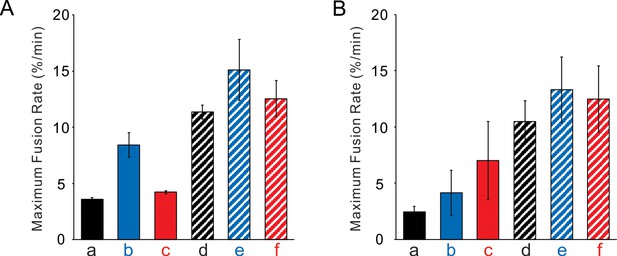
Sec17/Sec18/ATP do not inhibit fusion from the HOPS:R:QaQb or HOPS:R:QbQctm intermediates.
(A) Proteoliposomes bearing Ypt7 (1:8000 molar ratio to lipids) and either R- or QaQb- SNAREs (1:16,000 molar ratio to lipids) were mixed with 50 nM HOPS at t = 0, and 100 nM Qc was either added at t = 0 (a–c) or at t = 30 (d–f). Sec17 (300 nM), Sec18 (300 nM) and 1 mM ATP were added at t = 0 (b, e) or at t = 25 (c, f). Kinetics shown are representative of n ≥ 3 experiments. The average and standard deviations of maximum fusion rates from three independent experiments are in Figure 4—figure supplement 1. (B) The analogous experiment was performed with Ypt7/R and Ypt7/QbQctm proteoliposomes with HOPS, sQa, and Sec17/Sec18/ATP as indicated.
-
Figure 4—source data 1
Source data file (Excel) for Figure 4A and B.
- https://cdn.elifesciences.org/articles/53559/elife-53559-fig4-data1-v2.xlsx
Rapid-fusion intermediates in the presence of Sec17, Sec18 and ATP.
(A) The intermediate of HOPS:R:QaQb was not disassembled by Sec17/Sec18/ATP. The maximum rates of fusion after Qc addition are shown. All proteoliposomes had SNAREs and Ypt7-tm at 1:16,000 and 1:8000 protein:lipid molar ratios, respectively. Fusion assays were as described in Figure 4A, with R and QaQb SNARE proteoliposomes. Average and standard deviations of fusion rates from three independent experiments are shown. (B) The intermediate of HOPS:R:QbQctm was not disassembled by Sec17, Sec18 and ATP. The analogous experiment was performed, as in Figure 4B, with Ypt7/R and Ypt7/QbQctm proteoliposomes plus HOPS, sQa, and Sec17, Sec18, and ATP as indicated in Figure 4B.
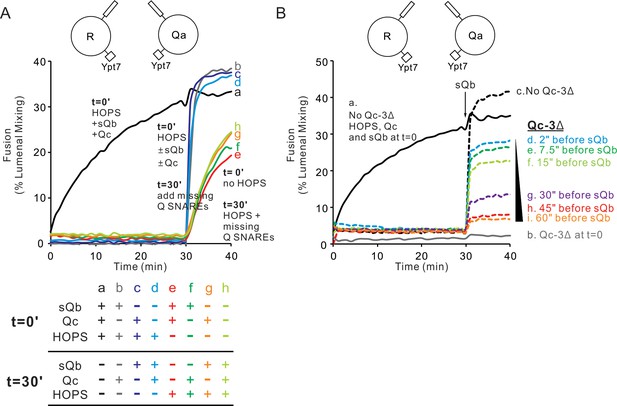
HOPS supports the assembly of a sudden-fusion intermediate between Ypt7/R-SNARE and Ypt7/Qa-SNARE proteoliposomes in the absence of Qb or Qc.
(A) Mixed proteoliposomes bearing Ypt7 (1:8000 molar ratio to lipids) and either R- or Qa-SNARE (1:16,000 molar ratio to lipids) were mixed with 50 nM HOPS at t = 0 (a–d) or t = 30 min (e–h). Soluble Q SNAREs were added: (a,e) sQb and Qc at t = 0, (b,f) sQb at t = 0, and (c,g) Qc at t = 0. At t = 30’, all missing soluble SNAREs were added. (B) When Qc is present, it engages reversibly with the HOPS:R:Qa sudden-fusion complex. Proteoliposomes bearing Ypt7 (1:8000 molar ratio to lipids) and either R- or Qa- SNARE (1:16,000 molar ratio to lipids) were mixed and given 100 nM Qc, 4 μM Qb, 50 nM HOPS and/or 4 μM QcΔ3, then assayed for fusion as follows: (a) Qc, Qb and HOPS were added at t = 0, (b) Qc, Qb, HOPS and QcΔ3 (Schwartz and Merz, 2009) were added at t = 0, (c–i) Qc and HOPS were added at t = 0 and Qb was added at t = 30 min. For d-i, QcΔ3 was added 60 s (d), 45 s (e), 30 s (f), 15 s (g), 7.5 s (h) or 2 s (i) before Qb addition. Kinetics shown are representative of n ≥ 3 experiments. Average and standard deviations of maximum fusion rates from three independent experiments are in Figure 5—figure supplement 1.
-
Figure 5—source data 1
Source data file (Excel) for Figure 5A and B.
- https://cdn.elifesciences.org/articles/53559/elife-53559-fig5-data1-v2.xlsx
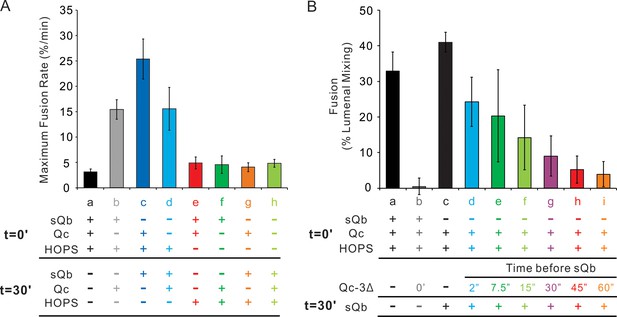
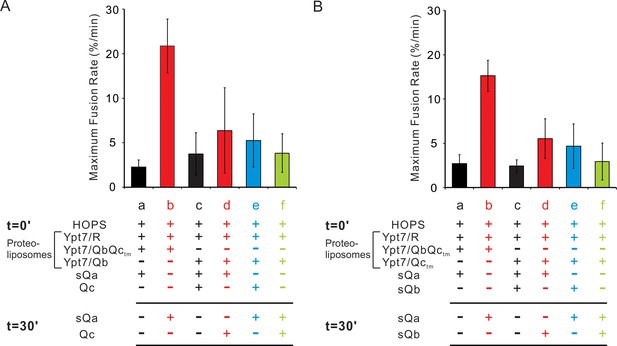
A sudden-fusion intermediate with R and Qa proteoliposomes.
(A) Pre-incubation R and Qa proteolipoosome with HOPS allows rapid-fusion intermediate assembly without sQb or Qc. (B) Qc, when present, forms an unstable complex with the HOPS:R:Qa rapid-fusion intermediate. Fusion assays were as described in Figure 5 with R and Qa proteoliposomes. All proteoliposomes had SNAREs and Ypt7-tm at 1:16,000 and 1:8000 protein:lipid molar ratios, respectively. Average and standard derivations of (A) maximum fusion rates and (B) fusion after 40 min incubation are from three independent experiments.
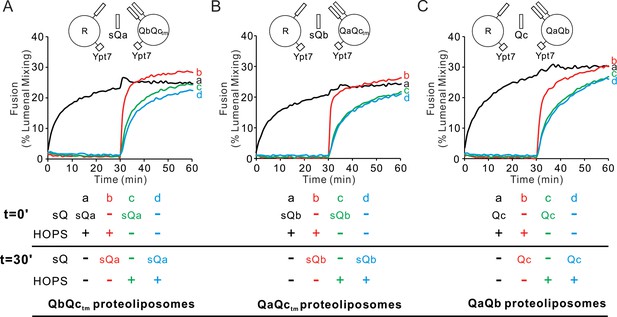
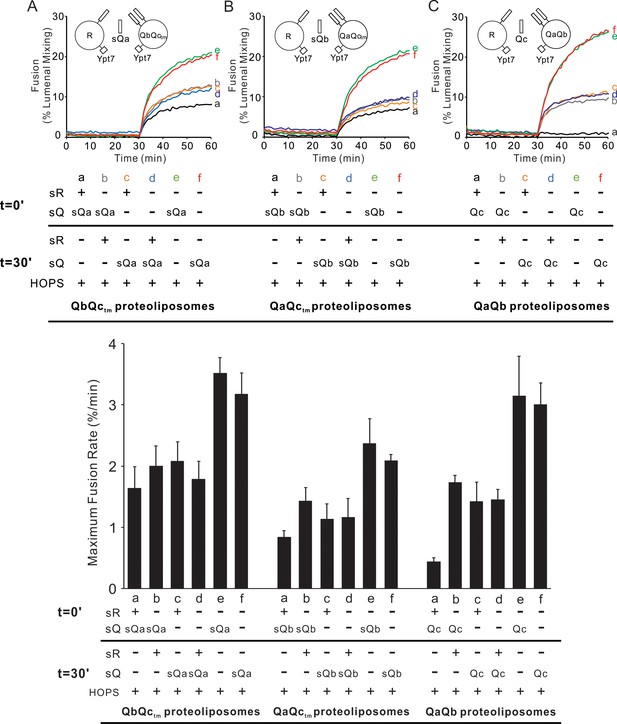
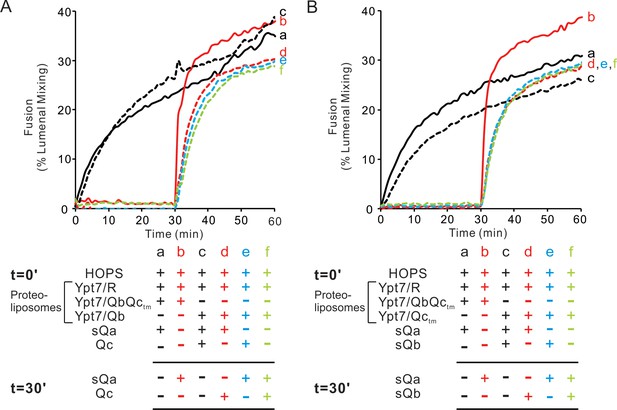
HOPS activates mixed proteoliposomes with R+Ypt7 and 2Q+Ypt7 for a burst of fusion when the missing sQ-SNARE is supplied.
(A–C) Fusion incubations received 50 nM HOPS at t = 0 min (a, b) or t = 30 min (c, d) with 4 μM soluble Q-SNARE at t = 0 min (a, c) or t = 30 min (b, d) as indicated. Fusion reactions had proteoliposomes bearing R mixed with proteoliposomes bearing either (A) QbQctm, (B) QaQctm, or (C) QaQb SNAREs at 1:16000 SNARE:lipid molar ratios. All proteoliposomes had Ypt7-tm at a 1:8000 protein: lipid molar ratio. Content mixing assays in this figure are representative of n ≥ 3 experiments; means and standard deviations for each experiment are presented in Figure 6—figure supplement 1.
-
Figure 6—source data 1
Source data file (Excel) for Figure 6A,B and C.
- https://cdn.elifesciences.org/articles/53559/elife-53559-fig6-data1-v2.xlsx
Preincubation of R- and 2Q-SNARE proteoliposomes with HOPS gives more rapid fusion when the third soluble SNARE is added than when all components are mixed without preincubation.
Fusion assays were as described in Figure 6, with R and QbQctm, QaQctm or QaQb SNARE proteoliposomes. All proteoliposomes had SNAREs at a 1:16000 protein: lipid molar ratio and Ypt7-tm at a 1:8000 protein: lipid molar ratio. Average and standard deviations of maximum fusion rate from three independent experiments.
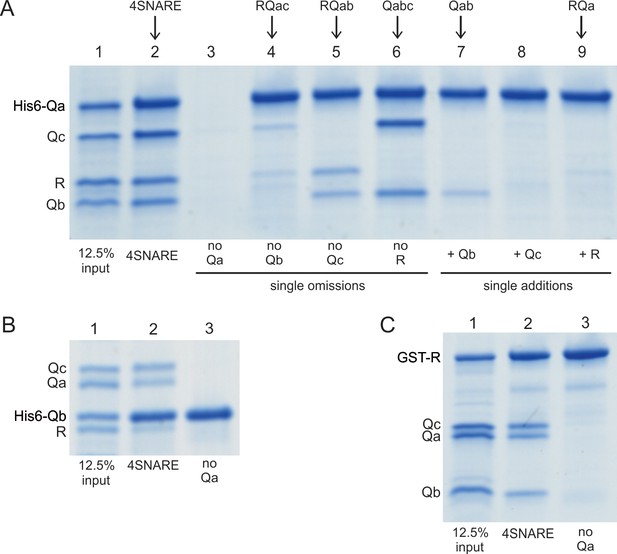
Spontaneous formation of SNARE complexes in detergent.
His6-tagged Qa SNARE (A), his6-tagged Qb SNARE (B) or GST-tagged R SNARE (C) were mixed at 4 µM with 4 µM of the other indicated full-length SNAREs, in a total volume of 50 µl in pulldown buffer (20 mM HEPES-NaOH, pH 7.4, 150 mM NaCl, 10% glycerol, 100 mM ß-octylglucoside), plus 20 mM imidazoleCl, pH 7.0 for incubations with a his6-tagged SNARE. After nutation for 1 hr at 4 °C, a portion (40 µl) was transferred to tubes containing either 20 µl of a 50% slurry of (A, B) nickel-NTA agarose (Qiagen, Hilden, Germany) or (C) glutathione agarose 4B (Genesee Scientific, San Diego, CA). Each was nutated at 4 °C for 1 hr, diluted with 0.5mls of pulldown buffer (C) or pulldown buffer plus imidazole (A, B), and centrifuged (500xg, 6 min, 4 °C). Supernatants were removed, and the beads were washed three more times with 0.5 ml portions of the same buffer. Proteins were eluted with 50 µl of SDS sample buffer with ß-mercaptoethanol by heating (95°C, 5 min). Eluates were analyzed by Coomassie-stained gel. The substantial increase in molecular weight for his6-Qa and his6-Qb is caused by the presence of a 36 amino acyl linker between the his6 tag and the N-terminus of each of these SNAREs (Izawa et al., 2012).
HOPS mediates the assembly of R- and QbQctm- SNAREs into a shared trans-complex in the absence of Qa.
Mixed proteoliposomes bearing Ypt7 (1:8000 molar ratio to lipids) and either R- or QbQctm-SNAREs (1:16,000 molar ratio to lipids) were mixed with 50 nM HOPS and 4 μM sQa where indicated, added either at the start or after 30-min incubation. (A) Fusion was measured by FRET signal. (B) Samples were withdrawn at 33 min, solubilized in RIPA buffer, and the R SNARE that co-immunoprecipitated with 1.25 μg anti-Qb antibody was assayed as a measure of trans-complex, as described in Figure 3 with 1.25 μg of antibody to Qb. (C) The average and standard deviation of the Nyv1 band intensity from three independent experiments are shown, normalized to sample d (HOPS and sQa added at t = 0).
-
Figure 8—source data 1
Source data file (Excel) for Figure 8A and C.
- https://cdn.elifesciences.org/articles/53559/elife-53559-fig8-data1-v2.xlsx
Both the Qb- and Qc-SNAREs are needed for the assembly of a rapid fusion intermediate without Qa.
(A) Fusion reactions had mixed proteoliposomes bearing the R-SNARE and either Qbctm (lines) or Qb (dotted lines)-SNAREs (1:8000 molar ratio to lipids). These were mixed with 50 nM HOPS at t = 0. Also at t = 0, soluble SNAREs were added: sQa (a,c,d) and/or Qc (c and e). After 30 min, sQa (b, e, f) and Qc (d, f) were added as indicated in the reaction scheme. (B) Fusion reactions had proteoliposomes bearing R-SNARE and proteoliposomes with either Qbctm (solid lines) or Qctm (dotted lines)-SNAREs (1:8000 molar ratio to lipids). These were mixed with 50 nM HOPS at t = 0. Soluble SNAREs were also added at t = 0 as indicated: sQa (a, c, d) and sQb (c, e). After 30 min, sQa (b, e, f) and sQb (d, f) or sQa and sQb were added. All proteoliposomes had Ypt7-tm at a 1:8000 protein:lipid molar ratio. Content mixing assays in this figure are representative of n ≥ 3 experiments; means and standard deviations from four independent experiments are in Figure 9—figure supplement 1.
-
Figure 9—source data 1
Source data file (Excel) for Figure 9A and B.
- https://cdn.elifesciences.org/articles/53559/elife-53559-fig9-data1-v2.xlsx
The assembly of a rapid-fusion intermediate without Qa needs membrane-bound Qb and Qc SNAREs.
Fusion assays were as described in Figure 9, with R and Qb (A) or R and Qctm (B) proteoliposomes. All proteoliposomes had SNAREs and Ypt7-tm at 1:8000 protein:lipid molar ratios. Average and standard derivations of maximum fusion rate from three independent experiments.
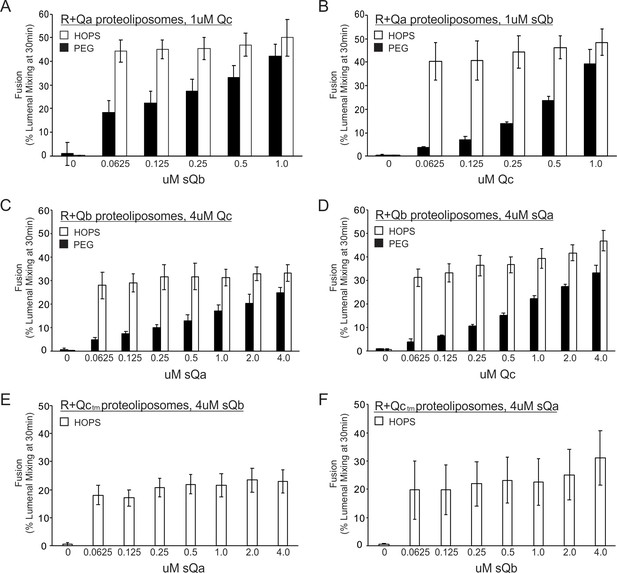
Fusion with HOPS is saturable for each vacuolar Q-SNARE.
Reconstituted proteoliposomes of VML composition were prepared with wild-type Ypt7 at 1:4000 protein to lipid molar ratio and either R-SNARE or each single Q-SNARE at a 1:2500 protein to lipid ratio, employing a transmembrane version of Qc. Fusion assays were performed in RB150. Ypt7/R-SNARE proteoliposomes and Ypt7/Q-SNARE proteoliposomes were separately incubated at 1 mM (lipid) with 20 µM streptavidin, 2 mM EDTA, 0.5 mM MgCl2, and 1 mM GTP for 10 min at 27 °C. MgCl2 was then added to bring the concentration to 2.5 mM. The nucleotide-exchanged R- and Q- proteoliposomes were then combined and portions were added to tubes containing one half volume of either 0.16 µM HOPS or 8% PEG. Aliquots of each (16 µl) were pipetted into a 384-well plate. During the nucleotide exchange process, a mixture of the missing soluble Q-SNAREs was prepared in RB150, containing 4 µM of each soluble Q-SNARE (A and B) or 16 µM of each soluble Q-SNARE (C), (D), (E), and (F). Two dilution curves were then prepared, keeping one soluble SNARE at the starting concentration while diluting the other twofold. A portion (5 µl) of each dilution was pipetted into empty wells of a 384-well plate, which then received 15 µl of the mixtures of proteoliposome with HOPS or PEG. Final concentrations of HOPS or PEG in the 20 µl reaction were 40 nM and 2%, respectively.
-
Figure 10—source data 1
Source data file (Excel) for Figure 10A,B,C,D,E and F.
- https://cdn.elifesciences.org/articles/53559/elife-53559-fig10-data1-v2.xlsx
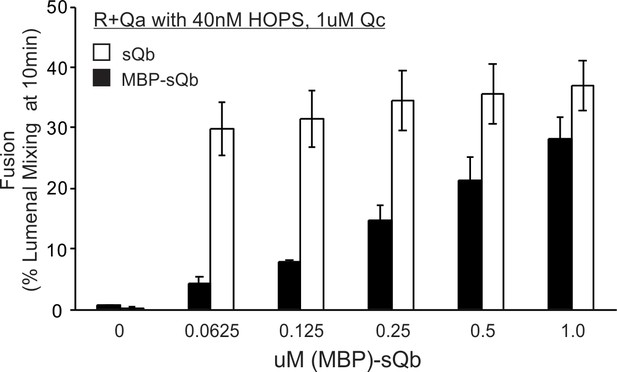
An MBP tag on the soluble Qb-SNARE interferes with its recruitment by HOPS.
Fusion assays were conducted as described in Figure 10, using R-SNARE+Ypt7 and Qa-SNARE+Ypt7 RPLs, 40 nM HOPS, a constant (1 µM) level of Qc, and twofold decreasing concentrations of MBP-tagged soluble Qb that either had or had not been cleaved to remove its MBP domain by a 2 hr incubation with TEV protease.
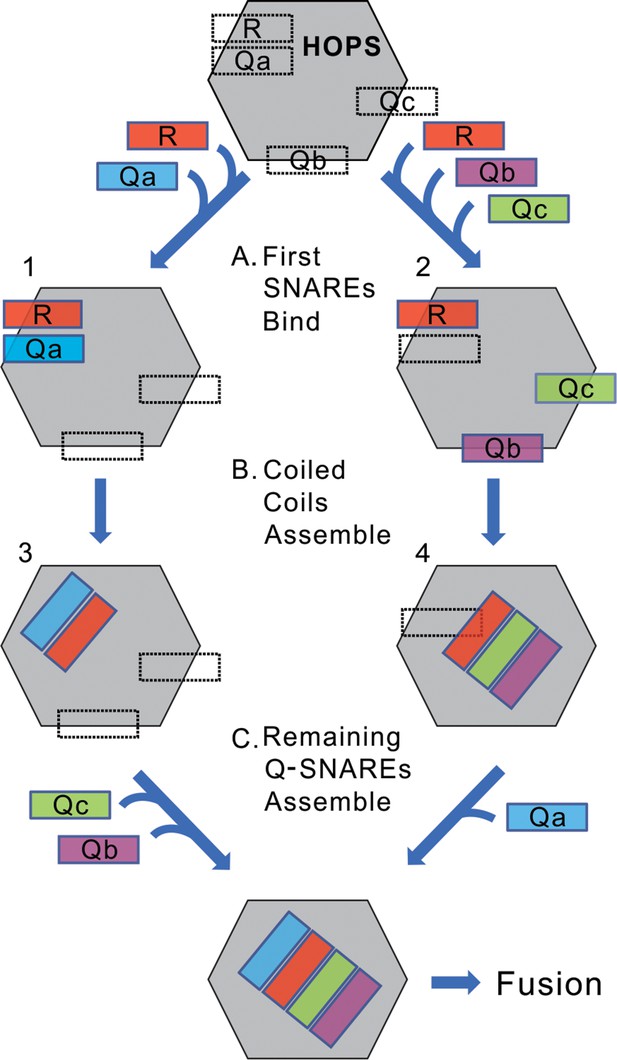
A conceptual model of rapid-fusion intermediates.
HOPS has binding sites for each of the four vacuolar SNAREs, indicated in dotted lines. We propose that (A) R, and either Qa (left) or Qb and Qc (right), bind to HOPS at their high-affinity sites, then (B) partially or wholly reorient to begin their coiled-coils assembly. (C) The binding sites for the remaining Q-SNAREs catalyze their rapid transfer to the nascent coiled-coil, triggering rapid fusion. Our data do not establish when SNAREs leave their initial binding sites to begin coiled-coils association; the rapid-fusion intermediates might be represented by 1 and 2, or 3 and 4, or all 4 SNAREs might remain bound to their initial sites before switching to coiled-coils association.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Saccharomyces cerevisiae) | Nyv1 | Saccharomyces Genome Database | SGD:S000004083 | |
| Gene (Saccharomyces cerevisiae) | Vam3 | Saccharomyces Genome Database | SGD:S000005632 | |
| Gene (Saccharomyces cerevisiae) | Vti1 | Saccharomyces Genome Database | SGD:S000004810 | |
| Gene (Saccharomyces cerevisiae) | Vam7 | Saccharomyces Genome Database | SGD:S000003180 | |
| Gene (Saccharomyces cerevisiae) | Ypt7 | Saccharomyces Genome Database | SGD:S000004460 | |
| Gene (Saccharomyces cerevisiae) | Sec17 | Saccharomyces Genome Database | SGD:S000000146 | |
| Gene (Saccharomyces cerevisiae) | Sec18 | Saccharomyces Genome Database | SGD:S000000284 | |
| Peptide, recombinant protein | GST-R (Nyv1) | PMID: 18650938 | purified from E. coli. | |
| Peptide, recombinant protein | GST-Qa (Vam3) | PMID: 18650938 | purified from E. coli. | |
| Peptide, recombinant protein | GST-Qb (Vti1) | PMID: 18650938 | purified from E. coli. | |
| Peptide, recombinant protein | GST-sR (soluble) | PMID: 15241469 | purified from E. coli. | |
| peptide, recombinant protein | GST-sQa (soluble) | PMID: 28637767 | purified from E. coli. | |
| Peptide, recombinant protein | MBP-sQb (soluble) | PMID: 24088569 | purified from E. coli. | |
| Peptide, recombinant protein | Vam7-tm | PMID: 23071309 | purified from E. coli. | |
| Peptide, recombinant protein | Ypt7-tm | PMID: 31235584 | purified from E. coli. | |
| Peptide, recombinant protein | His6-Qa | PMID: 22174414 | purified from E. coli. | |
| Peptide, recombinant protein | his6-Qb | PMID: 22174414 | purified from E. coli. | |
| Peptide, recombinant protein | Vam7 | PMID: 17699614 | purified from E. coli. | |
| Peptide, recombinant protein | TEV protease | PMID: 18007597 | purified from E. coli. | |
| Peptide, recombinant protein | HOPS | PMID: 18385512 | purified from Saccharomyces cerevisiae. | |
| Peptide, recombinant protein | GST-PX | PMID: 23071309 | purified from E. coli. | |
| Antibody | anti-Vam3 (rabbit polyclonal) | PMID: 12566429 | Wickner lab stock | WB: 0.67 μg/ml IP: 5 μg |
| Antibody | anti-Nyv1 (rabbit polyclonal) | PMID: 10385523 | Wickner lab stock | WB: 1 μg/ml |
| Antibody | anti-Vti1 (rabbit polyclonal) | PMID: 18007597 | Wickner lab stock | WB: 2 μg/ml IP: 1.25 μg |
| Antibody | anti-Vps16 (rabbit polyclonal) | PMID: 18007597 | Wickner lab stock | WB: 0.92 μg/ml |
| Antibody | anti-Vps33 (rabbit polyclonal) | PMID: 10944212 | Wickner lab stock | WB: 0.5 μg/ml |
| Chemical compound,drug | Cy5-derivatized streptavidin | SeraCare Life Sciences | 5270–0023 | |
| Chemical compound,drug | Biotinylated PhycoE | Thermo Fisher Scientific | p811 | |
| Chemical compound,drug | streptavidin | Thermo Fisher Scientific | 434302 | |
| Chemical compound,drug | 1,2-dilinoleoyl-sn-glycero-3-phosphocholine | Avanti polar lipids | 850385 | |
| Chemical compound,drug | 1,2-dilinoleoyl-sn-glycero-3-phospho-L-serine | Avanti polar lipids | 840040 | |
| Chemical compound,drug | 1,2-dilinoleoyl-sn-glycero-3-phosphoethanolamine | Avanti polar lipids | 850755 | |
| Chemical compound,drug | 1,2-dilinoleoyl-sn-glycero-3-phosphate | Avanti polar lipids | 840885 | |
| Chemical compound,drug | L-α-phosphatidylinositol | Avanti polar lipids | 840044 | |
| Chemical compound,drug | 1,2-dipalmitoyl-sn-glycerol | Avanti polar lipids | 800816 | |
| Chemical compound,drug | ergosterol | Sigma | 45480 | |
| Chemical compound,drug | PI(3)P diC16 | Echelon Bioscience | P-3016 | |
| Chemical compound,drug | rhodamine DHPE | Invitrogen | L1392 | |
| Chemical compound,drug | NBD-PE | Invitrogen | N360 | |
| Chemical compound,drug | Marina-blue | Invitrogen | M12652 | |
| Software and Algorithms | UN-SCAN-IT | Silk Scientific |





