Systematic detection of horizontal gene transfer across genera among multidrug-resistant bacteria in a single hospital
Figures
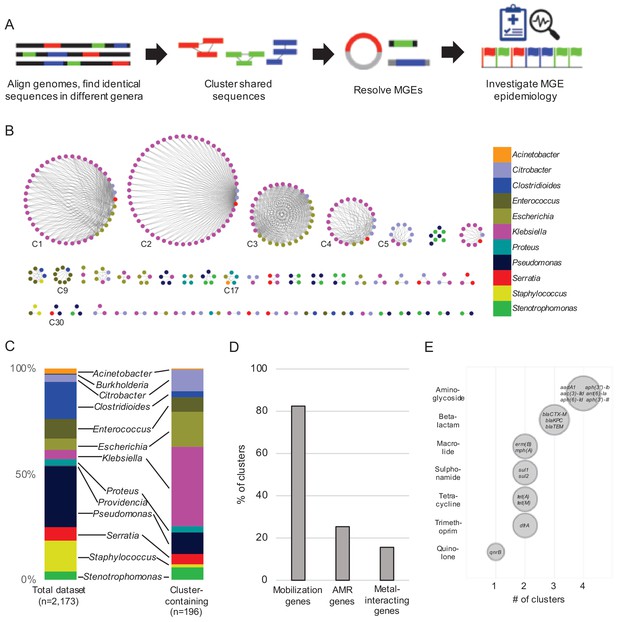
Identification of nucleotide sequences shared across bacterial genera in a single hospital.
(A) Approach to identify shared sequence clusters, and then resolve the MGEs that carry them. (B) 51 clusters of shared sequences found in distinct genera visualized with Cytoscape. Nodes represent bacterial isolates and are color-coded by genus. Edges connect nodes from different genera sharing >5 kb of sequence at 100% nucleotide sequence identity. Clusters examined more closely in subsequent figures are labeled. (C) Genus distribution of all 2173 genomes in the dataset (left) and the 196 isolates encoding one or more shared sequence clusters (right). (D) Prevalence of mobilization, antimicrobial resistance (AMR) and metal-interacting genes among 51 shared sequence clusters. (E) Summary of AMR genes identified in shared sequence clusters. Genes are grouped by antibiotic class, and bubble sizes correspond to prevalence among the clusters shown in (B). AMR gene names are listed inside each bubble. To generate (D) and (E) the longest sequence in each cluster was examined.
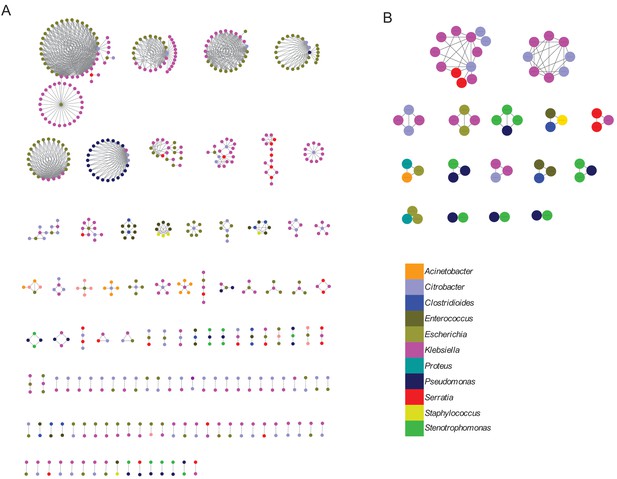
Shared sequence clustering based on alternate sequence length parameters.
Clusters of shared sequences identified by the alignment-based method depicted in Figure 1A, with different cutoffs for identical alignment length (3 kb in panel A, 10 kb in panel B), and visualized with Cytoscape. Nodes represent bacterial isolates and are color-coded by genus. Edges connect nodes from different genera that share an identical sequence longer than the length cut-off. (A) 120 clusters of sequences sharing at least 3 kb with 100% identity to at least one other sequence in another genus. (B) 16 clusters of sequences sharing at least 10 kb with 100% identity to at least one other sequence in another genus.
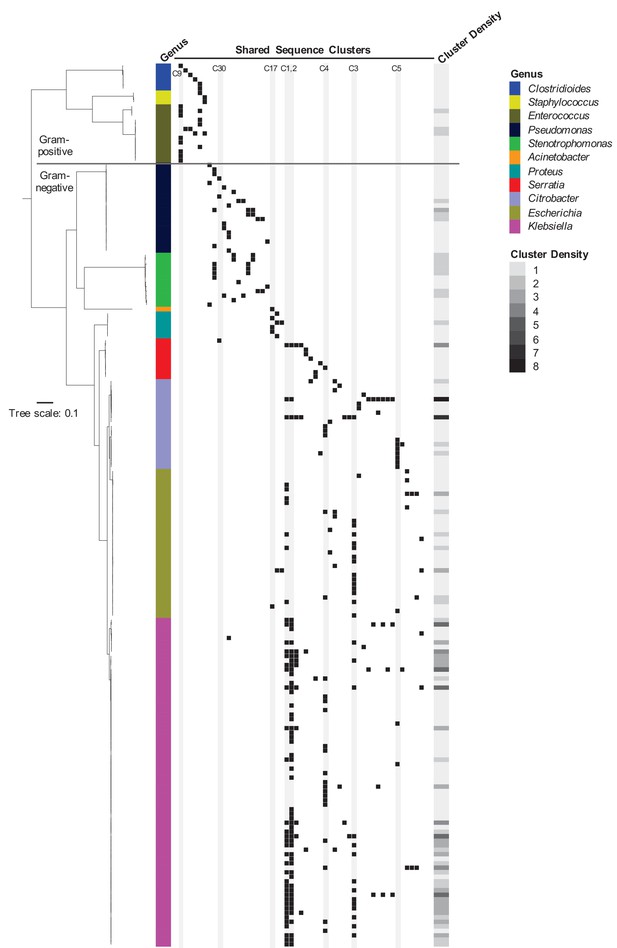
Phylogenetic distribution of shared sequence clusters across 196 genomes.
A phylogeny was made by aligning amino acid sequences of 120 ubiquitous protein coding genes from the Genome Taxonomy Database Tool Kit. The scale bar shows the number of amino acid substitutions per site. Black squares mark the presence of one or more clusters in each genome, with each column corresponding to a different cluster. The heat map to the right shows cluster density (i.e. total number of cross-genus shared sequence clusters) in each bacterial genome. Clusters examined more closely in subsequent figures are labeled and shaded in gray.
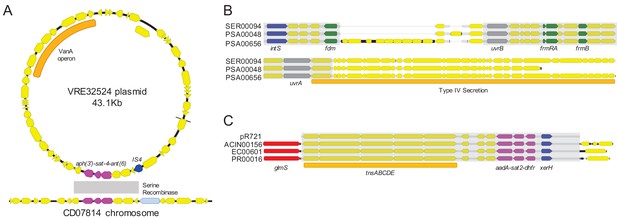
Examples of sequence sharing across genera.
(A) Genes shared between a vancomycin-resistant E. faecium (VRE) plasmid and a C. difficile chromosome (cluster C9). The VanA operon, conferring vancomycin resistance, is marked with an orange bar. Shared drug resistance genes are colored magenta, and mobilization genes are colored blue. Gray shading marks DNA sequence that is 100% identical between isolates. (B) Identical portions of an integrated conjugative element (cluster C30) shared between an S. marcescens genome (SER00094) and two P. aeruginosa genomes (PSA00048 and PSA00656). Blue = intS integrase; green = formaldehyde resistance genes; gray = UvrABC system genes. Type IV secretion machinery is marked with an orange bar, and gray shading marks sequences that are 100% identical between isolates. (C) Identical Tn7 transposons shared between A. baumannii, E. coli, and P. mirabilis (cluster C17). The Tn7 sequence of the pR721 plasmid is shown at the top. The tnsABCDE transposon machinery is marked with an orange bar, and the glmS gene, which flanks the Tn7 insertion site, is colored red. Shared drug resistance genes are colored magenta, and an xerH tyrosine recombinase is colored blue. Gray shading marks sequences that are 100% identical between isolates.
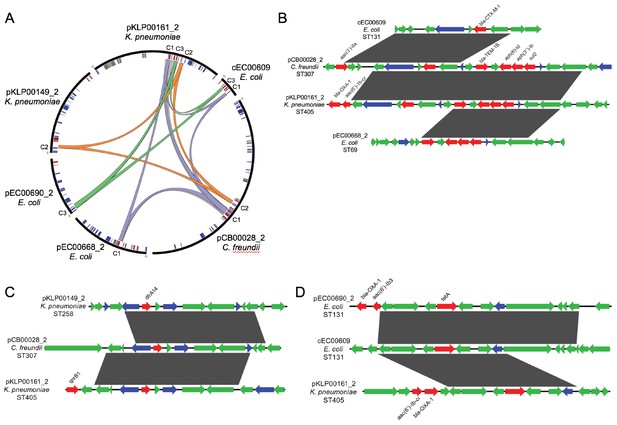
Mosaicism of shared sequence clusters present on diverse MGEs.
(A) Circos plot of six distinct MGEs (black bars) that encode shared sequence clusters C1, C2, and C3. Lowercase letters in sequence names indicate element type (c = chromosome, p=plasmid). Homologous cluster sequences are connected to one another with colored links (purple = C1, orange = C2, green = C3, gray = other). Inner circle depicts genes involved in mobilization (blue), antibiotic resistance (red) and metal interaction (gray). (B–D) Alignments of sequences grouped into shared sequence clusters C1 (B), C2 (C), and C3 (D) from the MGEs displayed in (A). ORFs are colored by function (blue = mobilization, red = antibiotic resistance, green = other/hypothetical). Antibiotic resistance genes are labeled above and dark gray blocks connect sequences that are identical over at least 5 kb.
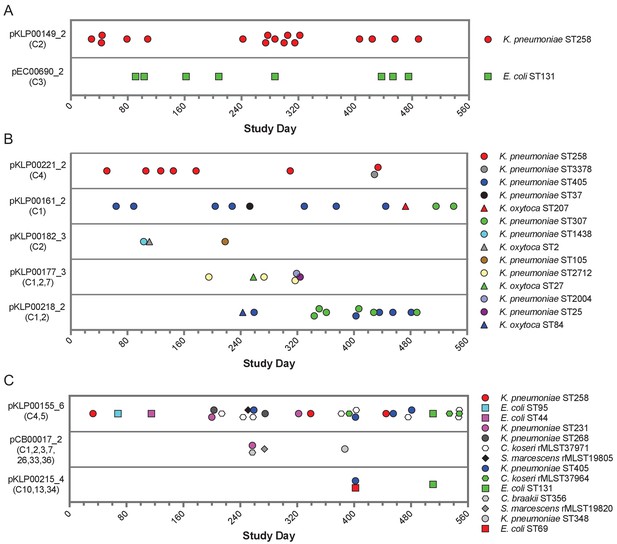
Timelines of plasmid occurrence among isolates of the same ST (A), same genus (B), or different genera (C).
Illumina contigs of all study isolate genomes were mapped to the reference plasmid sequences indicated to the left of each panel, and plasmids were called 'present' if an isolate genome of any genus contained >90% of the reference sequence (based on mapping coverage). Timelines show the study date of each isolate, and the shared sequence clusters carried by each plasmid are listed in parentheses below the plasmid names. Shape and color of data points correspond to bacterial species and ST, respectively. More information about each plasmid is provided in Table 2.
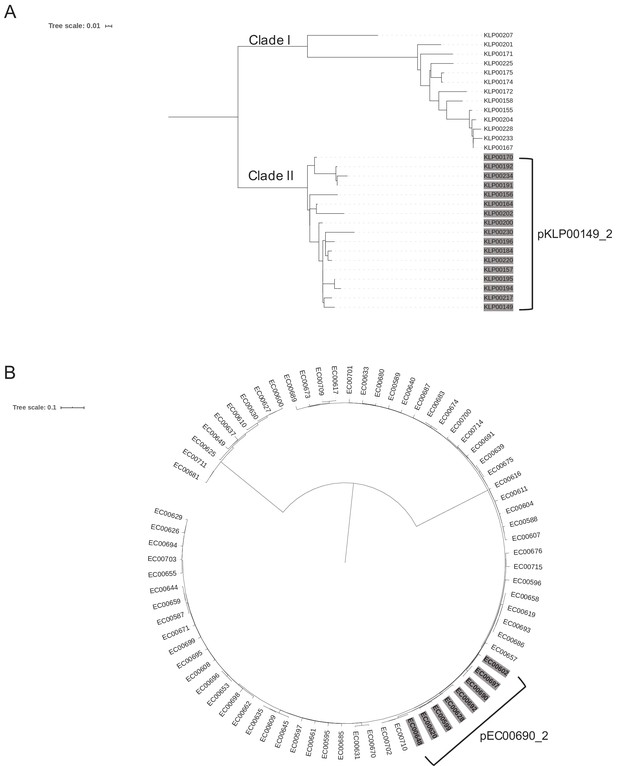
Phylogenetic context of hospital-associated infection isolates carrying the same plasmids.
(A) Core genome phylogeny of 30 K. pneumoniae ST258 isolates in the dataset. Shading indicates the presence of the pKLP00149_2 plasmid, which was maintained among all Clade II isolates. (B) Core genome phylogeny of 74 E. coli ST131 isolates in our dataset. Shading indicates the presence of the pEC00690_2 plasmid in a subset of closely related isolates.
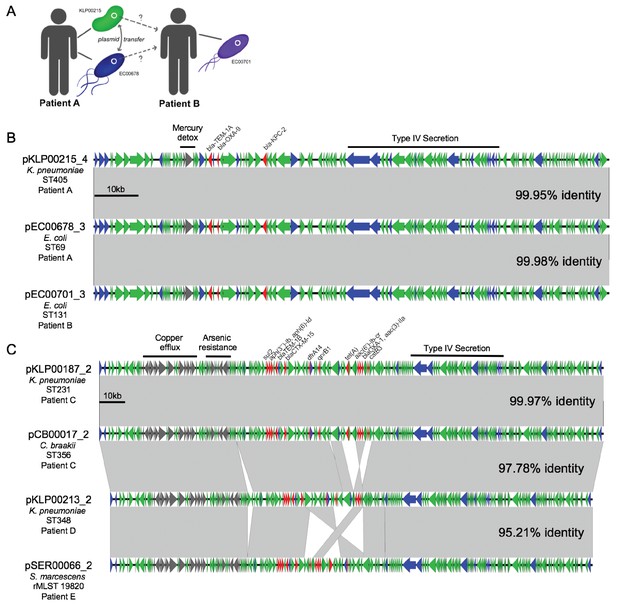
Cross-genus transfer of plasmids within and between patients.
(A) Schematic diagram showing K. pneumoniae and E. coli isolates bearing the same plasmid sampled from two patients. (B) Nucleotide alignment of the plasmid presumably transferred within and between the patients shown in (A). A 113.6 kb IncF carbapenemase-encoding plasmid was resolved from two genomes of different bacterial isolates from the same clinical specimen from Patient A. A nearly identical plasmid was also identified in an isolate from Patient B, who occupied a hospital room adjacent to Patient A. (C) Alignment of a 196.8 kb IncF multidrug-resistance plasmid resolved from two genomes of different bacterial isolates from the same clinical specimen from Patient C. Similar plasmids were also found in isolates from two additional patients (Patient D and Patient E), who had no identifiable epidemiologic links with Patient C. ORFs are colored by function (blue = mobilization, red = antibiotic resistance, gray = metal interacting, green = other/hypothetical). Antibiotic resistance genes, metal-interacting operons, and Type IV secretion components are labeled. Gray blocks between sequences indicates regions > 5 kb with >99.9% identity, and pairwise identities across the entire plasmid are noted to the right.
Tables
Demographics and co-morbidities of study patients.
| All isolates | Shared sequence isolates | p-value† | |
|---|---|---|---|
| Total number of isolates | 2173 | 196 | |
| Number of unique patients | 1533 | 172 | |
| Demographics*: | n = 1445 | n = 157 | |
| Median age, years (range) | 62 (17–98) | 63 (19–89) | 0.89 |
| Male gender | 738 (51%) | 81 (52%) | 0.93 |
| Co-morbidities: | |||
| Median Charlson Co-morbidity Index (range) | 3 (0–15) | 4 (0–13) | 0.03 |
| Solid organ transplant | 180 (12%) | 29 (18%) | 0.02 |
| Diabetes mellitus | 369 (26%) | 42 (27%) | 0.7 |
| Cystic fibrosis | 31 (2%) | 5 (3%) | 0.37 |
-
*Demographics and co-morbidities are reported for patients for whom information was available.
†p-values were calculated using Fisher’s Exact test for categorical variables and Wilcoxon rank-sum test for continuous variables. Shared sequence isolates were removed from the ‘all isolates’ group to assess the significance of differences between groups.
Resolved MGEs and associated antibiotic resistance and metal interaction gene contents.
| MGE* | Length (kb) | % GC | Replicons† | MOB Family‡ | Antibiotic resistance Genes§ | Metal interaction Genes¶ |
|---|---|---|---|---|---|---|
| cEC00609 | 39.1 | 52.6 | None | None | aac(3)-IIa, aac(6')-Ib-cr, blaCTX-M-1, blaOXA-1, catB3, tet(A) | None |
| pCB00017_2 | 196.8 | 51.7 | FIB, FIIK | MOB-F | aac(6')-Ib-cr, aph(3'')-Ib, aph(6)-Id, blaCTX-M-15, blaOXA-1, blaTEM-1B, catB3, qnrB1, tet(A), sul2 | copD operon, pcoE, silE, silP, ars operon |
| pCB00028_2 | 383.1 | 47.5 | HI2, HI2A | MOB-H | aac(3)-IIa, aac(6')-Ib-cr, aadA1, aph(3'')-Ib, aph(6)-Id, blaCTX-M-15, blaOXA-1, baTEM-1B, catA1, catB3, dfrA14, sul2, tet(A) | pcoE, merR, merB |
| pEC00668_2 | 145.4 | 55.9 | FIA, FII | MOB-F | aac(6)-Id, aph(3'')-Ib, dfrA14, blaTEM-1B, mph(A), sul2 | efeU, merA, merC, merP, merR, merT |
| pEC00690_2 | 106.8 | 54.7 | FIA, FII | MOB-F | aac(6')-Ibcr, blaOXA-1, catB3, tet(A) | efeU |
| pKLP00149_2 | 165.2 | 52.9 | FIIY | MOB-F | aac(6')-Ib, aac(6')-Ib-cr, aadA1, aph(3'')-Ib, aph(6)-Id, blaKPC-3, blaOXA-9, blaSHV-182, blaTEM-1A, dfrA14, sul2 | csoR |
| pKLP00155_6 | 9.5 | 54.9 | ColRNAI | MOB-C | None | None |
| pKLP00161_2 | 236.5 | 55.1 | FIB, FIIK | MOB-F | aac(6')-Ib-cr, aph(3'')-Ib, aph(6)-Id, blaCTX-M-15, blaOXA-1, blaTEM-1B, dfrA14, qnrB1, sul2, tet(A) | copD operon, pcoC, pcoE, silE, silP, ars operon |
| pKLP00177_3 | 170.8 | 52.0 | FIB | MOB-F | aac(3)-IIa, aac(6')-Ib-cr, aph(3'')-Ib, aph(6)-Id, blaCTX-M-15, blaOXA-1, blaTEM-1B, catB3, dfrA14, qnrB1, sul2, tet(A) | copD operon, pcoC, pcoE, silE, silP, ars operon |
| pKLP00182_3 | 15.8 | 51.2 | A/C | MOB-H | aac(6')-Ib-cr, blaOXA-1, catB3, dfrA14, tet(A) | None |
| pKLP00215_4 | 113.6 | 53.9 | FIB, FIIK | MOB-F | blaKPC-2, blaOXA-9, blaTEM-1A | merB, merR |
| pKLP00218_2 | 164.7 | 54.9 | FIB, FIIK | MOB-F | aph(3'')-Ib, aph(6)-Id, blaCTX-M-15, blaTEM-1B, dfrA14, sul2 | copD operon, pcoC, pcoE, silE, silP, ars operon |
| pKLP00221_2 | 242.3 | 53.2 | ColRNAI, FIB, FII | MOB-C, MOB-F | aac(6')-Ib, aada2, aph(3')−1a, blaKPC-2, blaOXA-9, blaTEM-1A, catA1, dfrA12, mph(A), sul1 | copD operon, pcoC, pcoE, silE, silP, ars operon |
-
*MGE names include location (c = chromosome, p=plasmid), name of the reference isolate sequenced, and assembly contig number (_2, _3, _4, _6).
†Replicons were identified by querying Plasmid MLST and PlasmidFinder databases.
-
‡MOB families were assigned with MOBscan.
§Antibiotic resistance genes were identified by querying the ResFinder database.
-
¶Metal interaction genes were identified by examining annotations assigned by Prokka.
Additional files
-
Supplementary file 1
List of isolates and accession information for Illumina sequence data.
- https://cdn.elifesciences.org/articles/53886/elife-53886-supp1-v1.xlsx
-
Supplementary file 2
Genus distribution and gene content of shared sequence clusters.
- https://cdn.elifesciences.org/articles/53886/elife-53886-supp2-v1.xlsx
-
Supplementary file 3
Accession information for hybrid assembled genomes.
- https://cdn.elifesciences.org/articles/53886/elife-53886-supp3-v1.xlsx
-
Supplementary file 4
Plasmid coverage among all isolates with >90% coverage of at least one plasmid.
- https://cdn.elifesciences.org/articles/53886/elife-53886-supp4-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/53886/elife-53886-transrepform-v1.docx





