Distinct regulatory ribosomal ubiquitylation events are reversible and hierarchically organized
Figures
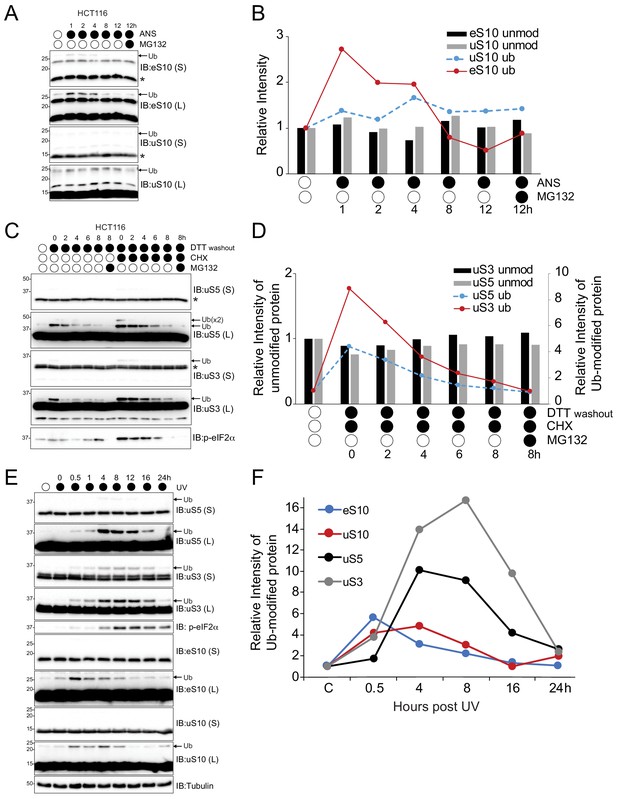
Stress-induced RRub events are reversible.
(A) HCT116 cells were treated with anisomycin (ANS, 5 μg/ml) alone or with MG132 (10 μM) for the indicated times. Whole-cell extracts were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies. The ubiquitin-modified and unmodified ribosomal protein is indicated by the arrow and asterisk, respectively. S and L denote short and long exposures (n = 1). (B) The amount of ubiquitylated eS10 (red line) and uS10 (blue dashed line) and unmodified eS10 (black column) and uS10 (gray column) after the indicated treatments compared to untreated cells quantified from panel A. (C) HCT116 cells were treated with DTT (5 mM) alone or with cycloheximide (CHX, 100 μg/ml) for 4 hr followed by DTT washout in media with or without CHX alone or with MG132 (10 μM) for the indicated times. Whole-cell extracts were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies. The ubiquitin-modified and unmodified ribosomal protein is indicated by the arrow and asterisk, respectively. S and L denote short and long exposures (n = 1). (D) The amount of ubiquitylated uS3 (red line) and uS5 (blue dashed line) and unmodified uS3 (black column) and uS5 (gray column) after the indicated treatments compared to untreated cells quantified from panel C. (E) 293 T cells were exposed to UV and allowed to recover for the indicated times. Whole-cell extracts were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies. The ubiquitin-modified ribosomal protein is indicated by the arrow. S and L denote short and long exposures (n = 1). (F) The amount of ubiquitylated eS10 (blue line), uS10 (red line), uS5 (black line) and uS3 (gray line) after UV exposure compared to untreated cells quantified from panel E.
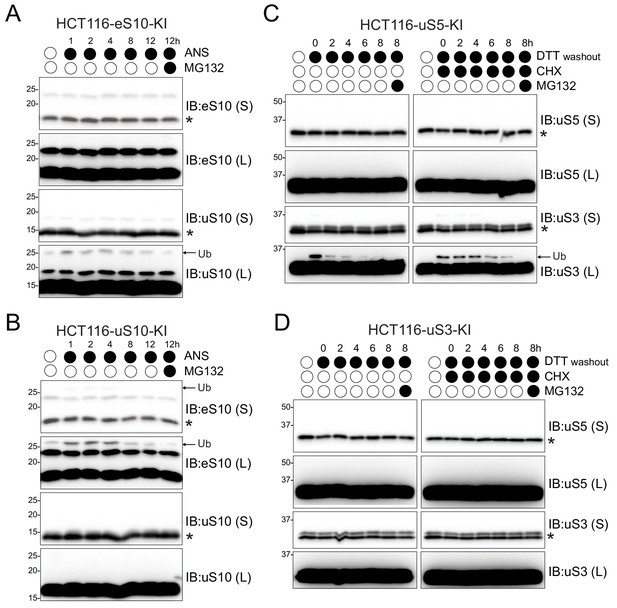
Distinct sets of RRub events are hierarchically organized.
(A,B) HCT116 mutant RRub knock-in (KI) cell lines eS10-KI (K138RK139R) or uS10-KI (K4RK8R) (A,B) were treated with ANS (5 μg/ml) and MG132 (10 μM) continuously for the indicated times. Whole-cell extracts were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies. (C,D) HCT116 mutant uS5-KI (K54RK58R) or uS3-KI (K214R) were either untreated or pre-treated with DTT (5 mM) alone or with CHX (100 μg/ml) for 4 hr followed by DTT washout in media with or without CHX (100 μg/ml) and MG132 (10 μM). Cells were collected at the indicated times post DTT washout. Whole-cell extracts were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies. The ubiquitin-modified and unmodified ribosomal protein is indicated by the arrow and asterisk, respectively. S and L denote short and long exposures (n = 1).
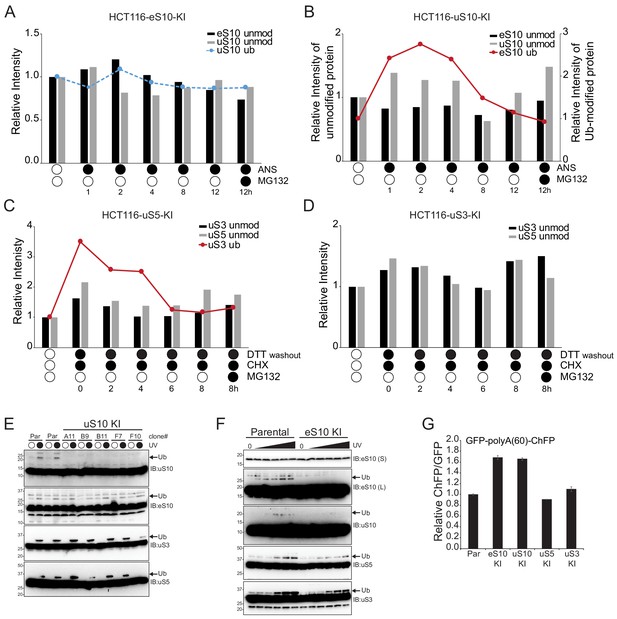
Quantification of site-specific RRub demodification upon exposure to stress.
(A,B) The amount of ubiquitylated eS10 (red line) and uS10 (blue dashed line) and unmodified eS10 (black column) and uS10 (gray column) in the indicated cell lines after the indicated treatments compared to untreated cells quantified from immunoblots in Figure 2A,B. (C,D) The amount of ubiquitylated uS3 (red line) and unmodified uS3 (black column) and uS5 (gray column) after the indicated treatments compared to untreated cells quantified from immunoblots in Figure 2C,D. (E) Whole cell extracts from individual clones of HCT116 uS10 knock-in (KI) cells or control parental (par) HCT116 cells exposed to UV were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies. The ubiquitin-modified ribosomal protein is indicated by the arrow (n = 1). (F) HCT116 eS10 knock-in cells or control parental (par) HCT116 cells were exposed to increasing doses of UV radiation. Whole cell extracts were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies. The ubiquitin-modified ribosomal protein is indicated by the arrow. S and L denote short and long exposures (n = 1). (G) The ChFP:GFP ratio from HCT116 parental, eS10-KI, uS10-KI, uS5-KI and uS3-KI cells transfected with the poly(A)-stall reporter plasmid relative to parental cells. Error bars denote SEM for triplicate transfections.
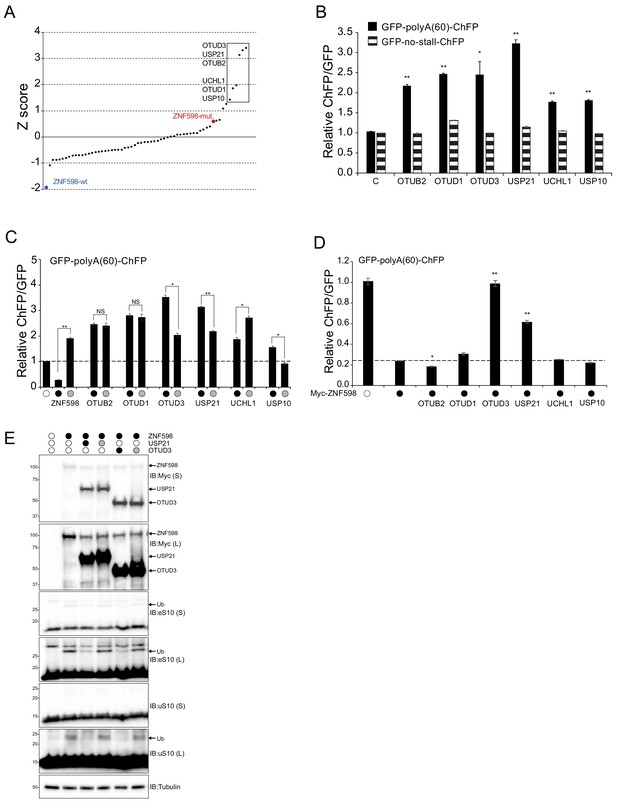
Identification of deubiquitylating enzymes that allow for readthrough of poly(A)-mediated ribosome stalls.
(A) 60 human deubiquitylating enzyme (Dub) expression plasmids were individually co-transfected with the poly(A)-stall reporter plasmid and the resulting ChFP and GFP fluorescence intensities were measured by flow cytometry. The Z-score value for each Dub is depicted (n = 1). The six Dubs with the highest Z-score are boxed to indicate candidate Dubs that induce increased readthrough of poly(A)-stall reporter. Expression of wild type (blue dot) and catalytically inactive (red dot) ZNF598 are shown as controls. (B) The ChFP:GFP ratio from cells transfected with the poly(A)-stall reporter (solid bars) or a reporter containing no stall sequence (striped bars), along with expression plasmids for the indicated Dubs relative to a control plasmid. Error bars denote SEM for triplicate transfections. **p<0.0001, *p<0.01 using Student’s t-test comparing Dub expression to control transfections. (C) The ChFP:GFP ratio from cells transfected with expression plasmids for either wild type (black circle) or catalytically inactive (gray circle) Dubs and the poly(A)-stall reporter relative to a control plasmid. Control transfections with the poly(A) reporter plasmid alone are indicated by the open circle. Error bars denote SEM for triplicate transfections. **p<0.0001, *p<0.001 using Student’s t-test comparing the wild type to the catalytically inactive mutant for each Dub or ZNF598. NS = not significant. (D) The ChFP:GFP ratio from cells transfected with the poly(A)-stall reporter plasmid and ZNF598 alone or in combination with the indicated wild type Dubs (black circle) relative to a control plasmid. Control transfections with the poly(A) reporter plasmid alone are indicated by the open circle. Error bars denote SEM for triplicate transfections. **p<0.0001, *p<0.01, using Student’s t-test comparing wild type ZNF598 alone to samples with ZNF598 and the indicated Dub. (E) Whole-cell extracts from ZNF598- knockout (KO) cells transfected with expression plasmids for wild type (black circles) ZNF598, USP21, or OTUD3 and their respective inactive mutants (gray circles) were analyzed by SDS-PAGE and immunoblotted using the indicated antibodies. The ubiquitin-modified ribosomal protein is indicated by the arrow. S and L denote short and long exposures (n = 1).
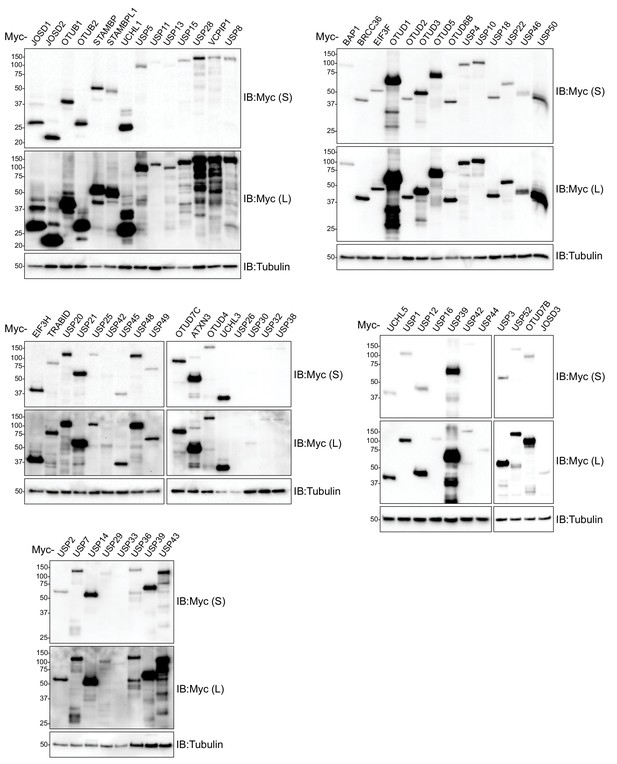
Validation of human Dub expression plasmids.
Whole-cell extracts from cells transiently transfected with the indicated Myc-tagged Dub expression plasmids were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies. S and L denote short and long exposures (representative immunoblots (n = 2)).
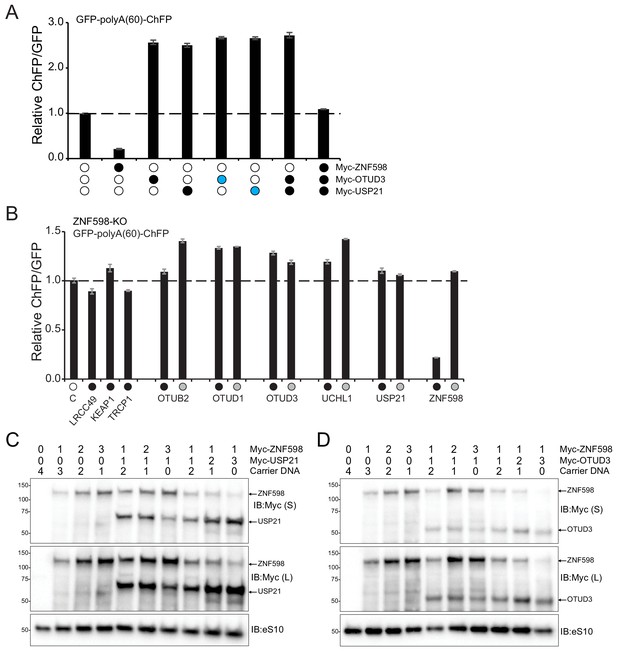
OTUD3 and USP21 enhance poly(A)-stall readthrough in a non-synergistic and ZNF598 dependent manner.
(A) The ChFP:GFP ratio from 293T cells transfected with the poly(A)-stall reporter plasmid and expression plasmids for USP21, OTUD3 or wild type ZNF598 alone (black circle) or with twice the amount of an individual expression plasmid (blue circle) relative to control transfection. Error bars denote SEM for triplicate transfections. (B) The ChFP:GFP ratio from ZNF598 knockout (KO) cells transfected with the indicated expression plasmids and the poly(A)-stall reporter. Wild type (black circles) or inactive (gray circles) versions of each candidate Dub, ZNF598 or control plasmid is indicated. Error bars denote SEM for triplicate transfections. (C,D) Whole-cell extracts from cells transfected as indicated in Figure 4F and G were analyzed by SDS-PAGE and immunoblotted for the indicated antibodies. S and L denote short and long exposures (representative immunoblots (n = 2)).
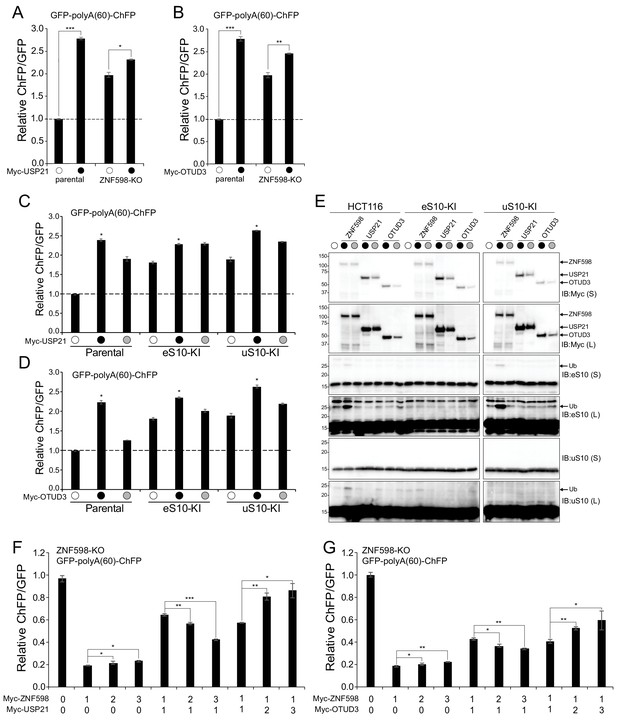
USP21 and OTUD3 antagonize ZNF598-mediated RRub events.
(A,B) Parental HCT116 cells and ZNF598 knock-out (KO) cells were transfected with USP21 (A) or OTUD3 (B) expression plasmids and the poly(A)-stall reporter (black circles) or the poly(A)-stall reporter alone (open circles). Fluorescence intensities were measured by flow cytometry and the relative ChFP:GFP ratio is depicted. Error bars denote SEM for triplicate transfections. ***p<0.0001, **p<0.001, *p<0.05, using Student’s t-test comparing Dubs to control transfection. (C,D) The ChFP:GFP ratio from parental HCT116 cells or point mutant knock-in (KI) eS10 or uS10 cell lines transfected with the poly(A)-stall reporter alone (open circles) or with expression plasmids for wild type (black circles) or inactive mutant (gray circles) USP21 (C) or OTUD3 (D) relative to control transfections in the indicated cell lines. Error bars denote SEM for triplicate transfections. *p<0.0001 using Student’s t-test comparing wild type Dub transfections to control transfection in the indicated cell lines. (E) Whole-cell extracts from cells transfected as indicated in panels C and D were analyzed by SDS-PAGE and immunoblotted for the indicated antibodies. Black and gray circles denote expression of wild type or inactive versions, respectively. The ubiquitin-modified ribosomal protein is indicated by the arrow. S and L denote short and long exposures (n = 1). (F,G) The ChFP:GFP ratio from HCT116 ZNF598 knockout (KO) cells transfected with increasing amounts of plasmid DNA for either wild type ZNF598 and USP21 (F) or OTUD3 (G) and the poly(A)-stall reporter. Numbers indicate the ratio of transfected DNA for each plasmid. Error bars represent SEM of triplicate replicates. ***p<0.0001, **p<0.001, *p<0.05 using Student’s t-test comparing the different ZNF598 to Dub DNA ratios as indicated.
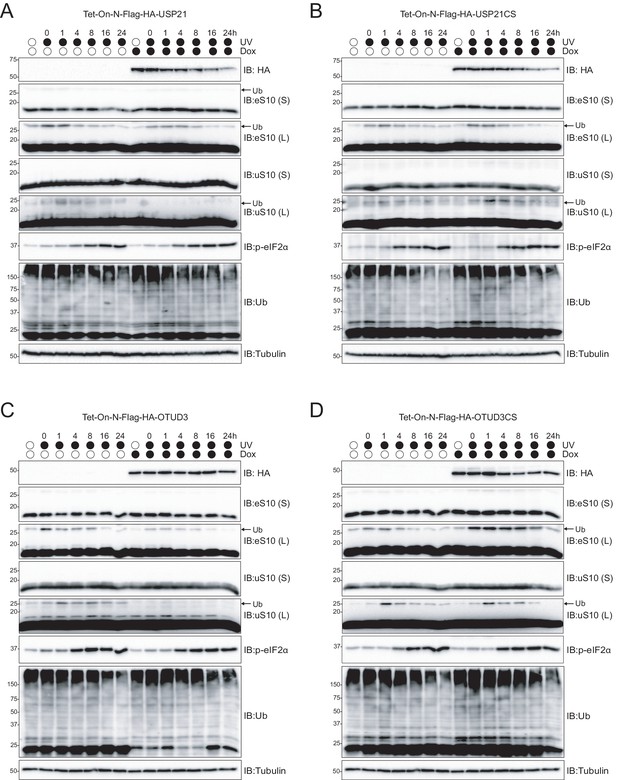
USP21 and OTUD3 expression accelerates RRub demodification following UV exposure.
(A–D) Cells with stable inducible expression of wild type USP21 (A), OTUD3 (C) or inactive versions (B, D, respectively) were induced or uninduced with doxycycline (Dox, 2 μg/ml) for 16 hr followed by UV exposure. Whole cell extracts from cells collected at the indicated times after UV exposure were analyzed by SDS-PAGE and immunoblotted using the indicated antibodies. The ubiquitin-modified ribosomal protein is indicated by the arrow. S and L denote short and long exposures (n = 1).
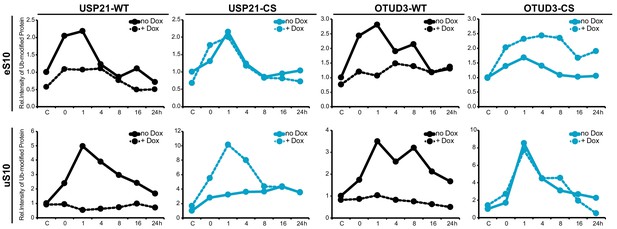
Quantification of site-specific RRub demodification upon Dub overexpression.
Quantification of the relative amount of ubiquitylated eS10 and uS10 during the UV treatment time course from the immunoblots in Figure 5A–D.
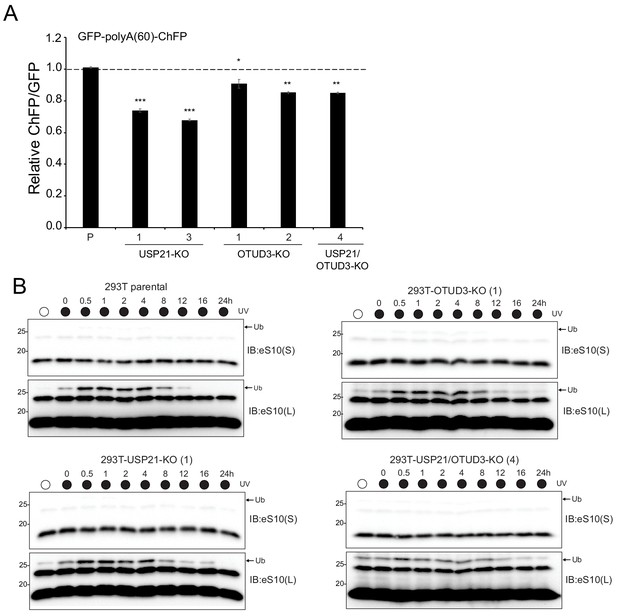
Loss of USP21 or OTUD3 expression results in enhanced ribosome stalling on poly(A) sequences and delayed eS10 ubiquitylation following RQC activation.
(A) Parental 293 T cells (P), USP21 knockout (KO) cells, OTUD3-KO cells and the combined double-KO cells were transfected with the poly(A)-stall reporter. Fluorescence intensities were measured by flow cytometry and the relative ChFP:GFP ratio is depicted. Numbers represent distinct knockout clones for OTUD3 or USP21. Error bars denote SEM for triplicate transfections. ***p<0.0001, **p<0.001, *p<0.05 using Student’s t-test comparing the different KO clones to the parental control transfection. (B) Parental 293T cells, USP21-KO (clone 1), OTUD3-KO (clone 1), and USP21/OTUD3 double-KO (clone 4) cells were exposed to UV and allowed to recover for the indicated times. Whole-cell extracts were analyzed by SDS-PAGE and immunoblotted with the indicated antibody. The ubiquitin-modified ribosomal protein is indicated by the arrow. S and L denote short and long exposures (representative immunoblots shown (n = 2).
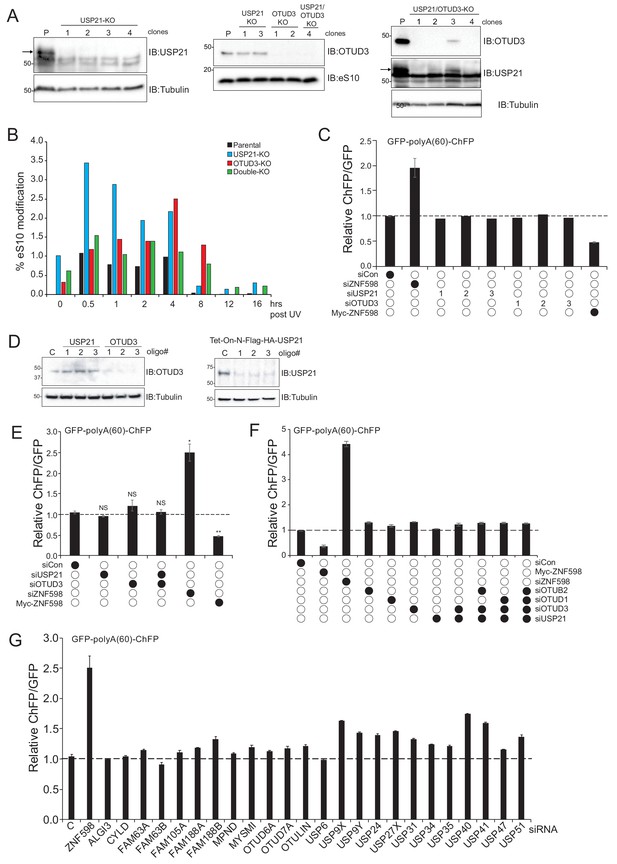
Knockdown of OTUD3 or USP21 does not result in enhanced resolution of poly(A)-induced RQC.
(A) Whole-cell extracts from USP21 knockout (KO), OTUD3 KO, or USP21/OTUD3 double KO cell lines were generated and analyzed by SDS-PAGE and immunoblotted with the indicated antibodies. The numbers represent individual cell line clones. The band for USP21 is indicated by the arrow. S and L denote short and long exposures (representative immunoblots of n = 2 independent experiments). (B) Quantification of the percent modification of eS10 during the UV treatment time course from the immunoblots in Figure 6B. (C) The ChFP:GFP ratio from cells transfected with either control siRNA oligos or siRNA oligos targeting OTUD3, USP21 or ZNF598 followed by poly(A)-stall reporter transfection. Numbers represent distinct siRNA oligos used to target OTUD3 or USP21. Error bars denote SEM for triplicate transfections. (D) Parental 293T (top) or inducible 293 cells expressing Flag-HA-tagged USP21 (bottom) were transfected with either control siRNA oligos or three separate siRNA oligos targeting USP21 or OTUD3. Whole-cell lysates were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies (n = 1). (E) The ChFP:GFP ratio from cells transfected with control siRNA oligos or siRNA oligos targeting USP21, OTUD3, or ZNF598 as indicated followed by poly(A)-stall reporter transfection. Error bars denote SEM for triplicate transfections. **p<0.001, *p<0.01, using Student’s t-test comparing the different siRNA knockdowns or wild type ZNF598 overexpression to control siRNA. (F) The ChFP:GFP ratio from cells transfected with control siRNA oligos or siRNA oligos targeting OTUB2, OTUD1, OTUD3, USP21 or ZNF598 individually or in combination followed by poly(A)-stall reporter transfection. Error bars denote SEM for triplicate transfections. (G) The ChFP:GFP ratio from cells transfected with control siRNA oligos or pools of siRNA oligos (four oligos each) targeting 24 individual Dubs or ZNF598 followed by poly(A)-stall reporter transfection. Error bars denote SEM for triplicate transfections.
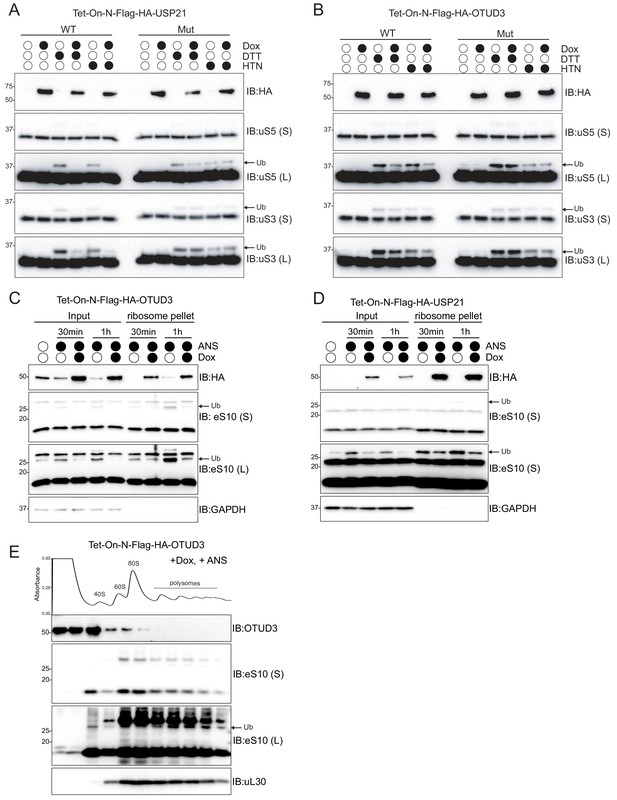
OTUD3 preferentially demodifies RQC RRub sites and is present within ribosome enriched fractions.
(A,B) Cells with stable inducible expression of wild type USP21 (A), or OTUD3 (B) were induced or uninduced with doxycycline (2 μg/ml) for 16 hr and then treated with dithiothreitol (DTT, 5 mM) or harringtonine (HTN, 2 μg/mL). Whole-cell extracts were analyzed by SDS-PAGE and immunoblotted using the indicated antibodies. The ubiquitin-modified ribosomal protein is indicated by the arrow. S and L denote short and long exposures (n = 1). (C,D) Cells with stable inducible expression of wild type OTUD3 or USP21 were induced or uninduced and then treated with anisomycin (ANS 5 μg/ml) for the indicated times. Ribosomes were pelleted through a sucrose cushion and whole-cell extracts (input) and pelleted material were analyzed by SDS-PAGE and immunoblotted using the indicated antibodies. The ubiquitin-modified ribosomal protein is indicated by the arrow. S and L denote short and long exposures (n = 1). (E) Cells with stable inducible expression of wild type OTUD3 were induced for 16 hr followed by ANS (5 μg/ml) treatment for 1 hr. Whole-cell extracts were separated by sucrose density gradient centrifugation and fractions were collected. The UV absorbance across the fractions is depicted above the immunoblots. Individual fractions were TCA precipitated and analyzed by SDS-PAGE and immunoblotted using the indicated antibodies. The ubiquitin-modified ribosomal protein is indicated by the arrow. S and L denote short and long exposures (n = 1).
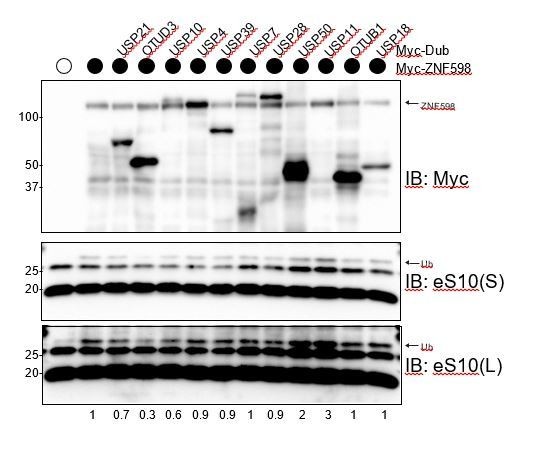
Specific Dub overexpression is required to antagonize ZNF598-mediated eS10 ubiquitylation.
Myc-tagged ZNF598 was expressed alone or in combination with the indicated myc-tagged Dubs and whole cell extracts were blotted for myc or eS10. The intensity of the ubiquitylated eS10 band (normalized to unmodified eS10 intensity) relative to ZNF598 expression alone is depicted under the immunoblots.
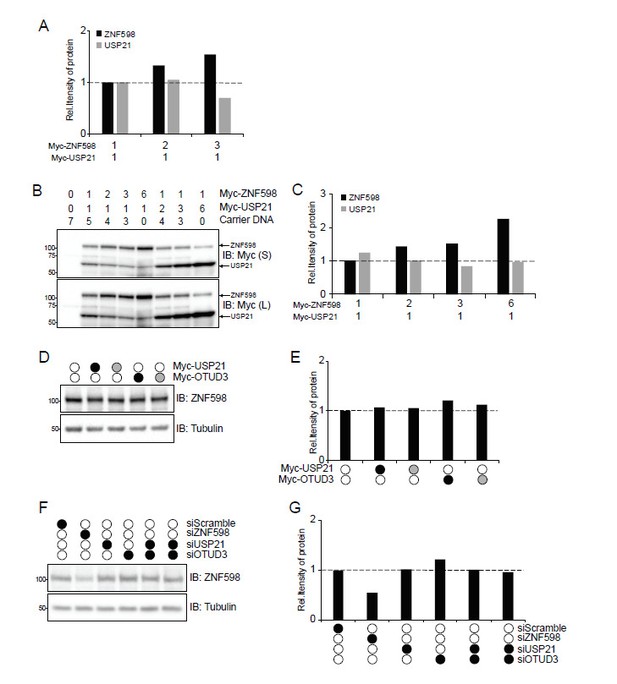
Analysis of ZNF598 protein levels upon Dub overexpression.
(A) Quantification of the relative amount of Myc-tagged ZNF598 or USP21 from immunoblots in Figure 3—figure supplement 2C. (B) Whole-cell extracts from HCT116 ZNF598 knock-out (KO) cells transiently co-transfected with increasing amounts of plasmid DNA for either wild type ZNF598 or USP21. Numbers indicate the ratio of transfected DNA for each plasmid. Extracts were analyzed by SDS-PAGE and immunoblotted for the indicated antibodies. S and L denote short and long exposures, respectively. (C) Quantification of the relative amount of Myc-tagged ZNF598 or USP21 from immunoblots in panel B. (D) HCT116 cells were co-transfected with expression plasmids for wild type (black circles) USP21, or OTUD3 and their respective catalytically-inactive mutants (grey circles). Cells were exposed to UV and allowed to recover for 4h. Whole-cell extracts were analyzed by SDS-PAGE and immunoblotted using the indicated antibodies. (E) Quantification of the relative amount of endogenous ZNF598 from immunoblot in panel D. (F) 293T cells were transfected with either control siRNA oligos or siRNA oligos targeting OTUD3, USP21 or ZNF598 as indicated. Whole-cell extracts were analyzed by SDS-PAGE and immunoblotted using the indicated antibodies. (G) Quantification of the relative amount of endogenous ZNF598 from immunoblot in panel F.
Additional files
-
Supplementary file 1
Key resources table.
- https://cdn.elifesciences.org/articles/54023/elife-54023-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/54023/elife-54023-transrepform-v2.docx





