Nodal and planar cell polarity signaling cooperate to regulate zebrafish convergence and extension gastrulation movements
Figures
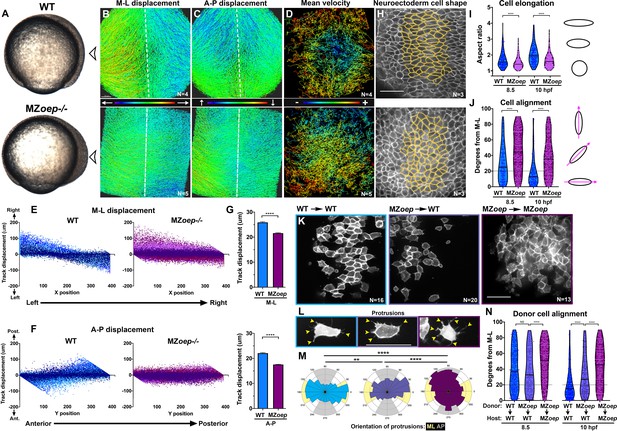
Nodal signaling regulates convergence and extension gastrulation cell behaviors.
(A) Bright-field images of live WT and MZoep–/– embryos at 80% epiboly (8.5 hpf). The arrowheads indicate the point of view (dorsal side) for all fluorescent confocal micrographs. (B–D) Representative images of automated tracking of fluorescently labeled nuclei in the dorsal hemisphere of WT (top) and MZoep–/– (bottom) gastrulae. Tracks represent cell movements over three hours of time-lapse confocal imaging, beginning at 8.5 hpf, and are colored according to their displacement in the mediolateral (B) and anteroposterior (C) dimensions or the mean velocity of cell movement (D). Dotted lines indicate dorsal midline. (E, F) Displacement of cell tracks in the mediolateral (E) and anteroposterior (F) dimensions in WT (blue) and MZoep–/– (purple) gastrulae [as shown in (B–D]). Each dot represents a single cell track, each color represents an individual embryo, N = 4 WT and 5 MZoep–/–. (G) Absolute displacement of cell tracks in ML (top) and AP (bottom) dimensions. Bars are mean with 95% confidence interval, p<0.0001, Kolmogorov-Smirnoff (K-S) tests. (H) Representative images of membrane-labeled neuroectoderm in live WT (top) and MZoep–/– (bottom) gastrulae with cells outlined in yellow. (I, J) Neuroectoderm cell elongation (I) and alignment (J) at 8.5 hpf (left) and 10 hpf (right). Each dot represents a single cell, black bars are mean values in (I), and median values in (J). N = 3 embryos of each genotype, p<0.0001, Mann-Whitney test in (I), K-S test in (J). (K) Representative images of membrane-labeled donor cells of the indicated genotypes within the neuroectoderm of unlabeled host gastrulae. N indicates the number of embryos analyzed from three independent trials. (L) Representative images of protrusions (arrowheads) made by transplanted neuroectoderm cells of the genotypes/conditions indicated in (K). (M) The orientation of all protrusions between 8.5 and 10 hpf is shown in radial histograms divided into 20° bins, with 0 and 180 representing the ML axis. Yellow and gray quadrants represent ML- and AP-oriented protrusions, respectively. **, p=0.0053; ****, p<0.0001; Chi-square. (N) Alignment of donor cells as in (J). The number of embryos in each condition is indicated in the corresponding panels in (K). Anterior is up in all images, scale bars are 50 μm. Dotted lines in (J, N) show 20 degrees from ML for reference.
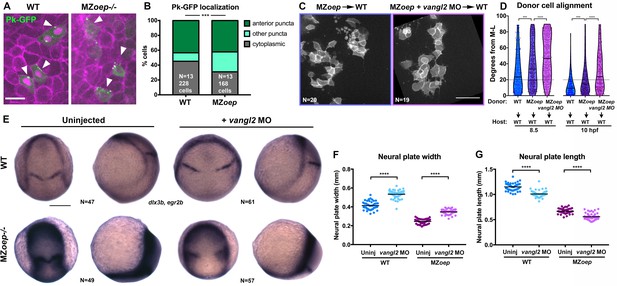
PCP signaling is active and contributes to C and E in Nodal signaling mutants.
(A) Representative images of transplanted Prickle (Pk)-GFP donor cells (co-expressing H2B-RFP) within the neuroectoderm of membrane-labeled WT and MZoep–/– host gastrulae. Arrowheads indicate puncta at anterior edges. (B) Pk-GFP localization in the genotypes indicated. N indicates the number of embryos and cells analyzed for each condition from four independent trials, p<0.001, Chi-square test. (C) Representative images of membrane-labeled MZoep–/– donor cells without (left) and with (right) 2 ng MO4-vangl2 transplanted into the neuroectoderm of unlabeled host gastrulae from five independent trials. (D) Donor cell alignment as in Figure 1. The number of embryos in each condition is indicated on the corresponding panels in (C), WT→WT control N = 10. ***, p<0.001; ****, p<0.0001; K-S tests. (E) Whole mount in situ hybridization (WISH) for dlx3b and egr2b in WT (top) and MZoep–/– (bottom) gastrulae at 9.5 hpf, uninjected or injected with 2 ng MO4-vangl2. Dorsal views on the left, lateral views on the right. (F, G) Width (F) and length (G) of neural plates in the embryos depicted in (E). Each dot represents a single embryo, black bars are mean values. Number of embryos in each condition is indicated on the corresponding panel in (E), p<0.0001, Unpaired T-tests. Anterior is up in all images, scale bar is 20 μm in (A), 50 μm in (C), and 200 μm in (E).
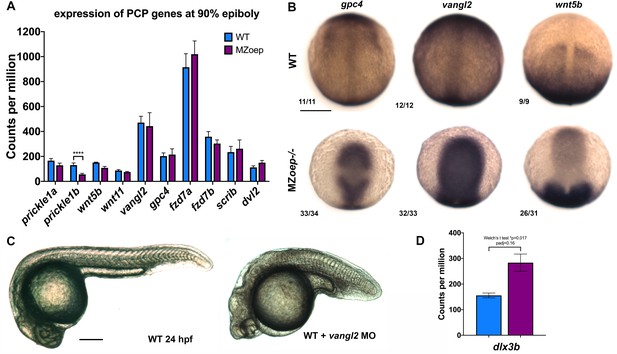
PCP signaling in Nodal-deficient embryos.
(A) Expression of PCP signaling genes in WT and MZoep–/– gastrulae at 90% epiboly stage (~9 hpf) as measured by RNA-seq from three biological replicates with 50 pooled embryos each. Bars are mean with SD. ****padj = 4.02−6. (B) Representative images of WISH for the transcripts indicated in WT (top) and MZoep–/– (bottom) gastrulae at 9.5 hpf. Fractions indicate the number of embryos with the pictured phenotype over the number of embryos examined. (C) Representative images of live WT embryos uninjected (left) or injected with 2 ng MO4-vangl2 (right) at 24 hpf. (D) Expression of dlx3b in WT and MZoep–/– gastrulae as measured by RNA-seq. p=0.017, Welch’s T-test, padj = 0.16. Anterior is up in (B), to the left in (C). Scale bars are 200 μm.
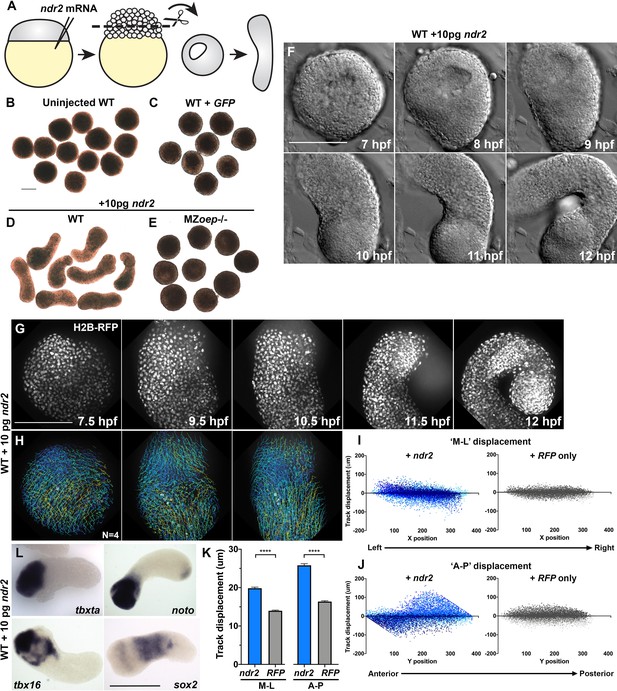
Nodal ligands promote ex vivo C and E of blastoderm explants.
(A) Diagram of injection and explantation of zebrafish embryos. (B–E) Representative bright-field images of live blastoderm explants of the indicated conditions/genotypes at the equivalent of the 2–4 somite stage. (F) Time-lapse DIC series of a representative explant from a WT embryo injected with 10 pg ndr2 RNA. (G, H) Time-lapse series of H2B-RFP labeled nuclei (G) and automated cell tracking (H) within a representative explant from a WT embryo injected with 10 pg ndr2 RNA. Tracks represent cell movements over 3.5 hr of time-lapse confocal imaging beginning at 7.5 hpf and are colored according to mean track displacement. (I, J) Displacement of cell tracks in the ‘mediolateral’ (I) and ‘anteroposterior’ (J) dimensions in explants from ndr2-injected (blue) and control RFP-injected (gray) WT embryos (as in Figure 1). Each dot represents a single cell track, each color represents an individual explant. N = 4 explants of each condition from two independent trials. (K) Absolute displacement of cell tracks in the ML and AP dimensions. (L) Representative images of WISH for the transcripts indicated in explants from WT embryos injected with 10 pg ndr2 RNA. Scale bars are 200 μm.
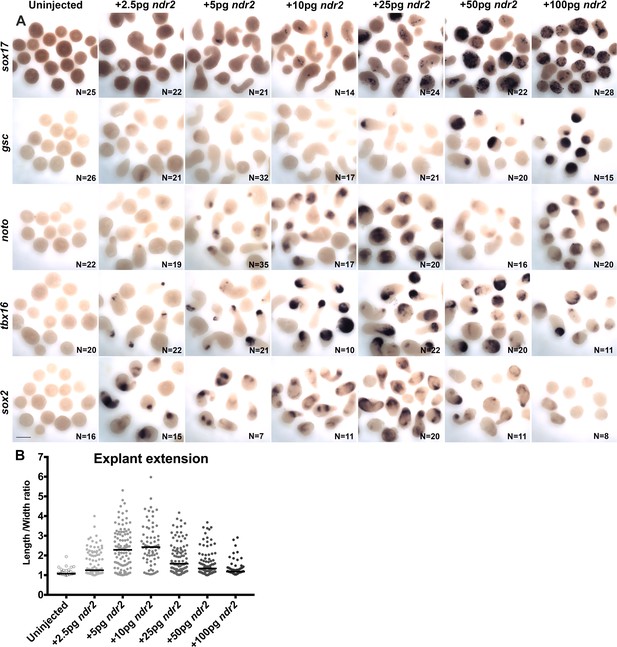
Nodal ligand levels regulate cell fate and extension of explants.
(A) Representative images of WISH for the transcripts indicated in explants from WT embryos injected with 0–100 pg ndr2 RNA and fixed at the equivalent of the 2–4 somite stage from two independent trials. (B) Length/width ratios of the explants depicted in (A). Each dot represents a single explant, black bars are median values. Scale bar is 200 μm.

Cell divisions within ndr2-expressing explants.
(A) Manually annotated cell divisions (per explant) within four live explants from WT embryos co-injected with H2B-RFP and 10 pg ndr2 RNA. Measurements were taken at 5 min intervals from 7.5 to 11 hpf. Boxes represent the 25th to 75th percentiles, whiskers are minimum and maximum vales, bars are median values. (B) Left: representative image of cell divisions (arrowheads) within an explant co-expressing H2B-RFP and ndr2. The inset is enlarged from the region in the yellow square. Right: radial histogram of locations of all cell divisions (with respect to the center of each explant) detected within four ndr2-expressing explants between 7.5 and 11 hpf, with 90°/270° representing the axis of extension.
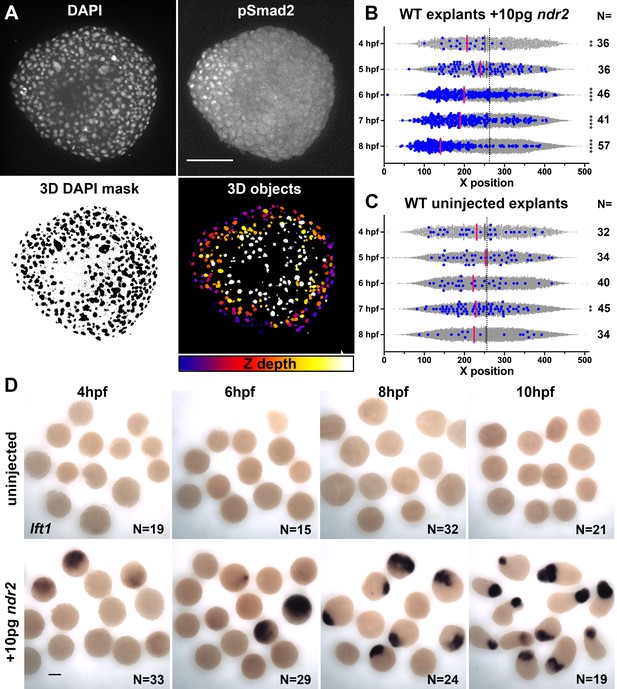
Nodal-expressing explants exhibit asymmetric Nodal signaling activity.
(A) Representative confocal images of immunofluorescent staining for phosphorylated Smad2 and DAPI-labeled nuclei in 8hpf explants from WT embryos injected with 10 pg ndr2. DAPI z-stacks were used to create a three-dimensional mask from which nuclear pSmad intensities were detected and measured in an automated fashion. (B, C) Axis position of pSmad2-positive nuclei (blue) and all nuclei (gray) in explants from WT embryos injected with 10 pg ndr2 (B) or uninjected (C) at the time points indicated. Each dot represents a single nucleus, pink bars are median values among pSmad2+ nuclei. N indicates the number of explants in each condition from five independent trials. Kolmogorov-Smirnov tests were used to compare the distribution of pSmad+ nuclei to all nuclei; ****, p<0.0001; **, p<0.01. (D) Representative images of WISH for lefty1 in uninjected (top) and ndr2-injected (bottom) explants fixed at the time points indicated. Scale bars are 100 μm.
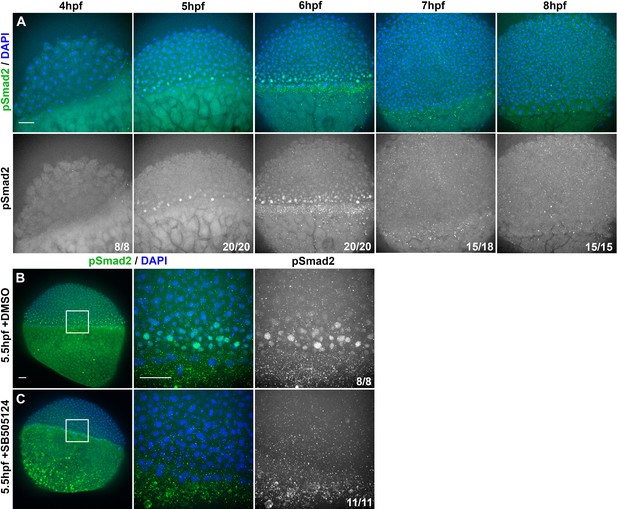
Nodal signaling activity in intact embryos.
(A) Representative confocal z-projections of immunofluorescent (IF) staining for phosphorylated Smad2 (bottom) overlaid with DAPI-labeled nuclei (top) in WT embryos fixed at the time points indicated. (B) Representative confocal z-projections of pSmad2 IF and DAPI staining in 5.5 hpf WT embryos treated with DMSO (top) or SB505124 (bottom), starting at 3 hpf. Panels to the right are magnified views of the regions within the white squares. Fractions indicate the number of embryos with the depicted phenotype over the total number of embryos examined for each condition and/or timepoint. The animal pole is at the top of all images. Scale bars are 50 μm.
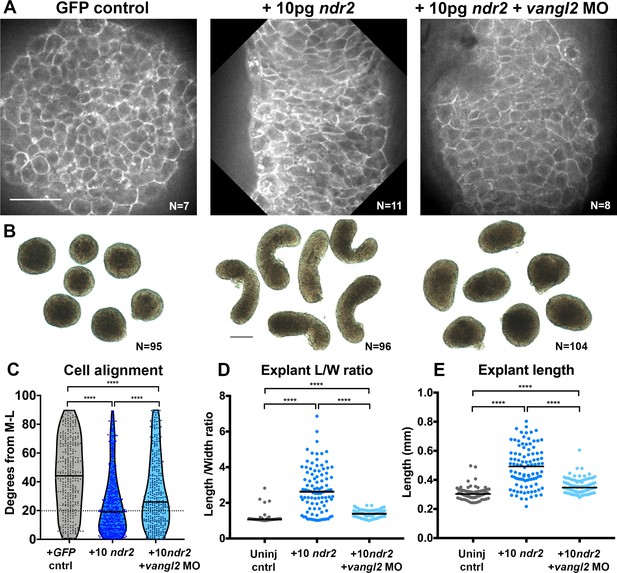
Disrupted PCP reduces Nodal-induced cell polarization and C and E within explants.
(A) Representative confocal micrographs of live membrane-labeled explants of the indicated conditions at the equivalent of the 2–4 somite stage from two independent trials. (B) Representative bright-field images of blastoderm explants at 2–4S from four independent trials. (C) Explant cell alignment as in Figure 1. The number of explants in each condition is indicated on the corresponding panel in (A). Mediolateral (ML) is defined as orthogonal to the axis of extension. (D, E) Length/width ratios (D) and length (E) of explants depicted in (B). Each dot represents a single explant, black bars are median values. The number of explants in each condition is indicated on the corresponding panel in (B). p<0.0001; Kruskal-Wallis test. Scale bar is 50 μm in (A), 200 μm in (B).
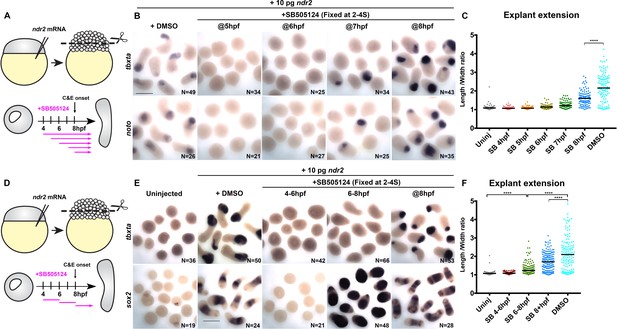
Nodal promotes ex vivo C and E independent of mesoderm.
(A) Diagram of the time course of SB-505124 (SB) treatment of ndr2-expressing explants. (B) Representative images of WISH for the transcripts indicated in explants from WT embryos injected with 10 pg ndr2 RNA, treated with SB at the indicated time points, and fixed at the equivalent of the 2–4 somite stage from four independent trials. (C) Length/width ratios of explants shown in (B). Each dot represents a single explant, black bars are median values; p<0.0001, Mann-Whitney test. (D) Diagram of the time course of SB treatment of ndr2-expressing explants followed by washout. (E) Representative images of WISH for the indicated transcripts in explants from WT embryos injected with 10 pg ndr2 RNA, treated with SB at the indicated time points, and fixed at the equivalent of the 2–4 somite stage from four independent trials. (F) Length/width ratios of explants shown in (E), as in panel (C). ****p<0.0001, Mann-Whitney test. Scale bars are 300 μm.
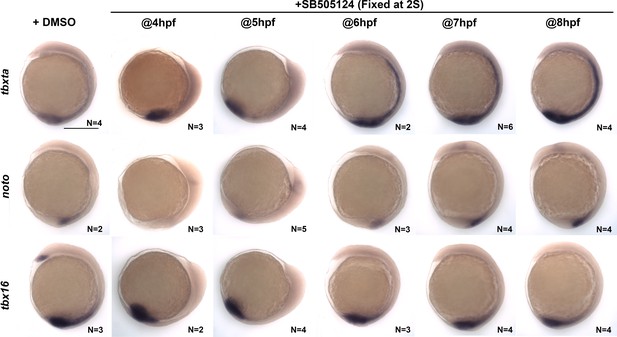
Nodal inhibitor treatment of intact embryos.
Representative images of WISH for the indicated transcripts in WT embryos treated with DMSO or SB-505124 beginning at 4–8 hpf and fixed at the two-somite stage. The scale bar is 300 μm.
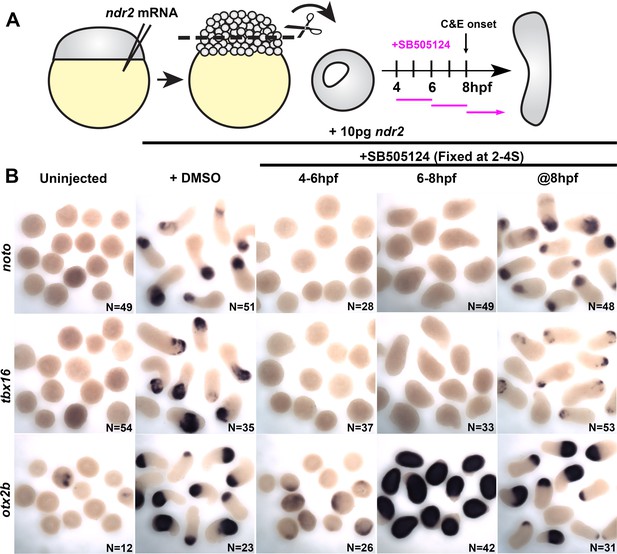
Time-course of Nodal inhibition in ndr2-expressing explants.
(A) Diagram of the time course of SB-505124 (SB) treatment of ndr2-expressing explants, as in Figure 6. (B) Representative images of WISH for the transcripts indicated in explants from WT embryos injected with 10 pg ndr2 RNA, treated with SB at the indicated time points, and fixed at the equivalent of the 2–4 somite stage from four independent trials.
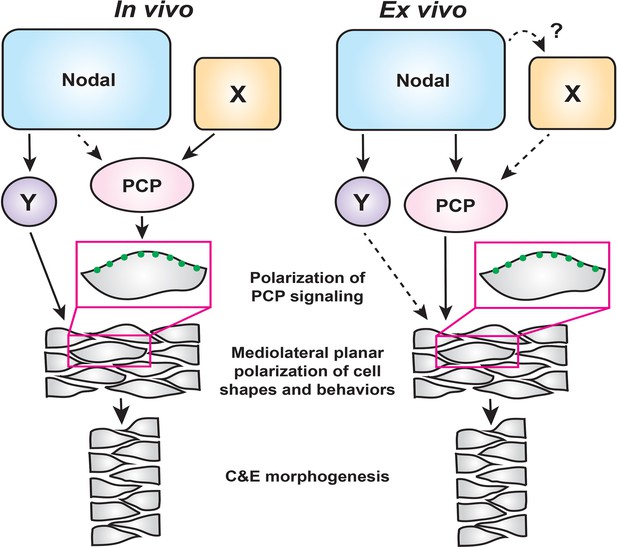
A model for the roles of PCP and Nodal signaling in C and E gastrulation movements.
In intact embryos (left), Nodal signaling acts largely in parallel with PCP signaling to regulate the ML cell polarization that underlies C and E. PCP signaling activity and localization of its components are regulated by an additional unknown signal(s) (X), and maintains residual polarizing activity in the absence of Nodal. In embryonic explants (right), PCP signaling activity and C and E cell behaviors are regulated wholly downstream of Nodal signaling.
Videos
Automated tracking of fluorescent nuclei in live zebrafish gastrulae.
Time-lapse confocal series from approximately 8.5 to 11.5 hpf in representative WT (left) and MZoep–/– (right) gastrulae injected with H2B-RFP RNA. Cell tracks shown below are colored according to track displacement, with warmer colors indicating higher displacement.
Ex vivo extension of zebrafish blastoderm explants.
Time-lapse differential interference contrast (DIC) series from 7 hpf to 12.5 hpf of representative explants from an uninjected WT embryo (left) and a WT embryo injected with 10 pg ndr2 RNA (right).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Danio rerio) | tdgf1 (oep) | ZFIN | RRID:ZFIN_ZDB-GENE-990415-198 | |
| Gene (Danio rerio) | ndr2 (cyc) | ZFIN | RRID:ZFIN_ZDB-GENE-990415-181 | |
| Strain, strain background (Danio rerio) | AB* | ZIRC | RRID:ZFIN_ZDB-GENO-960809-7 | |
| Genetic reagent (Danio rerio) | oeptz257 | Hammerschmidt et al., 1996 | RRID:ZFIN_ZDB-GENO-130130-2 | Point mutation |
| Recombinant DNA reagent | pJZoepFlag1-2 in pcDNA3 (plasmid) | Zhang et al., 1998 | Template for in vitro transcription | |
| Recombinant DNA reagent | ndr2 in PCS2+ (plasmid) | Sampath et al., 1998 | Template for in vitro transcription | |
| Recombinant DNA reagent | membrane Cherry in PCS2+ (plasmid) | Gift from Dr Fang Lin | Template for in vitro transcription | |
| Recombinant DNA reagent | membrane eGFP in PCS2+ (plasmid) | Wallingford and Harland, 2002 | Template for in vitro transcription | |
| Recombinant DNA reagent | H2B-RFP in PCS2 (plasmid) | Gift from Dr John Wallingford | Template forin vitro transcription | |
| Recombinant DNA reagent | Drosophila Prickle-GFP (plasmid) | Jenny et al., 2003 | Template for in vitro transcription | |
| Antibody | Anti-phospho Smad2/3 | Cell Signaling Technology #8828 | RRID:AB_2631089 | IF (1:1000) |
| Antibody | Invitrogen AlexaFluor 488 goat anti-rabbit IgG | Thermo Fisher #A-11008 | RRID:AB_143165 | IF (1:1000) |
| Antibody | Roche Anti-digoxigenin-AP Fab fragments | Millipore Sigma #11093274910 | RRID:AB_2734716 | 1:5000 |
| Commercial assay or kit | Roche Digoxigenin RNA labelling mix | Millipore Sigma #11277073910 | ||
| Other | Roche BM Purple AP staining solution | Millipore Sigma #11442074001 | ||
| Sequenced-based reagent | MO4-vangl2 Morpholino antisense oligonucleotide | GeneTools (Williams et al., 2012) | AGTTCCACCTTACTCCTGAGAGAAT | |
| Commercial assay or kit | Invitrogen mMessage mMachine SP6 kit | Thermo Fisher # AM1340 | ||
| Commercial assay or kit | RNeasy Mini kit | Qiagen #74104 | ||
| Chemical compound, drug | Ambion Trizol reagent | Thermo Fisher #15596018 | ||
| Chemical compound, drug | SB-505124 | Millipore Sigma # S4696 | 50 mM | |
| Peptide, recombinant protein | Roche Pronase | Millipore Sigma #10165921001 | ||
| Other | New-born calf serum | Invitrogen #26010–066 | ||
| Software, algorithm | Imaris | Oxford Instruments | RRID:SCR_007370 | Live cell tracking |
| Software, algorithm | ImageJ/FIJI | ImageJ/FIJI | RRID:SCR_002285 | Image analysis |
| Software, algorithm | Prism 8 | Graphpad | RRID:SCR_002798 | Statistics and graphs |
Additional files
-
Source data 1
Differentially expressed genes between WT and MZoep–/– gastrulae detected by RNA-seq at the 90% epiboly stage.
Values shown in columns H through M are counts per million reads mapped (CPM) for each biological replicate.
- https://cdn.elifesciences.org/articles/54445/elife-54445-data1-v2.xlsx
-
Source data 2
CPM values for all genes within WT and MZoep–/– gastrulae detected by RNA-seq at the 90% epiboly stage.
- https://cdn.elifesciences.org/articles/54445/elife-54445-data2-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/54445/elife-54445-transrepform-v2.docx





