The evolutionary history and genomics of European blackcap migration
Figures
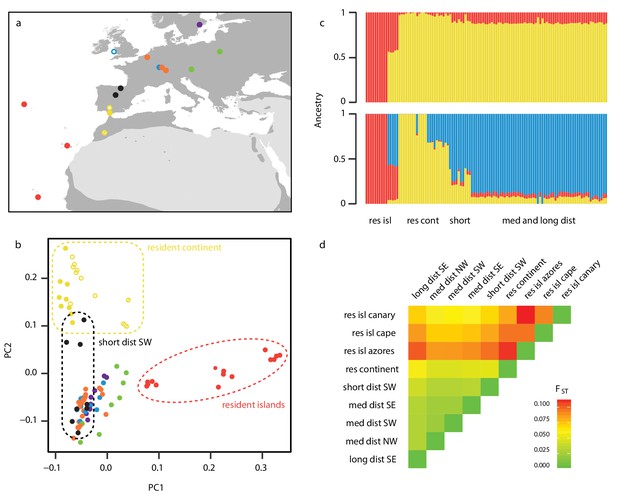
Sampling design and population structure.
(a) Sampling sites and migratory phenotypes. Samples were collected from the breeding grounds, except for a subset of NW migrants that were sampled during winter in the UK (open blue circle) (details in Supplementary file 4). (b–d) Population structure represented by a Principle Component Analysis (PCA) (b), NGSadmix (K = 2 and 3 shown) (c) and pairwise estimates of FST (d), showing differentiation between migrants and residents (as well as among residents themselves). Long dist SE = long distance migrants that orient SE in autumn (purple), med dist = medium distance migrants that orient in the corresponding heading during autumn migration (SE = green, SW = orange and NW = blue), res continent = residents found on the continent (yellow), short dist SW = short distance migrants that orient SW (black), res isl = resident birds on islands (cape = Cape Verde, canary = Canary Islands). Among continental residents, open circles indicate Cazalla de la Sierra, open circles with dash Asni, and filled circles Gibraltar. A PCA excluding islands can be found in Figure 1—figure supplement 1; results from NGSadmix at larger values of K can be found in Figure 1—figure supplement 2.
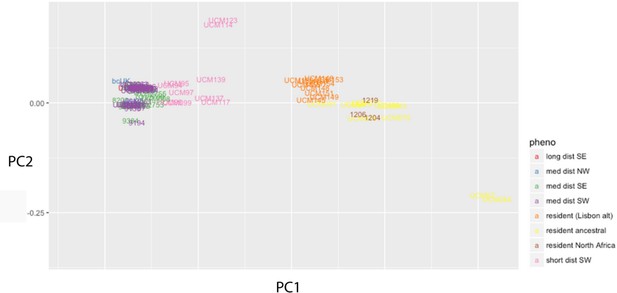
Principal component analysis matching that in Figure 1 but excluding island populations.
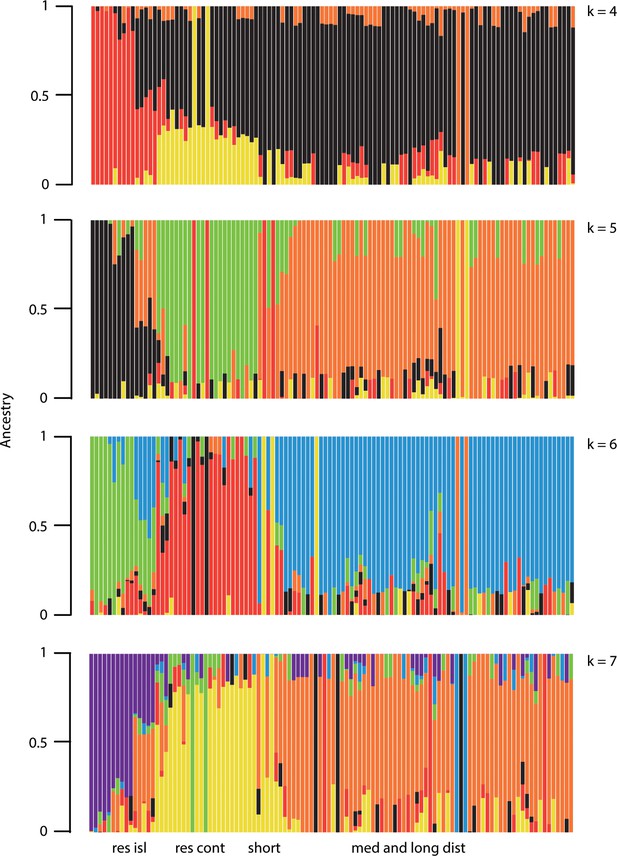
Complimentary figure to Figure 1c, showing ancestry proportions estimated by ADMIXTURE at larger cluster values (k = 4 through 7).
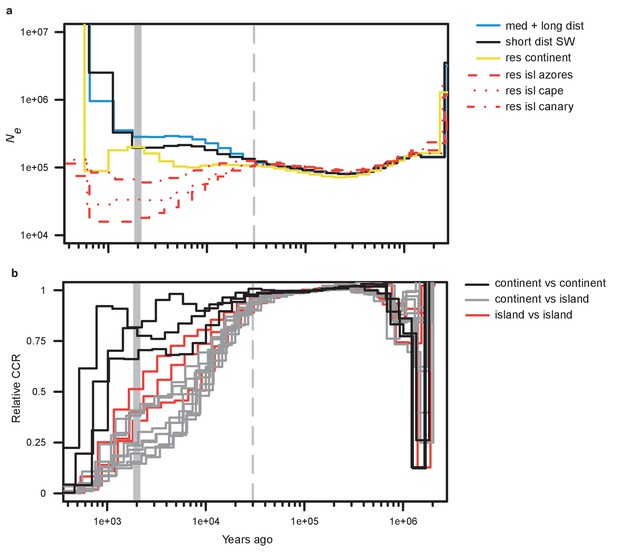
Demographic history.
(a) Effective population size by time estimated by MSMC2 using five individuals per blackcap phenotype. Note that the most recent time segment is regarded as being unreliable in MSMC2 results. (b) Relative cross-coalescence rate estimated by MSMC2. 15 lines with three colours indicate relative cross-coalescence rate for all pairwise combinations of the six populations (three for comparisons between populations on the continent [continent vs. continent], three for comparisons between populations on the islands [island vs. island], and nine for comparisons between continent and island populations [continent vs. island]). The dotted vertical line indicates the inferred time of population separation. Results from down-sampling can be found in Figure 2—figure supplements 1, 2 and 3; results for medium- and long-distance migrants run separately can be found in Figure 2—figure supplements 4, 5 and 6.
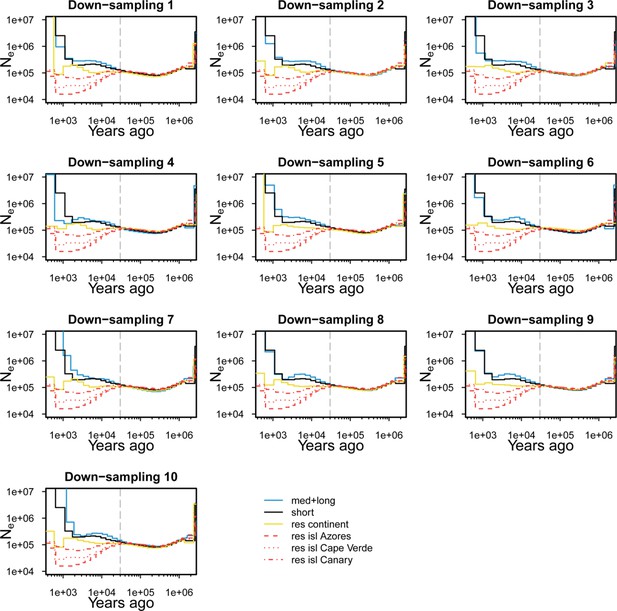
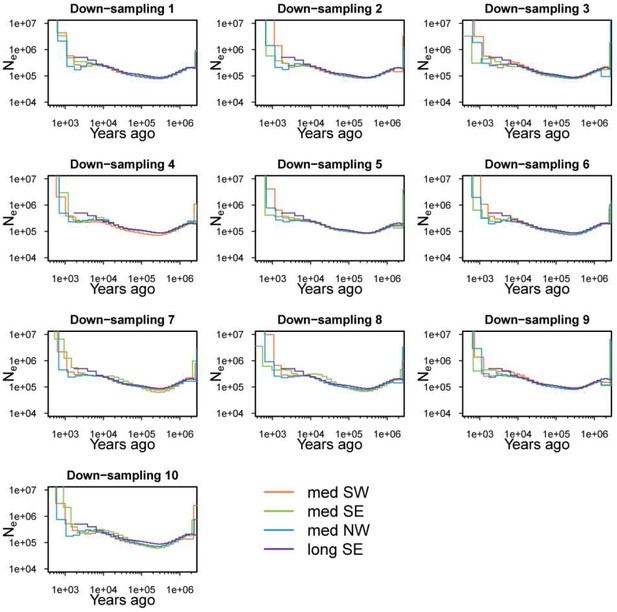
Down-sampling for demography analysis of effective population size.
Ten selections each containing five individuals were randomly sampled from the 44 med+long migrants and from the 19 continental residents (down-sampling 1 to 10). These selections were used for 10 runs of demography analysis with MSMC2. Because there were only five individuals for each of the other four groups (short, Azores, Cape Verde, and Azores), the same sample sets were used for all 10 runs of the demography analysis. The results of the 10 runs of demography analysis are shown separately. Note that demography estimates of three island populations (red) and short migrants (black) are same across the 10 panels.
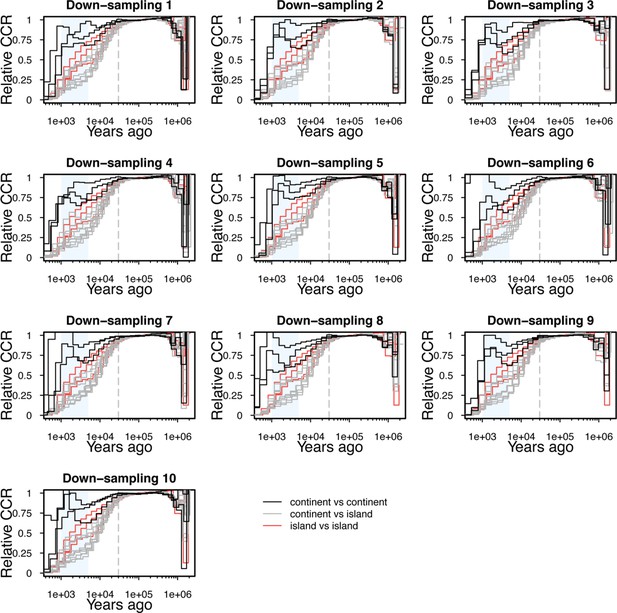
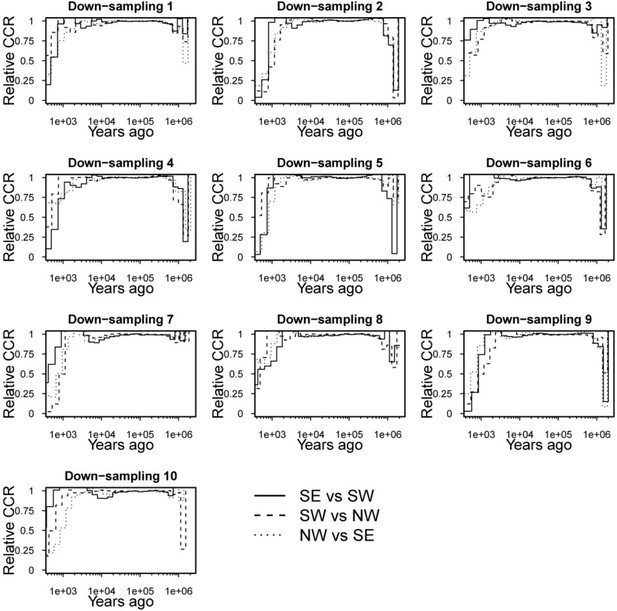
Down-sampling for demography analysis of relative cross-coalescence rate.
The same down-sampled individuals as those taken for effective population size analysis (Figure 2—figure supplement 1) were also used for down-sampling of relative cross-coalescence rate analysis. Although the exact inferences of relative cross-coalescence rate especially between two continental groups (continent vs continent, black) are variable across down-samplings, the general pattern of steeper decline of relative cross-coalescence rate between continental and island groups (continent vs island, grey) than that between continent vs continent is consistent across all 10 down-samplings. Note that some inferences (three of continent vs island and three (all) of island vs island) are same across the 10 down-samplings because the both two phenotypes had only five individuals (see Figure 2—figure supplement 3 ).
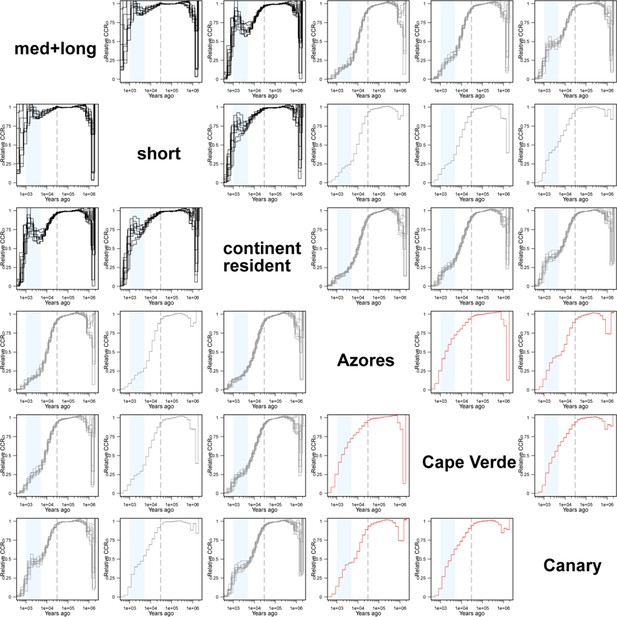
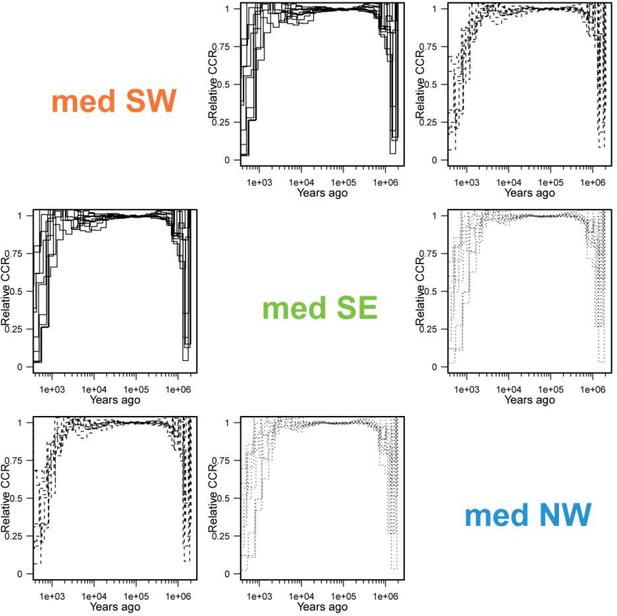
Demography analysis of relative cross-coalescence rate.
Relative cross-coalescence rate of all 15 possible combinations of six groups are shown. Each line represents the relative cross-coalescence rate inference of one down-sampling (five individuals per group). The three line colours correspond to those in Figure 2b and Figure 2—figure supplement 2. Relative cross-coalescence rate started to increase ~5000 years ago between med+long and continental resident populations (shaded with light blue). Note that there is only one inference for short vs islands (Azores, Cape Verde, Canary) and island vs island (Azores vs Cape Verde, Cape Verde vs Canary, Canary vs Azores) because there are only five individuals for these phenotypes. Also note that the top and bottom diagonals are identical.
Medium distance NW, SW and SE migrants and long distance migrants show similar demographic histories.
Effective population sizes show the same demographic trajectories. Five individuals were randomly sampled from each medium distance phenotype 10 times (down-sampling 1 to 10), and used for 10 runs of demography analysis with MSMC2. The results of the 10 runs are shown separately. Down-sampling was not done for long distance migrants as only two individuals met the coverage cutoff to be included in the analysis.
Medium distance NW, SW and SE migrants show similar demographic histories.
Relative cross-coalescence rates stay high in all three pairwise comparisons between medium distance migrants, suggesting no clear population split among medium distance migrants with different orientation.
Medium distance NW, SW and SE migrants show similar demographic histories.
Relative cross-coalescence rates stay high in all three pairwise comparisons between medium distance migrants, suggesting no clear population split among medium distance migrants with different orientation. Note that the top and bottom diagonals are identical.
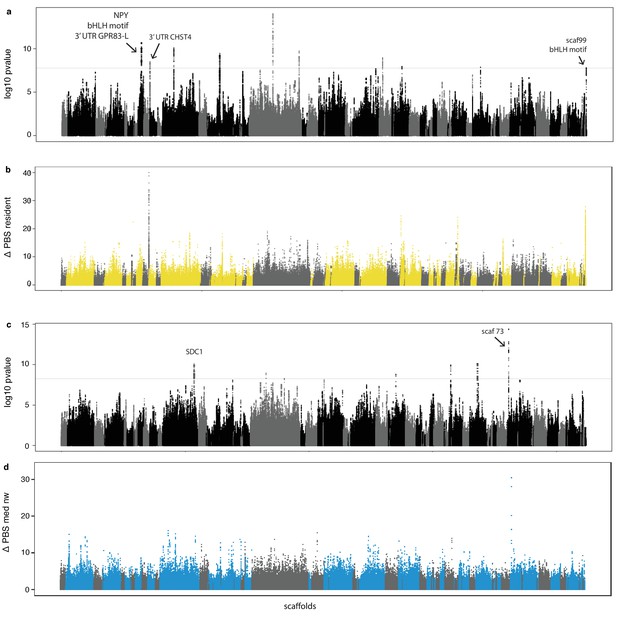
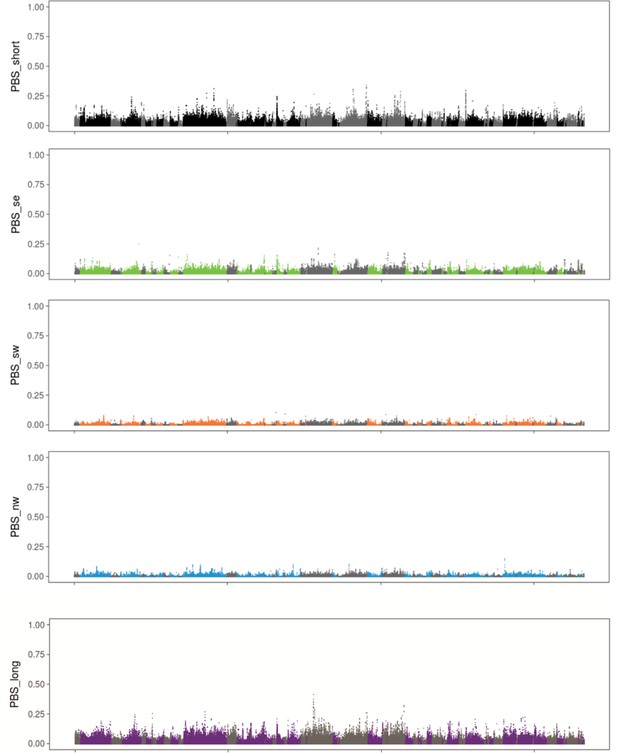
Genome-wide local estimates of population differentiation.
Results from hapFLK using haplotype frequencies (a,c) and ΔPBS using SNP frequencies (b,d; 2,500 bp windows). Estimates of ΔPBS for resident continent (b) and medium-distance NW migrants (d) are shown; results for the remaining populations can be found in Figure 3—figure supplement 1. Genetic elements, scaffolds and genes discussed in the text are highlighted.
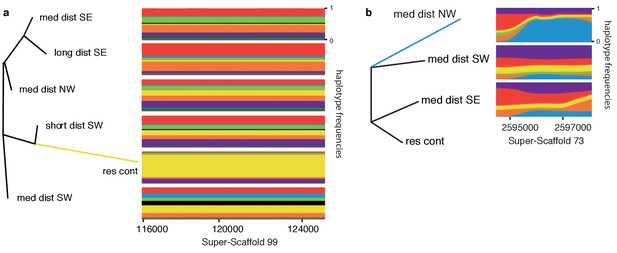
Exemplifying genomic regions under positive selection.
Local neighbour joining trees for regions under selection in (a) the resident continent population on Super-Scaffold 99, and (b) medium-distance NW population on Super-Scaffold 73. Selection is indicated by longer branch lengths in each population than is the case in global trees built using data from all genomic regions (Figure 4—figure supplement 1). Panels to the right of the trees show the corresponding frequency of haplotypes in each population of the tree. Haplotype clusters are colour coded (colours of haplotype clusters do not correspond to the population colour coding used in other figures), and frequencies are plotted along the Y axis. Haplotype frequency plots show the near fixation of a single dominating haplotype in (a) resident continent (yellow) and (b) medium-distance NW populations (blue). The location (in bp) of these regions on each Super-Scaffold is shown below these panels and the resident continent group is only included to root the tree in panel (b), and thus has no haplotype frequencies.
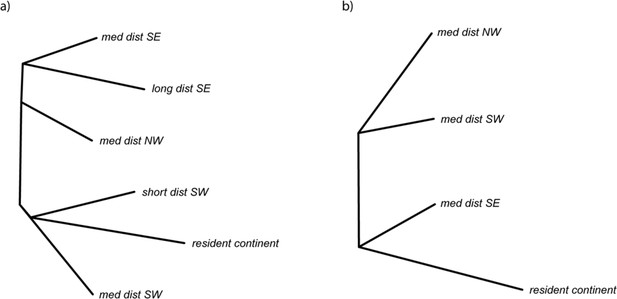
Global neighbor joining trees built using hapFLK data and data from all genomic regions, for comparison with local trees showing positive selection in Figure 4.
(a) For the analysis including all phenotypes and (b) for the analysis limited to medium-distance migrants. The resident continent group is only included to root the tree in panel (b) (i.e., it was not included in the analysis that focused only on medium-distance migrants).
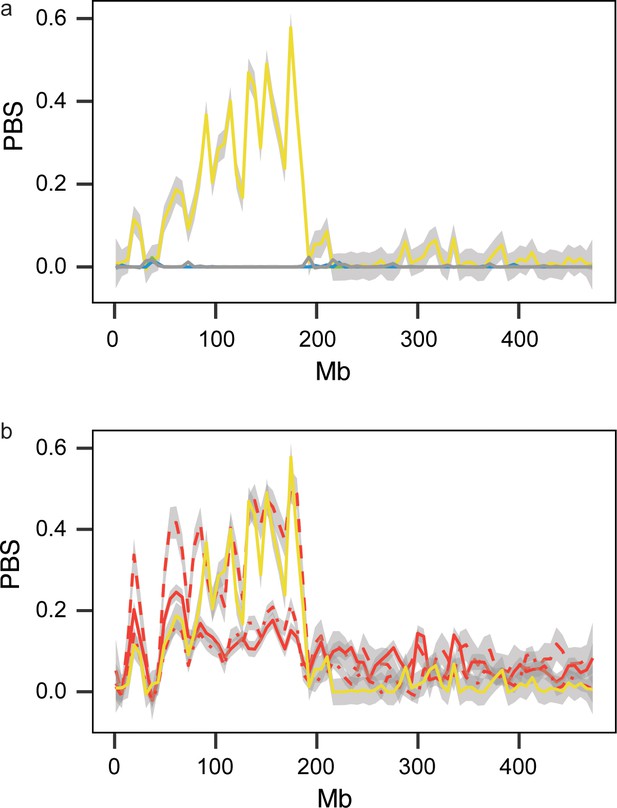
Estimates of ΔPBS on Super-Scaffold 99 corresponding with the region shown in Figure 4a (smoothed using the geom_smooth function in ggplot to summarize data in 2500-bp windows).
(a) Estimates for resident continent, medium-distance NW, SW and SE migrants, and short- and long-distance birds. These estimates are only elevated in the resident continent phenotype, ruling out a role for linked selection in generating this signature in residents. (b) Estimates for the resident continent and island birds (Azores, Canaries and Cape Verde), which are all elevated, implying that parallel selection is probably involved in the transition from migration to residency in this region. Colours correspond to Figure 1a with yellow showing data for resident continent birds.
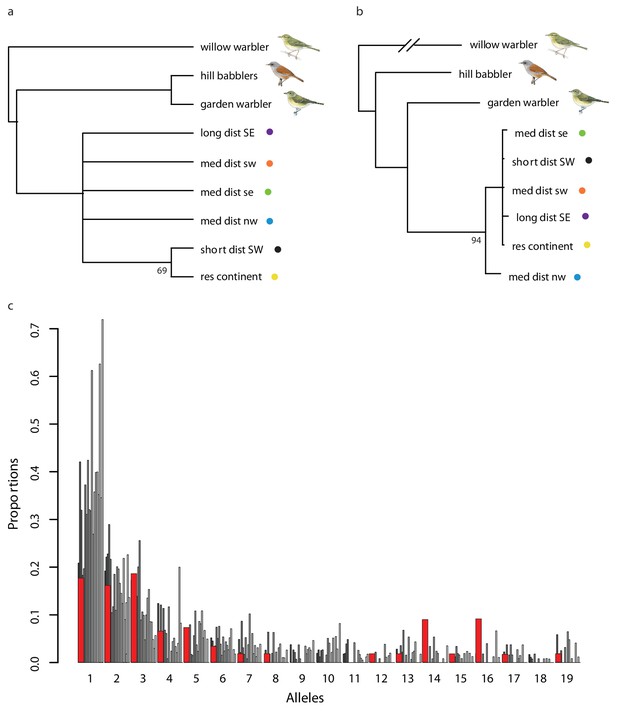
Evidence for the use of shared variation on Super-Scaffold 73.
(a) A rooted extended majority rule consensus tree summarizing maximum likelihood (ML) trees constructed for all scaffolds in the blackcap reference genome (96 scaffolds). Node numbers indicate the number of scaffolds in which populations were partitioned into two sets. (b) A ML tree constructed for the region on Super-Scaffold 73 with migratory garden warbler more closely related to blackcaps and medium-distance NW birds occurring at the base of this clade in Figure 4b. Nodes with bootstrap values <80 are collapsed; nodes without numbers have support values of 100. (c) The site frequency spectrum (SFS) for the region on Super-Scaffold 73 (red) compared to SFSs for 1000 random sequences from the genome (varying shades of gray).
Tables
Genetic variants underlying variation in migration.
(a) Results from analyses including all continental birds and (b) results from analyses limited to medium-distance migrants. Results from hapFLK include the size, the population where the signal was found and genes within the region. Estimates of ΔPBS and (PBS) in the same regions are shown; they are bolded if in the top 1% of the focal population’s distribution and new sizes are estimated using neighbouring windows above this threshold (if larger than the limits from hapFLK, additional genes are specified). Estimates of PBS were re-estimated using island populations (vs.continent resident populations). Regions in the top 1% of an island population’s distribution are indicated in section (a) (recorded as 'NA' if the initial population under selection was not resident). 'Scaf' refers to the scaffold within the blackcap genome where the region is found and 'chr' refers to the flycatcher chromosome that these scaffolds map to. For the number of strongly associated SNPs identified by CAVIAR and estimates of nSL, see Supplementary file 5.
| (a) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| hapFLK | Δpbs | ||||||||
| Scaf | Chr | Size (bp) | Log p-value | Population | Genes | Size (Mb) | ΔPBS (PBS) | Island replacement | Genes |
| 12 | 4A | 14,059 | 9.4 | Resident | LOC100859173 | 52 | 18.7 (0.40) | Azores | EDA2R |
| 13 | 11 | 29,195 | 8.3 | Resident | CHST4, TERF2IP, KARS | 303 | 41.0 (0.87) | Cape Verde | DHX38, DHODH, IST1, C2H2, ATXN1, AP1G1, PHLPP2, TAT, GABARAPL2, TMEM231, CHST6 |
| 17 | 3 | 7610 | 9.5 | Short SW | 316.5 | 0 (0.02) | NA | NKAIN1 | |
| 22 | 9 | 53,890 | 8.8 | Med SE | CLSTN2 | 1,005.5 | 21.9 (0.19) | NA | DUF4637, PIK3CB, FOXL2, MRPS22, COPB2, RBP2, NMNAT3 |
| 30 | 2 | 13,756 | 11.5 | Resident | 42.5 | 8.14 (0.19) | Cape Verde | ||
| 30 | 2 | 7902 | 8.8 | Resident | 1,029.5 | 19.1 (0.42) | Canaries, Cape Verde | ||
| 41 | 8 | 10,341 | 8.3 | Resident | 11.5 | 15.0 (0.33) | |||
| 46 | 1A | 412 | 7.9 | Med SE | 9.5 | 9.0 (0.03) | NA | ||
| 99 | 3 | 13,140 | 7.8 | Resident | TTBK1 | 192 | 28.6 (0.61) | Azores, Canaries, Cape Verde | LOC101820716, ACSS1, NEIL1, SLC22A7, TTL |
| (b) | |||||||||
| hapFLK | Δpbs | ||||||||
| Scaf | Chr | Size (bp) | Log p-value | Population | Genes | Size (Mb) | ΔPBS (PBS) | ||
| 17 | 3 | 3258 | 9.04 | Med NW | SDC1 | 5 | 14.49 (0.20) | ||
| 30 | 2 | 311 | 8.85 | Med NW | 7 | 11.31 (0.16) | |||
| 46 | 1A | 461 | 8.71 | Med NW | 3 | 8.14 (0.15) | |||
| 63 | 1A | 1088 | 9.55 | Med SE | 1.14 (0.05) | ||||
| 67 | 6 | 995 | 9.46 | Med SW | 5 | 9.03 (0.18) | |||
| 73 | 5 | 3611 | 11.81 | Med NW | ATG2B, BDKRB2 | 3 | 30.41 (0.35) | ||
Additional files
-
Supplementary file 1
Summary of sequencing data used for ALLPATHS-LG assembly.
Libraries designated a and b are from the same library preparation but sequenced on two separate lanes.
- https://cdn.elifesciences.org/articles/54462/elife-54462-supp1-v1.docx
-
Supplementary file 2
Assembly statistics at each stage.
The second ALLPATHS assembly follows the removal of duplicates and contaminants along with gap filling.
- https://cdn.elifesciences.org/articles/54462/elife-54462-supp2-v1.docx
-
Supplementary file 3
Results from satsuma showing which flycatcher chromosome each scaffold in the blackcap reference genome hit.
Mean position and orientation refer to the location and orientation of scaffolds on the flycatcher genome. The last six scaffolds did not hit any of the flycatcher chromosomes. Comparing the annotation of the blackcap and zebra finch genomes suggests they match the indicated chromosomes.
- https://cdn.elifesciences.org/articles/54462/elife-54462-supp3-v1.docx
-
Supplementary file 4
Samples used in the present study, including their locations and details on how phenotypes were determined.
- https://cdn.elifesciences.org/articles/54462/elife-54462-supp4-v1.xlsx
-
Supplementary file 5
Extension of Table 1 showing regions identified by hapFLK as being under selection but including the number of causal SNPs identified by CAVIAR and their location within genes.
Estimates of nSL are shown (bolded if in top 1% of values, bolded and italicised if in the top 5%).
- https://cdn.elifesciences.org/articles/54462/elife-54462-supp5-v1.xlsx
-
Supplementary file 6
Additional details on genome assembly and annotation.
- https://cdn.elifesciences.org/articles/54462/elife-54462-supp6-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/54462/elife-54462-transrepform-v1.docx