Deep evolutionary analysis reveals the design principles of fold A glycosyltransferases
Figures
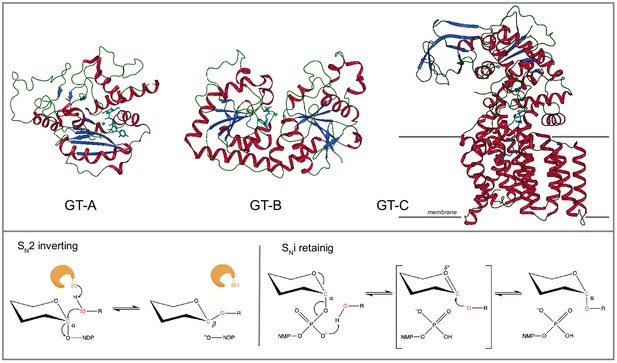
Glycosyltransferase (GT) folds and mechanisms.
Top: The three representative structural folds of GTs. The GT-A fold is characterized by a single globular domain that contains a α/β/α Rossmann nucleotide binding domain (shown 2rj7;GT6). The GT-B fold enzymes are usually metal independent and contain two α/β/α domains separated by a flexible linker region with the substrate binding cleft in between (shown 1jg7;GT63). The GT-C fold enzymes are hydrophobic integral membrane proteins, generally use lipid phosphate linked sugar donors and have multiple transmembrane helices (shown 6gxc; GT66). Bottom: The mechanism of sugar transfer employed by GTs. Inverting GTs follow a direct displacement SN-2-like mechanism that results in an inverted anomeric configuration. The mechanism for retaining GTs is still under debate although recently a same side SNi-type reaction has been proposed where the donor phosphate oxygen acts as a catalytic base and deprotonates the acceptor hydroxyl facilitating a same side attack, that results in the retention of anomeric configuration. The enzyme and catalytic base B are shown in orange. A generic hexose with α-linkage to a nucleoside diphosphate is used. Other mechanisms possibly employed by GTs is discussed in detail in M.
-
Figure 1—source data 1
List of CAZy GT families.
The structural fold and the number of sequences from each taxonomic group are shown. The number of sequences with structure or are characterized are also provided.
- https://cdn.elifesciences.org/articles/54532/elife-54532-fig1-data1-v2.xlsx
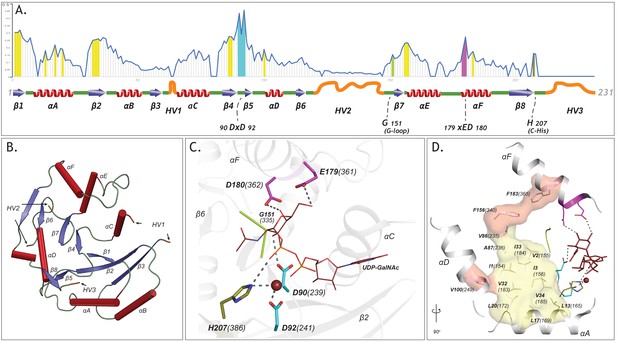
The GT-A common core and its elements.
(A) Plot showing the schematics of the GT-A common core with 231 aligned positions. Conserved secondary structures (red α-helices, blue β-sheets, green loops) and hypervariable regions (HVs)(orange) are shown. Conservation score for each aligned position is plotted in the line graph above the schematics. Evolutionarily constrained regions in the core: the hydrophobic positions (yellow) and the active site residues (DxD: Cyan, xED: Magenta, G-loop: green, C-His: olive) are highlighted above the positions. (B) The conserved secondary structures and the location of HVs are shown in the N-terminal GT2 domain of the multidomain chondroitin polymerase structure fromE. coli(PDB: 2z87) that is used as a prototype as it displays closest similarity to the common core consensus. (C) Active site residues of the prototypic GT-A structure. Metal ion and donor substrate are shown as a brown sphere and sticks, respectively. (D) Architecture of the hydrophobic core (Yellow: core conserved in all Rossmann fold containing enzymes, Red: core elements present only in the GT-A fold). Residues are labeled based on their aligned positions. Numbers within parentheses indicate their position in the prototypic (PDB: 2z87) structure.
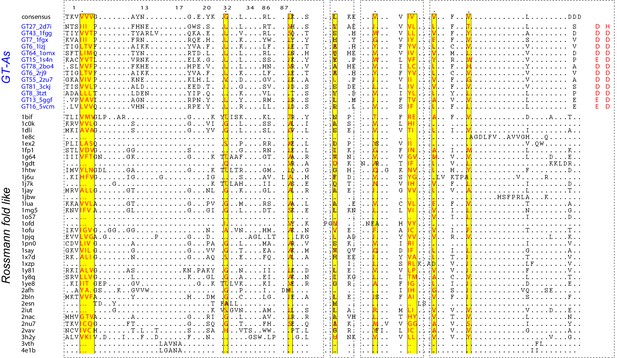
Structure based sequence alignment showing the hydrophobic residue positions present across a collection of Rossmann fold like enzymes.
The conserved hydrophobic positions are highlighted in yellow blocks. Aligned positions are indicated at the top that correspond to aligned positions in Figure 2D. The alignment extends until the DxD motif. Other regions were unaligned due to very low homology.
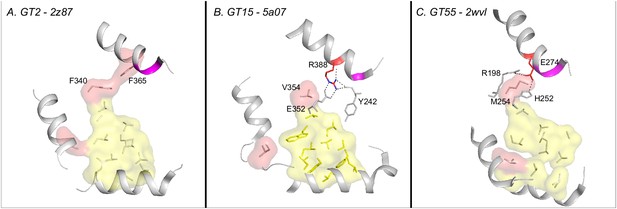
Changes in the extended hydrophobic core residues in selected retaining families.
(A) The conserved hydrophobic core in the prototypic GT (2z87). (B and C) Hydrophobic residue in the core is substituted by an Arginine and a Glutamate in GT15 and GT55 respectively. The charged residue replacing the hydrophobic residue of the core is highlighted in red sticks. The xED motif is shown in magenta.
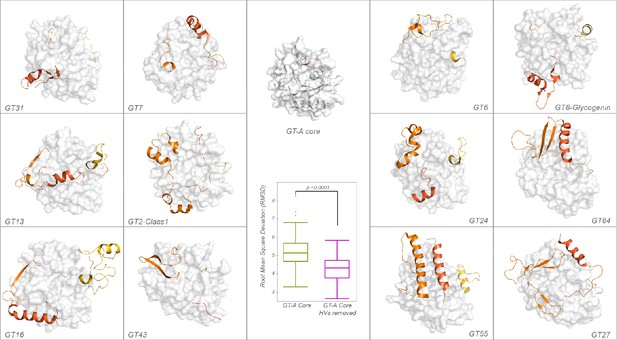
Comparison of structures for HV regions across GT-A families.
The GT-A common core is shown in surface in the middle. HVs are shown in shades of orange (HV1: light orange, HV2: dark orange, HV3: orange red). Root Mean Square Deviation (RMSD) was calculated by aligning the core GT-A domains of representative structures with and without the HVs. A significant reduction in the RMSD values was observed after removing HVs that is shown in the box plot in the center. *p-value<0.0001, t-test.
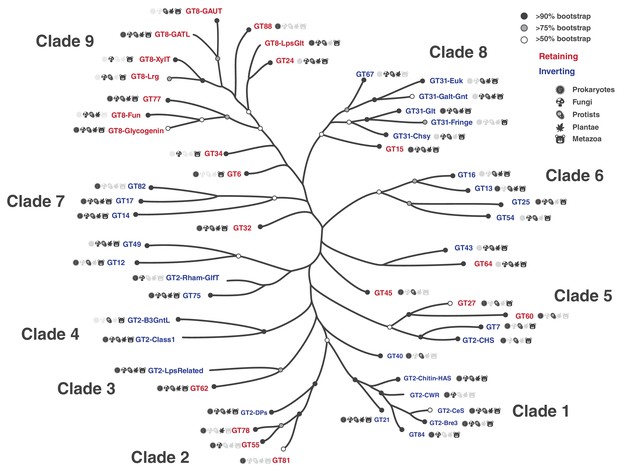
Phylogenetic tree highlighting the 53 major GT-A fold subfamilies.
Tips in this tree represent GT-A sub-families condensed from the original tree for illustration. Support values are indicated using different circles. Circles at the tips indicate bootstrap support for the GT-A family clade represented by that tip. Tips missing the circles represent GT-A families that do not form a single monophyletic clade. Nodes missing circles have a bootstrap support less than 50% and are unresolved. Icon labels indicate the taxonomic diversity of that sub clade. Colors indicate the mechanism for the families (blue: Inverting, red: Retaining). This condensed tree was generated by collapsing clades to the deepest node that includes sequences from the same family. For GT-A families that did not form a monophyletic clade, the clade that included the most sequences from that family was chosen. Branch lengths may approximate the original distances, but are not drawn to scale. Detailed tree with support values, expanded nodes and scaled branch lengths are provided in Figure 3—figure supplement 1 and in Newick format in Figure 3—source data 4. The family names are described in Figure 3—source data 1.
-
Figure 3—source data 1
List of GT-A fold families and subfamilies.
For each of these families, the groups obtained by the pattern based classification are provided in the ‘GT-A pattern based group’ column. Taxonomic distribution and a short description of these groups are also provided.
- https://cdn.elifesciences.org/articles/54532/elife-54532-fig3-data1-v2.xlsx
-
Figure 3—source data 2
The 993 representative GT-A domain sequences included in the phylogenetic analysis.
The GT-A family and the pattern based classification group for each sequence is indicated in the ‘GT-A family’ and the ‘GT-A pattern based group’ columns. The domain start and end positions are indicated. Sequence for the domain region and the full length sequences are also provided. An alignment of these sequences are available in Figure 3—source data 3.
- https://cdn.elifesciences.org/articles/54532/elife-54532-fig3-data2-v2.xlsx
-
Figure 3—source data 3
The trimmed FASTA alignment of the 231 positions of the GT-A core used for phylogeny.
- https://cdn.elifesciences.org/articles/54532/elife-54532-fig3-data3-v2.txt
-
Figure 3—source data 4
The phylogenetic tree file for the 993 GT-A fold sequences in Newick format.
- https://cdn.elifesciences.org/articles/54532/elife-54532-fig3-data4-v2.txt
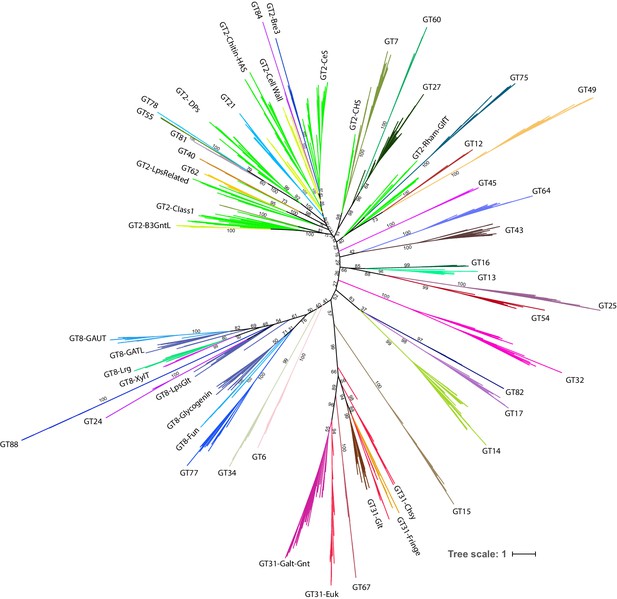
Complete phylogenetic tree of 993 representative GT-A sequences.
Sequences are provided in Figure 3—source data 2. Clades are colored for each of the 53 GT-A families and labeled. Values at nodes indicate bootstrap support with 1000 replicates. Values for all major nodes are indicated. This tree is also provided in Newick text format in Figure 3—source data 4.
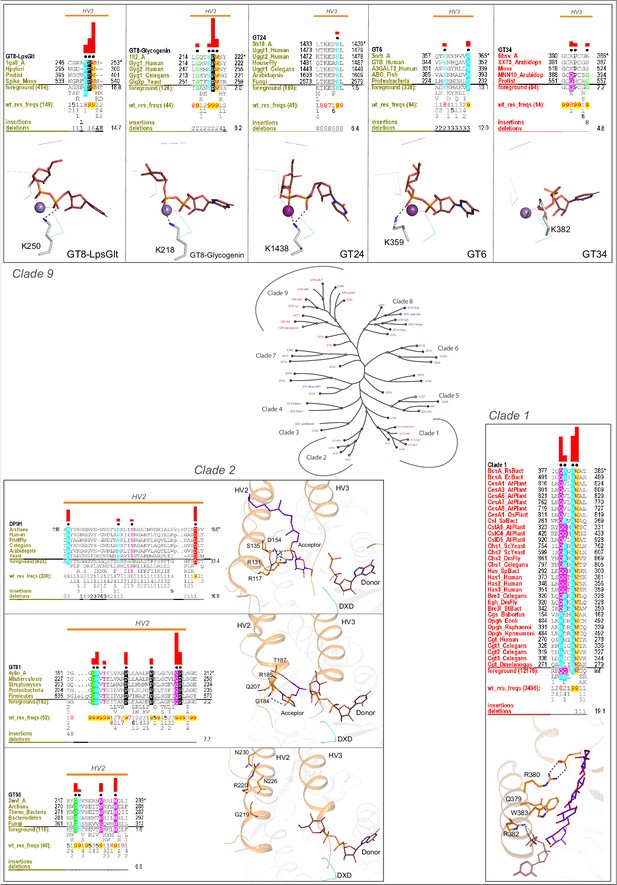
Clade specific conserved features in the HVs.
The conserved mode of donor binding in clade 9, conserved mode of acceptor binding in clade two and the conserved QXXRW motif in clade one are illustrated. HVs are shown in orange. Metal ions are shown as spheres. Red bars above the alignment indicate the extent of significance of conservation of residue in the column (Higher is more significantly conserved). Below every position in the alignment, numbers indicate the extent of conservation of residues at the position.
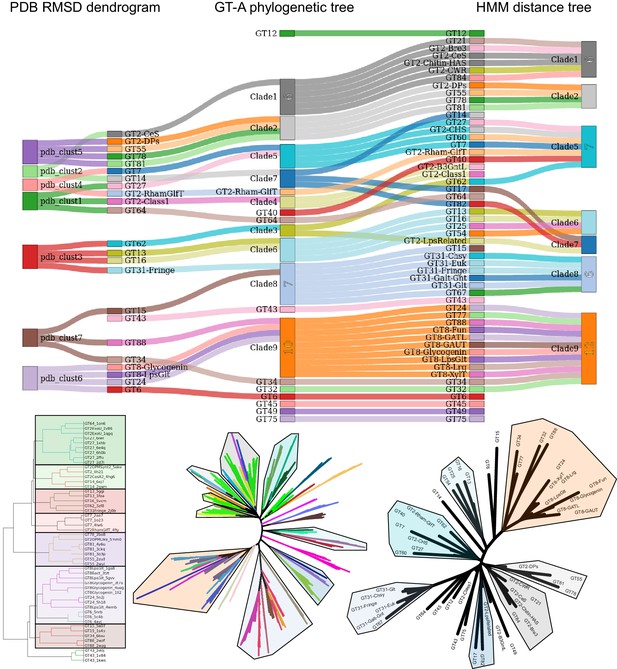
Sankey diagram comparing topologies of phylogenetic tree with pdb and hmm based clustering of GT-A families.
Each column highlights clusters of GT-A families obtained through different methods (from left to right: PDB structural alignment clustering, GT-A phylogeny and hmm-distance based tree). Corresponding GT-A families within clusters are connected through colored links. Non overlapping links indicate an agreement in the placement of families across methods. Full clusters and trees are shown below the columns.
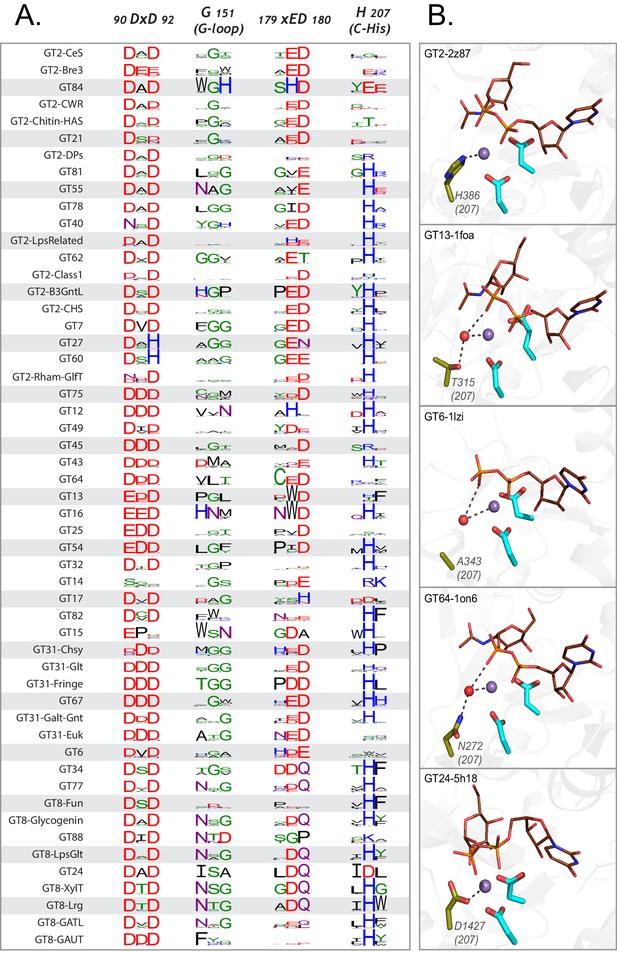
Variations in the GT-A conserved core.
(A) Weblogo depicting the conservation of active site residues in the common core are shown for each of the GT-A families. Residues are colored based on their physiochemical properties. (B) Variations in the C-His is compensated either using a water molecule (red sphere) or other charged residues (olive sticks) to conserve its interactions. The metal ion is shown as a purple sphere. The donor substrate is shown as brown lines. Interactions between the residues, metal ion and the donor are shown using dotted lines.
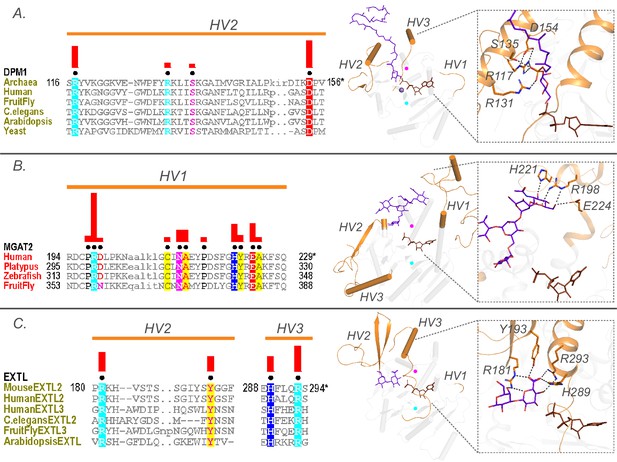
Family specific conserved features in the HV regions correlate with acceptor recognition and specificity.
Conserved residues in A) HV2 of the DPM1 sequences in the GT2-DP subfamily coordinate the phosphate group of the acceptor. (B) HV1 of GT16 MGAT1 provide acceptor specificity. (C) HV2 and HV3 of EXTL GT64 family (C-terminal GT domain of the multidomain sequences) coordinate the acceptor. Left: Alignments highlighting the constrained residues are shown for each family. The family specific conserved residues are shown using black dots above the alignment. Red bars above these dots indicate the significance of conservation (Higher bar corresponds to more significantly conserved position). Right: Representative pdb structures are shown for each family (GT2-DP:5mm1, GT16:5vcs, GT64:1on8); Donor substrates are colored brown. Acceptors are colored purple. HVs are highlighted in orange. The position of the conserved DxD and xED motif for each structure is shown as cyan and magenta circles respectively.
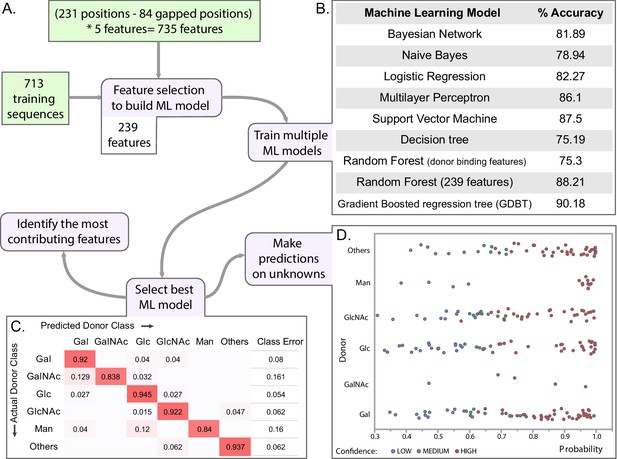
Machine learning (ML) approach for predicting donor class.
(A) Brief pipeline of the ML analysis. Training set input into the pipeline are shown in green boxes. Steps of the ML analysis in purple boxes are associated with different panels of the figure. (B) Percent accuracy based on 10-fold cross validation (CV) for each of the trained ML models. (C) Confusion matrix from the best model (GDBT using 239 features). (D) Scatter plot showing the probability scores assigned for each predicted sequence by the predicted donor type. Colors indicate the confidence level of the prediction based on probability of assignment to a given donor class as well as confidence intervals of the predicted class i.e. difference in probability values between the 1st prediction class and the 2nd prediction class. (Figure 6—source data 2).
-
Figure 6—source data 1
List of the 713 training dataset sequences used for machine learning.
The ‘Assigned Donor Class’ column indicates one of the six classes the donor belongs to.
- https://cdn.elifesciences.org/articles/54532/elife-54532-fig6-data1-v2.xlsx
-
Figure 6—source data 2
Results for donor prediction using the GDBT ML model for GT-A sequences from five model organisms.
The validation datasets (highlighted in blue rows) include GTs that have some experimental characterization but were not included in the characterized dataset. The validation set was used to compare the model predictions with the experimental results. The ‘Match Experimental’ column indicates whether the prediction matched experimental results. The prediction set includes predictions for GTs of unknown functions. The ‘Confidence’ column includes the confidence for prediction which was derived based on the probability for the 1 st class and its difference with the probability for the 2nd class. Probabilities for all the six classes are provided in the ‘Classwise Probablity’ columns.
- https://cdn.elifesciences.org/articles/54532/elife-54532-fig6-data2-v2.xlsx
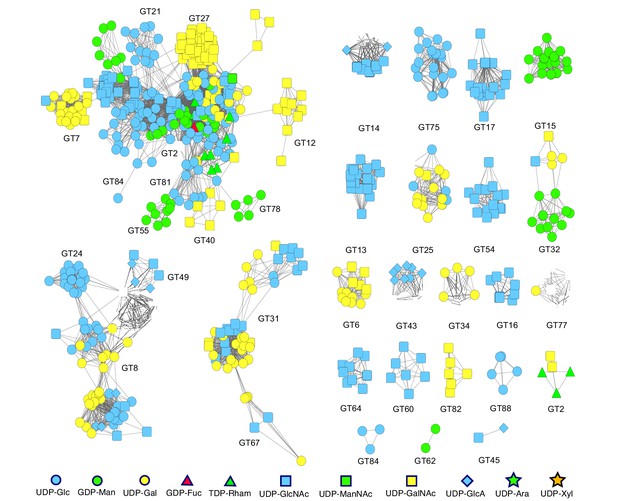
Sequence homology-based network of all the experimentally characterized sequences form the GT-A fold families.
Nodes represent the sequences that were annotated as characterized and collected from the CAZy database to be used in the training dataset for ML. The color and shape of the nodes indicate the donor specificity for that sequence. An edge between two nodes indicate that the sequences are homologous with an e-value better than 1e-5. Smaller edge distance indicates a higher similarity between nodes. An edge-weighted spring embedded layout from Cytoscape was implemented to minimize edge crossings and enhance visual interpretability. At multiple locations in the network, closely related sequences differ in donor specificity, rendering prediction through similarity alone difficult.
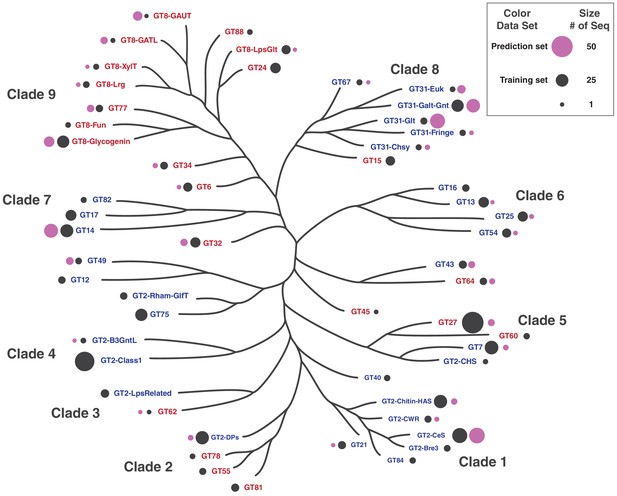
Distribution of training and prediction datasets used in machine learning.
The size of the bubbles next to GT-A family names indicates the number of sequences in the training and prediction set from that family. Color of the bubbles indicate training or prediction set.
-
Figure 6—figure supplement 2—source data 1
Distribution of sequences across different families.
The counts in this table were mapped in to the phylogenetic tree in Figure 6—figure supplement 2.
- https://cdn.elifesciences.org/articles/54532/elife-54532-fig6-figsupp2-data1-v2.xlsx
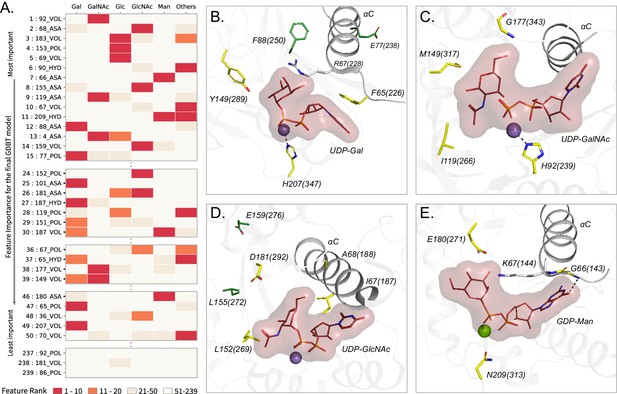
Top Contributing features from the GDBT model associated with sugar donor specificity.
(A) Heatmap showing the contributions of representative features. Features are ordered based on their importance for the final GDBT model along the vertical axis. The heatmap colors indicate how important each feature is for a given sugar donor type with red indicating ranks 1–10 (highly important) (M). (B–E) Contributing features important for individual donor types are mapped onto representative structures. The amino acids at the feature positions are shown in yellow sticks and labelled. Feature positions distal from the donor binding site are shown in green sticks. Labels include the amino acid code, aligned residue position and the amino acid position in the crystal structure within parentheses. Donor substrate with the sugar is shown in lines with surface bounds. Divalent metal ions are shown as spheres. The αC helix is shown. (B) Gal features mapped to a bovine β−1,4 Gal transferase (PDB ID: 1o0r). (C) GalNAc features mapped to a human UDP-GalNAc: polypeptide alpha-N-acetylgalactosaminyltransferase (PDB ID: 2d7i). (D) GlcNAc features mapped to a rabbit N-acetylglucosaminyltransferase I (PDB ID: 1foa). (E) Man features mapped to a bacterial Mannosyl-3-Phosphoglycerate Synthase (PDB ID: 2wvl).
-
Figure 7—source data 1
Feature Importance comparison for the full GDBT model with its importance for each sugar donor type.
‘GDBT_full’ columns include the rank and score for the 239 features in the GDBT model. Remaining columns show the rank and score for that same feature for the classification of the respective donor sugars. Feature positions ranked from 1 to 10 (most important) are colored red and ranked 11–20 are colored orange.
- https://cdn.elifesciences.org/articles/54532/elife-54532-fig7-data1-v2.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Software, algorithm | CAZy database | doi: 10.1093/nar/gkt1178 | CAZy- Carbohydrate Active Enzyme, RRID:SCR_012909 | |
| Software, algorithm | mapgaps | doi: 10.1093/bioinformatics/btp342 | ||
| Software, algorithm | omcBPPS | doi: 10.1089/cmb.2013.0099 | ||
| Software, algorithm | GT-A family classification and sequences | This paper | doi:10.5061/dryad.v15dv41sh | |
| Software, algorithm | MAFFT v7.3 | doi: 10.1093/molbev/mst010 | MAFFT, RRID:SCR_011811 | |
| Software, algorithm | Expresso from the t-coffee suite | doi: 10.1093/nar/gkl092 | T-Coffee, RRID:SCR_011818 | |
| Software, algorithm | IQTree v1.6.1 | doi: 10.1093/molbev/msu300 | ||
| Software, algorithm | PyMOL v2.0.6 | Schrödinger | PyMOL, RRID:SCR_000305 | |
| Software, algorithm | Python v3 with package scikitlearn | Pedregosa, 2011 | scikit-learn, RRID:SCR_002577 | |
| Software, algorithm | R package ‘randomForest’ | Liaw and Wiener, 2002 | RandomForest Package in R, RRID:SCR_015718 | |
| Software, algorithm | WEKA version 3.8.3 | Witten et al., 2016 | Weka, RRID:SCR_001214 |
Additional files
-
Supplementary file 1
The CAZy GT-A families included in the analysis.
The ‘Alignment source’ column includes the CDD alignment identifiers used to build the seed profiles for a given GT-A family. If a suitable CDD profile was not available, the seed profiles were built by manually selecting and aligning representative sequences.
- https://cdn.elifesciences.org/articles/54532/elife-54532-supp1-v2.xlsx
-
Supplementary file 2
Mapping of the 231 aligned positions to representative crystal structures available for all GT-A fold families.
The ‘Position Description’ column includes the labels for conserved motifs and hypervariable regions for the aligned positions. GT family, PDB IDs and reference to PubMed IDs are indicated at the header columns.
- https://cdn.elifesciences.org/articles/54532/elife-54532-supp2-v2.xlsx
-
Supplementary file 3
The ancient archaeal and bacterial sequences that most closely resemble the GT-A core consensus.
These sequences were collected by running a BLAST search against a single consensus sequence generated from the seed profiles of all GT-A families. The hits were further filtered to keep sequences that only had the minimal GT-A core (Materials and methods).
- https://cdn.elifesciences.org/articles/54532/elife-54532-supp3-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/54532/elife-54532-transrepform-v2.pdf





