Transient intracellular acidification regulates the core transcriptional heat shock response
Figures
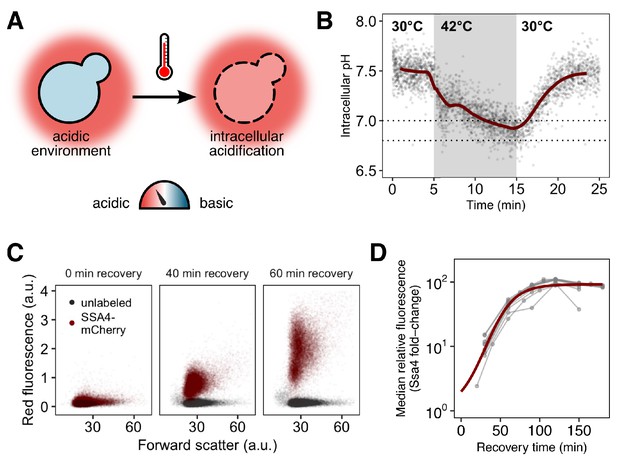
Yeast cells respond to heat shock with intracellular pH changes and production of heat-shock proteins, which can be tracked at the single-cell level.
(A) S. cerevisiae cells live in acidic environments but maintain a neutral or slightly basic intracellular pH. During heat stress the cell membrane becomes more permeable, leading to intracellular acidification. (B) Intracellular pH changes during stress measured with continuous flow cytometry. Each point is an individual cell. The gray region is the period during which cells were exposed to elevated temperature. A solid line shows a sliding-window average over all data; for visual clarity, only 2% of points are shown. Dashed lines represent the range we subsequently use as representative of the physiological pH drop. (C) Induction of labeled Hsp70 (Ssa4-mCherry) after heat shock. Each plot is a timepoint during recovery from 42°C, 20 min heat shock showing forward scatter pulse area, which correlates roughly with size, versus red fluorescence. Unlabeled cells are shown in black for comparison. (D) Summary of induction of Ssa4-mCherry after heat shock; each point represents the fold change, relative to unstressed cells, of the median fluorescence of >5000 cells expressed as a ratio to forward scatter; each gray line is an experiment (). Thick red curve is a sigmoid fit (see Materials and methods).
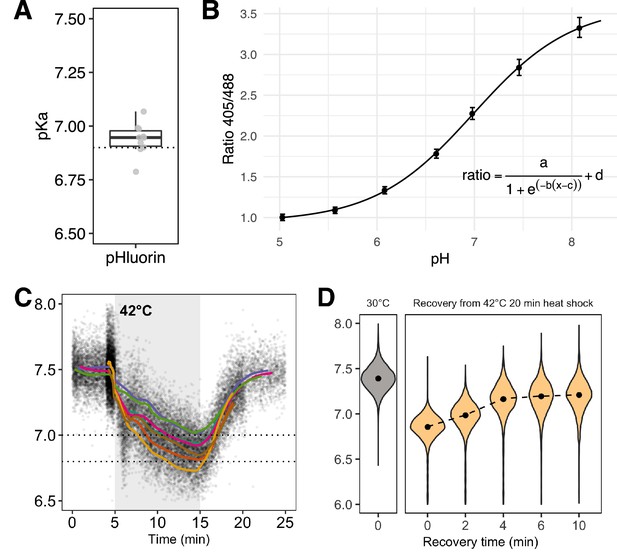
pHluorin calibration curve and replicates of B.
(A) Comparison between calibration curves taken on different days. The curves are compared by solving for the apparent pKa of pHluorin (see Materials and methods for equation); while absolute values of the ratio vary by day and instrument, the pKa should be constant. The in vitro pKa as calculated in Bagar et al., 2009 is shown with the dashed line. Each point is a separate experiment, . See Materials and methods for full details. (B) A representative pHluorin calibration curve showing the relationship between intracellular pH and fluorescence ratio. Error bars are the standard deviation of the population of cells measured. (C) Traces of intracellular pH as a function of time in cells expressing pHluorin and perturbed with a 42°C, 10 min heat stress. Each point is an observation of a single cell; colored lines are the moving average of one experiment. Individual points are sub-sampled for clarity; each experiment has at least 10,000 cells. (D) Intracellular pH drops in response to a 42°C, 20 min heat stress; the degree of acidification is the same as a 10 min heat stress.
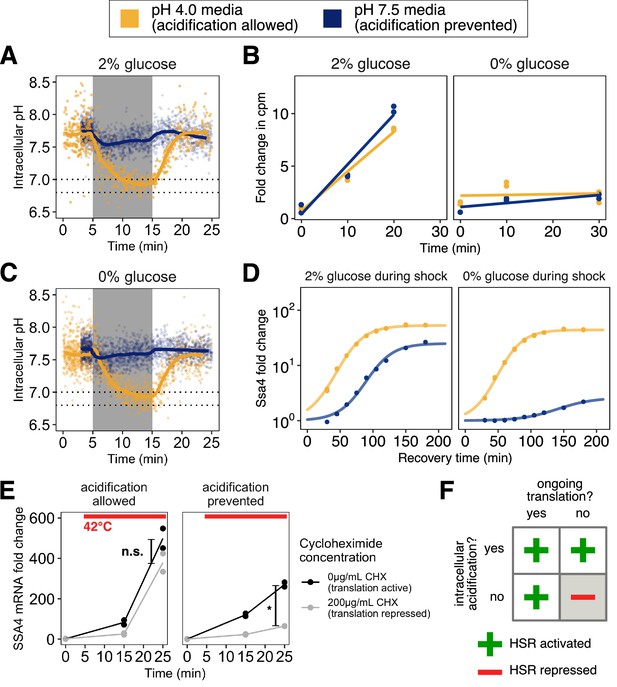
Preventing stress-associated acidification delays or impairs the heat shock response when cells are translationally inactive.
(A) Intracellular pH changes as a function of environmental pH. Cells stressed in acidic media (pH 4.0, yellow) acidify, whereas cells stressed in media at the resting pH (7.5, blue) do not. (B) Inhibition of translation by glucose withdrawal does not depend on environmental pH. Incorporation of radiolabeled amino acids into total cellular proteins in counts per minute (cpm) as a function of time after a switch from medium at pH 4 with 2% glucose to the indicated media. (C) Same as A, but in glucose-free medium. (D) Induction of Ssa4 in cells able (yellow) or unable (blue) to acidify during heat shock. All measurements are of cells recovering in media containing 2% glucose. (E) Acidification promotes the transcriptional heat shock response (production of SSA4 mRNA) when cells are treated with the translation inhibitor cycloheximide (200 μg/mL) prior to heat stress. *, ; n.s., , Welch two-sample t-test. (F) Acidification promotes the heat shock response, and is required when cells are translationally inactive.
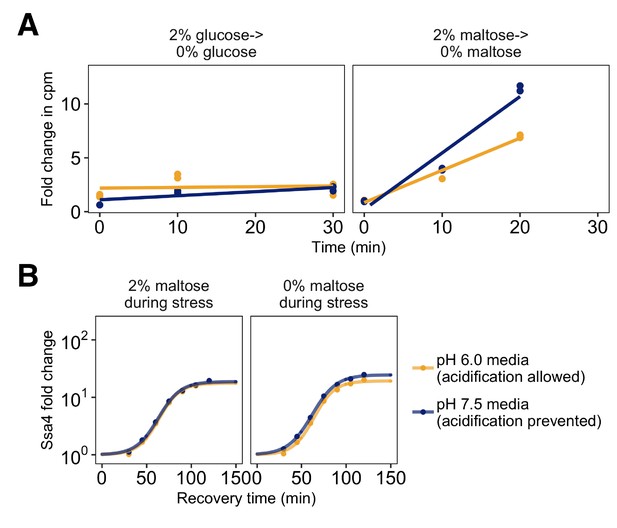
Translation and heat shock response after maltose withdrawal.
(A) Measurement of the incorporation of radiolabeled amino acids in counts per minute (cpm) into total cellular protein as a function of time after transfer to sugar-free medium. Translation abruptly ceases after withdrawal of glucose (left), but continues after maltose withdrawal (right). In both cases, acidification does not affect the translation rate. (B) Induction of Ssa4-mCherry for cells stressed after growth in maltose (left) or growth in maltose followed by brief maltose withdrawal (right). Yellow curves are data from cells in acidic media where acidification is prevented, blue are data from cells grown in media at the resting pH where acidification is prevented. Ssa4 induction after maltose withdrawal is pH-independent, demonstrating that translation attenuation rather than nutrient withdrawal explains the difference between induction in the right hand side of B.
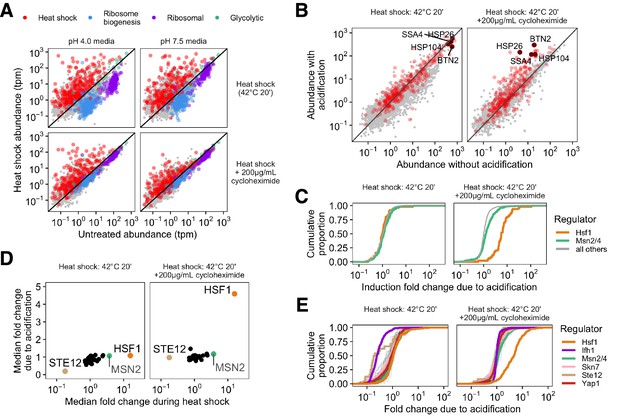
Failure to acidify during heat shock specifically represses Hsf1-activated genes.
(A) Transcript abundance (transcripts per million, tpm) in stressed versus unstressed populations of cells. Colors correspond to gene type; gray points show uncategorized genes. (B) Direct comparison of gene abundance after heat shock with and without acidification. Left, data for actively translating cells; right, with translation repressed by cycloheximide treatment. (C) Cumulative distribution of per-gene transcript abundance in cells heat shocked with acidification relative to cells shocked without acidification (induction fold change due to acidification). Genes regulated by Msn2/4 are in green, genes regulated by Hsf1 are in orange, and all detected genes are in gray. (D) Mean fold change during heat shock versus the median fold change due to acidification for the regulons of all transcription factors in the YeastMine database annotated under conditions of heat stress. (E) Cumulative distribution of acidification fold change during heat shock for a subset of stress-involved transcription factors.
Variation between replicates and induction of heat shock genes after brief acute glucose withdrawal.
(A) Correlation between gene abundances in replicate samples (RNA-seq data) (B) Gene abundances in cells heat shocked after acute glucose withdrawal – unstressed abundances are shown on the x-axis, abundances after stress on the y. Color corresponds to gene type.
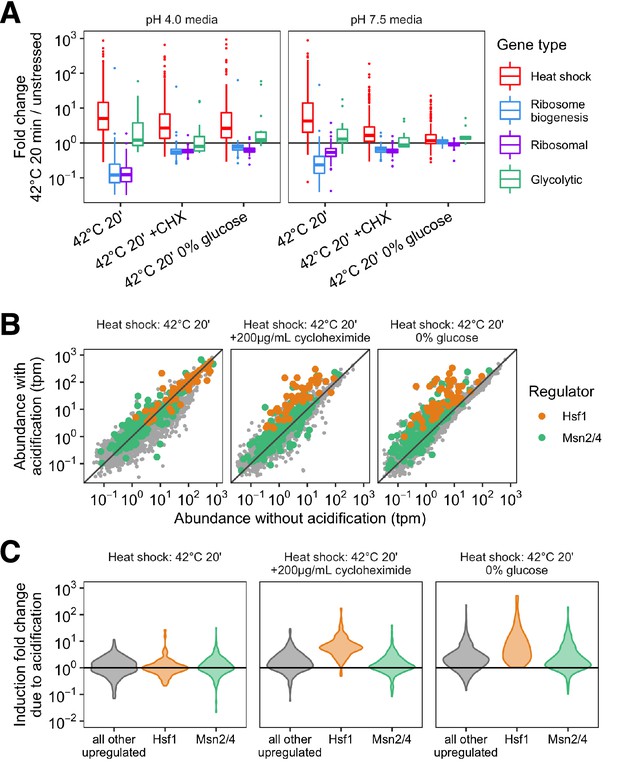
Summary of fold change after heat stress by gene type and transcription factor as a function of translation state and acidification.
(A) Fold change distribution for groups of genes (colored by gene type) after stress. Populations that were allowed to acidify are on the left (pH 4.0 media), and those where acidification was prevented are shown on the right (pH 7.5 media). (B) Gene abundance after stress without (x-axis) or with (y-axis) acidification. Panels correspond to populations translating proteins (left) or where translation has been arrested (center and right). Color corresponds to the transcription factor responsible for the induction of that gene. (C) A global analysis of the pH-sensitivity of all genes induced during acute 42°C heat shock. The threshold for upregulation was set using a > 4σ cutoff for fold change in unstressed replicates (∼2.5-fold).
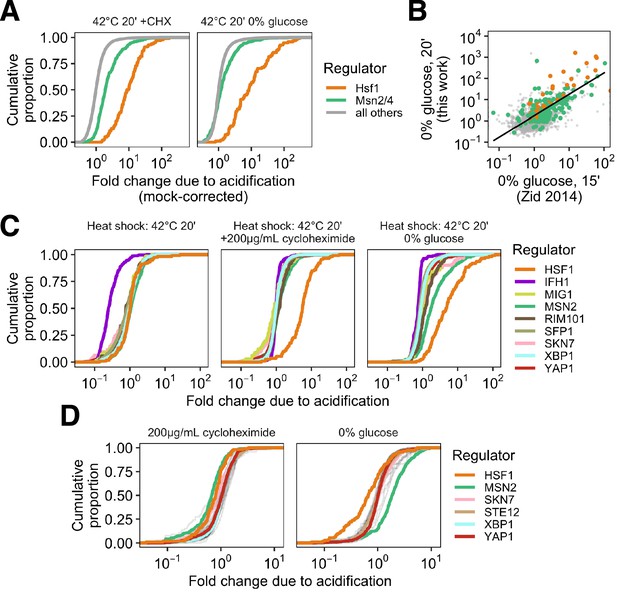
Normalization to mock treatment and distributions of pH sensitivity for additional transcription factor regulons.
(A) Cumulative distribution of pH-sensitivity values for cells heat shocked (42°C, 20’) with translation blocked either by cycloheximide treatment (left) or glucose withdrawal (right). Abundances were normalized to the abundance after equivalent mock treatment (rather than the untreated abundances as in the main text figure). (B) Per-gene fold change after acute glucose withdrawal colored by transcription factor as in A, compared to the same value as reported in Zid and O'Shea, 2014. (C) Distribution of pH-sensitivity values for genes belonging to regulons of all annotated stress-responsive transcription factors (grey). TFs of particular interest are shown in color. (D) Distribution of pH-sensitivity values for the same set of TFs as (B), but shown after manipulation of translation state followed by mock treatment (30°C, 20’).
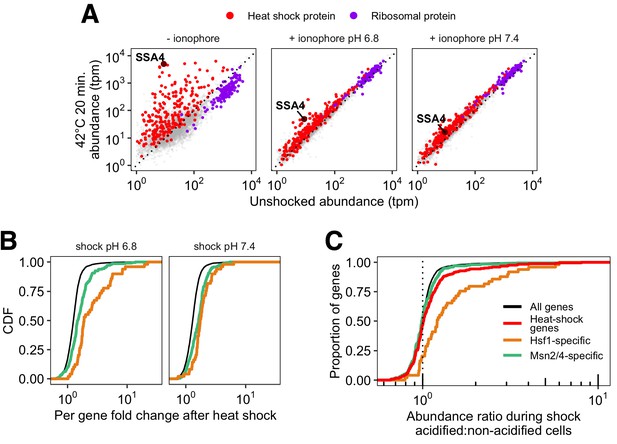
Gene induction and pH sensitivity after heat stress following pH manipulation with ionophore.
(A) Induction of heat shock genes in cells heat shocked without ionophore in acidic media (left panel), or in ionophore (center and right panels). (B) Induction (fold change in stressed cells relative to unstressed cells) for genes belonging to the Hsf1 (orange) and Msn2/4 (green) regulons. (C) Cumulative distribution of per-gene transcript abundance in cells heat shocked at pH 6.8 relative to cells shocked at pH 7.4 (induction ratio). The red line shows all heat shock proteins; this group is further divided into genes regulated by Msn2/4 (green) which show similar behavior to all detected transcripts (gray; , Wilcoxon rank sum test), and those regulated by Hsf1 (orange), which are significantly higher in acidified cells (, Wilcoxon rank sum test).
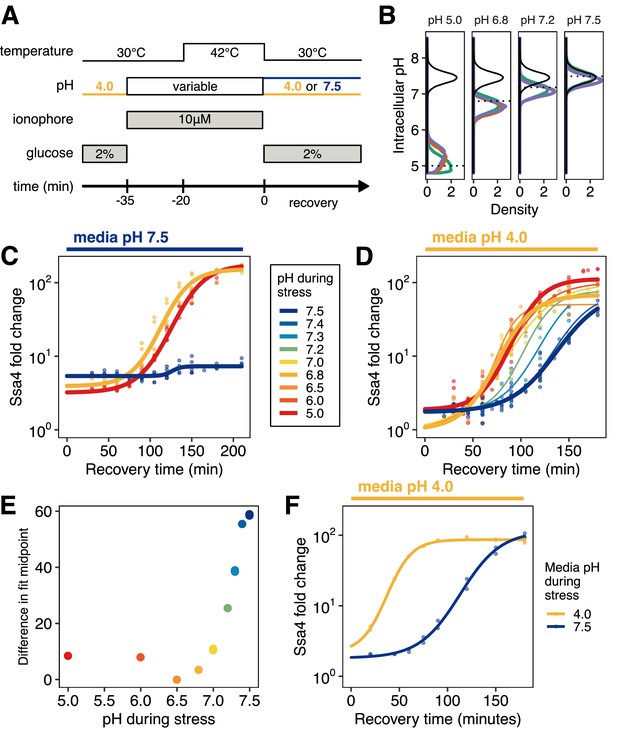
Quantitative control of intracellular pH reveals that, in the absence of translation, acidification is required for Ssa4 induction.
(A) Schematic of intracellular pH manipulation experiments. (B) Intracellular pH is accurately manipulated during stress. Intracellular pH distributions were measured to determine the efficacy of pH manipulation before (green), during (red), and after (purple) 42°C heat stress. Dashed lines indicate buffer pH, and the black distribution shows unmanipulated cells for comparison. (C) Manipulation of intracellular pH with ionophore reproduces the acidification-dependent induction of Ssa4. Compare to Figure 2B, right hand side. (D) Fold change in Ssa4 expression following stress at different intracellular pHs and recovery in acidic media. Points represent the median of individual measurements; at least three biological replicates were performed for each condition (see Materials and methods). Lines are sigmoid fits (see Materials and methods for fitting details). (E) pH dependence of the induction delay; points are the midpoint of the sigmoidal fits in D. (F) Dependence of the stress response on media pH, followed by recovery in acidic media, recapitulates the pH dependence of the stress response when ionophore treatment is used; compare to D.
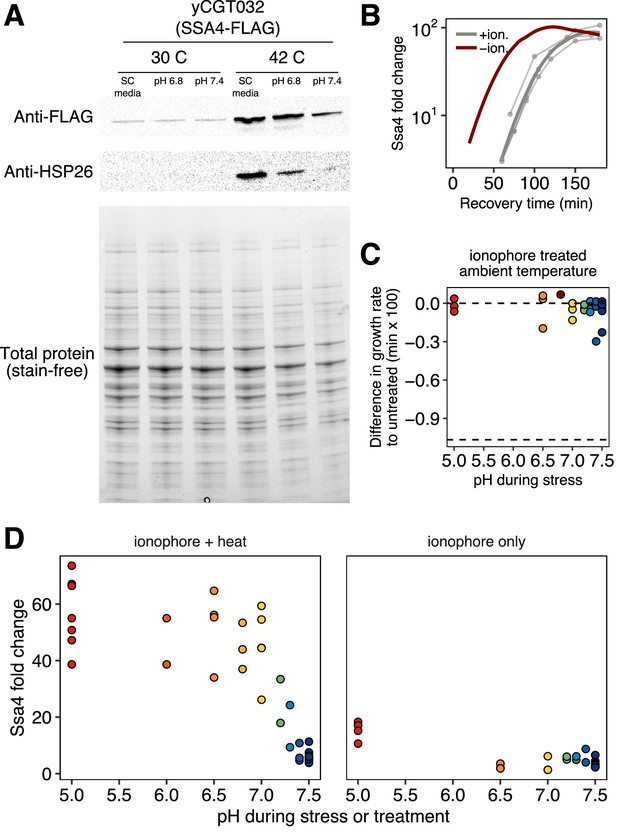
Western blot against Ssa4-mCherry and native Hsp26 after heat shock with and without acidification and stress protein production after acidification only.
(A) Western blot and total protein gels for yeast carrying a the genomic copy of SSA4 tagged with a FLAG tag heat stressed with and without ionophore treatment, as described in Growth Conditions in the Materials and methods section. Samples were taken 1 hr after stress. (B) Induction of Ssa4 during recovery from normal (red) or pH-manipulated (gray, pH 6.8) stress. Thin curves are individual experiments and thick curves are smoothed conditional means (see Materials and methods for details). The red curve is the same data from Figure 1D for comparison. Although pH manipulation causes a delay in Ssa4 production, it does not affect the ultimate level of induction. (C) Growth rate difference in cells treated with ionophore for 35 min at room temperature followed by return to ambient growth conditions. Competitor was untreated cells. The values cluster around zero, indicating little to no loss of fitness due to ionophore treatment. Bottom dashed line shows theoretical minimum of the growth rate difference, which would result if cells completely arrested growth. These data are the same as those in Figure 6B, light-colored points. (D) Comparison of Ssa4-mCherry induction in ionophore treated cells that were either heat stressed (left) or held at room temperature (right). Acidification artificially induced by ionophore treatment does not cause appreciable accumulation of stress protein.
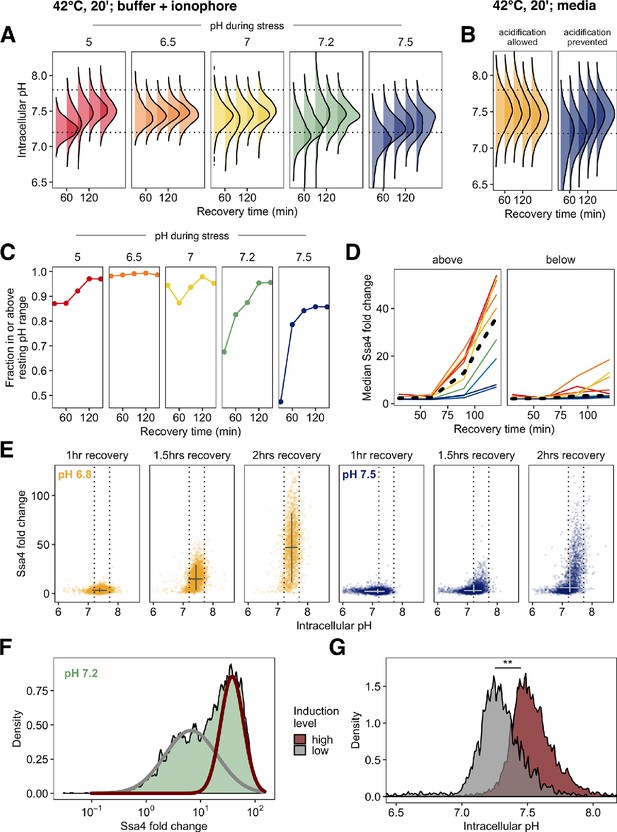
Post-stress acidification can rescue induction of the heat shock response.
(A) Intracellular pH distributions during recovery in cells held at various pH values during 42°C heat shock. Dashed horizontal lines represent the resting pH range for untreated cells. (B) Same as A, but measurement made following 42°C treatment in media without ionophore. (C) Fraction of cells that have entered the resting pH range during recovery. (D) Ssa4-mCherry fold change in cells above (left) or below (right) the lower bound of the resting pH range. Color is pH during stress, black line is the median of all cells. (E) Relationship between intracellular pH and Ssa4 fold change on the single cell level during recovery. Return to the resting pH, bounded by dotted lines, appears to precede Ssa4 induction, and is necessary but not sufficient for high expression levels. (F) Distribution of Ssa4 fold change during recovery from heat stress at pH 7.2. A two-component mixture model was used to classify cells into two groups: low (gray) and high (red) induction level (>0.90 posterior probability cutoff used for assignment). (G) Distribution of intracellular pHs in cells belonging to either the high-expression class (red) or the low-expression class (gray). ** , Wilcoxon rank sum test.
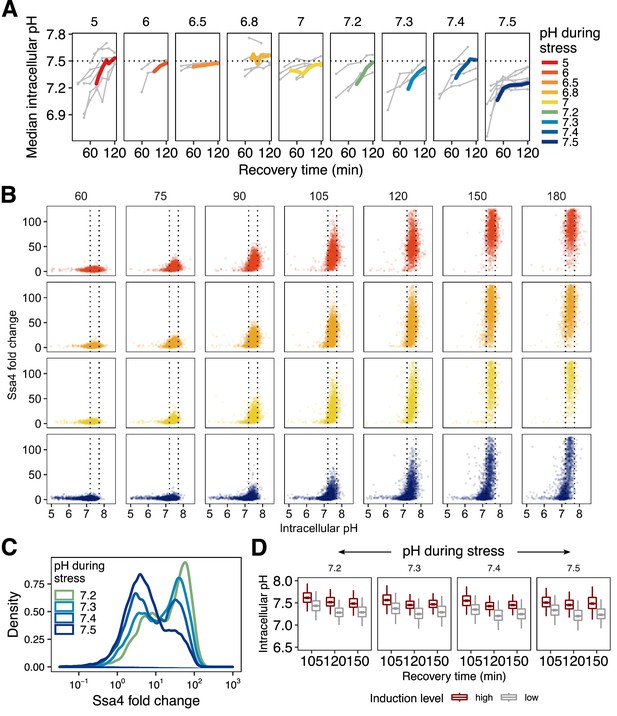
Mean pH during recovery for all pH values during stress, including averages and single-cell data.
(A) Intracellular pH recovery after stress at different intracellular pHs. Thick, colored lines are the moving average of individual experiments (thin gray lines). (B) Recovery of intracellular pH is correlated with high Ssa4 levels on the single-cell level. Cells that are stressed at the resting pH have a large proportion of cells that do not recover intracellular pH and do not produce high levels of Ssa4. (C) Bimodal distribution of Ssa4 fold-change in cells stressed close to or at the resting pH. (D) Intracellular pH distributions for both high-expressing (red) and low-expressing (gray) cells for all conditions shown in Figure 5C at multiple timepoints during recovery from heat shock.
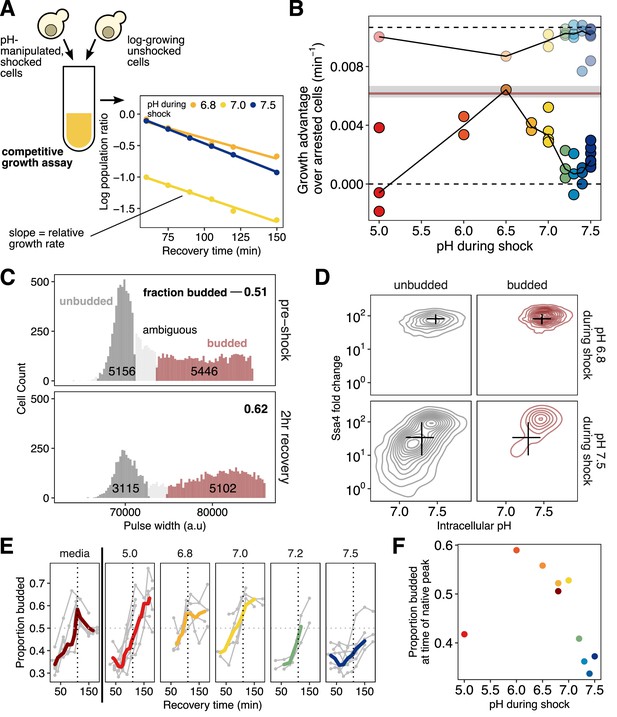
Intracellular acidification during heat shock promotes increased fitness during recovery on the population and single-cell levels.
(A) Schematic of the competitive growth assay which measures population fitness. (B) Intracellular pH during heat shock vs. relative growth rate expressed as the difference from the theoretical minimum for completely arrested cells. Each point is an independent experiment; opaque points are heat-shocked populations, transparent are control populations treated with ionophore at room temperature. Gray bar is the equivalent fitness loss for cells shocked without pH manipulation. See Materials and methods and Figure 6—figure supplement 1F for details and all fits. (C) Classification of cells: large/budded (red) and small/unbudded (dark gray). Classification was performed by fitting the forward-scatter pulse width to a two-component Gaussian mixture model and using a 90% confidence cutoff to classify cells into each category; cells that did not meet this criterion (shown in light gray) are not included in the analysis. Numeric labels show the number of cells in each category. (D) Ssa4 fold-change versus intracellular pH for budded and unbudded cells during recovery at three hours post-shock. Black lines are summary statistics of the entire population (budded and unbudded) and span the middle 50% of the data, crossing at the median. (E) Proportion of cells budded as a function of time during recovery. The characteristic shape of the curve derived from cells stressed without pH manipulation is shown in the left-most panel. The proportion budded peaks at approximately two hours of recovery (vertical dashed line). (F) Summary of E, showing the average proportion of cells budded between 90 and 120 min after heat shock.
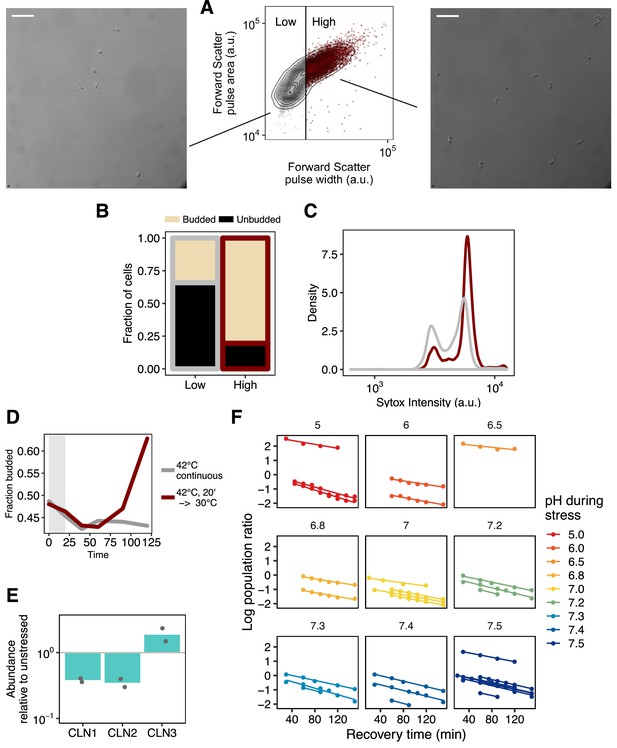
Confirmation of budding classifier by microscopy and DNA staining by flow, full fits for slopes plotted in B.
(A) Cells were partitioned into two categories in the forward scatter width channel and were sorted based on this partitioning. Sorted cells were fixed and visualized by microscopy. Representative images for each population are shown. Scale bar is 25 μm. (B) Quantification of microscopy data; N = 217 cells scored. (C) Fixed cells were stained with Sytox and analyzed by flow cytometry to assess DNA content. The relative heights of the two peaks reflect the proportion of cells in each population that have doubled their DNA, and are thus actively growing. (D) Proportion of budded cells as a function of time for cells moved from 30°C and held at 42°C (gray line) or cells that experienced a 42°C heat shock followed by recovery at 30°C (red line). Both populations initially show a dip in the proportion of budded cells, but populations returned to ambient growth temperature then rapidly and synchronously re-enter the cell cycle, as evidenced by an increased in the proportion of budded cells. (E) Relative mRNA abundance for three cell-cycle transcripts after a 42°C, 20 min heat stress. Degradation of CLN1 and CLN2 is characteristic of heat shock (Rowley et al., 1993). (F) Fits used to determine the relative growth rate for all data shown in Figure 6B. The log of the population ratio as a function of time was fit with a line using linear least squares.
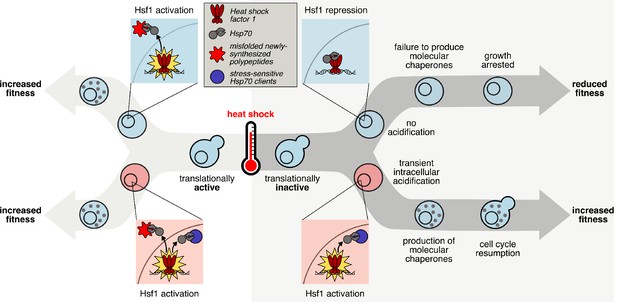
The transcriptional response to heat shock, chaperone production, and cellular fitness are promoted by intracellular acidification.
The key step in initiating the transcriptional heat shock response is the release of Hsf1 repression by Hsp70 through titration of the chaperone via client binding. When cells are translationally active (left hand side), newly synthesized polypeptides that misfold in response to elevated temperature can act as the trigger. However, cells that are not actively translating (right hand side) can still respond to heat shock, dependent on transient intracellular acidification, either during or after the temperature increase. We predict that pH-sensitive, stress-sensing proteins, similar to those already discovered, can act to titrate Hsp70, relieving Hsf1 repression and activating the transcriptional heat shock response.





