The copy-number and varied strengths of MELT motifs in Spc105 balance the strength and responsiveness of the spindle assembly checkpoint
Figures
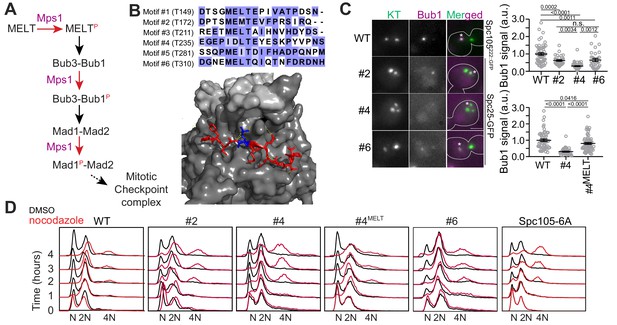
Only one MELT motif per Spc105 is sufficient for SAC signaling in nocodazole-treated cells.
(A) A simplified schematic of the signaling cascade of the SAC in budding yeast (Mad3/BubR1, Cdc20, etc. are not shown). (B) Top: amino acid sequence alignment for the six MELT motifs in the Saccharomyces cerevisiae Spc105. Bottom: Crystal structure of the MELpT-Bub3-Bub1 complex (adapted from PDB 4BL0 and 2I3S). Note that the phosphothreonine (shown in blue, inter-chain hydrogen bonds highlighted by dashed lines) buried in a grove on Bub3 surface. (C) Micrographs display representative images of nocodazole-treated yeast cells with fluorescently labeled kinetochores and Bub1-mCherry. Asterisks mark Bub1-mCherry colocalized with the unattached kinetochore clusters. Kinetochores in the other, larger cluster proximal to the spindle pole bodies do not recruit SAC proteins, presumably because they remain attached to short microtubule stubs at the spindle pole bodies. Scale bar ~3.2 µm. Scatterplots show the quantification of Bub1-mCherry fluorescence in wild-type (WT) and mutant strains (labeled by the number designation of the active MELT motif). Fluorescence values have been normalized to the average Bub1-mCherry fluorescence in wild-type cells. (mean+ s.e.m. Top: n = 55, 33, 51 and 32 for WT, #2, #4 and #6 respectively, accumulated from two technical replicates. Bottom: n = 55, 92 and 68 for WT, #2 and #6 respectively, pooled from two technical replicates. n.s. Not significant. (D) Quantification of DNA content in cells treated with nocodazole using flow cytometry. The assay was performed once.
-
Figure 1—source data 1
The data for two scattered plots shown in Figure 1C, right top and bottom.
- https://cdn.elifesciences.org/articles/55096/elife-55096-fig1-data1-v2.xlsx
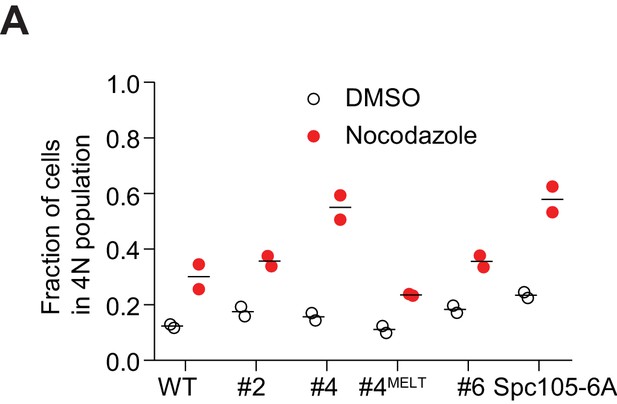
A The fraction of cells with 4N ploidy in the flow cytometry experiments shown in Figure 1D.
Data from 3 hr and 4 hr samples for each strain are pooled. Also see supplementary file 1.
-
Figure 1—figure supplement 1—source data 1
Data required to prepare the graph shown in Figure 1—figure supplement 1A.
- https://cdn.elifesciences.org/articles/55096/elife-55096-fig1-figsupp1-data1-v2.xlsx
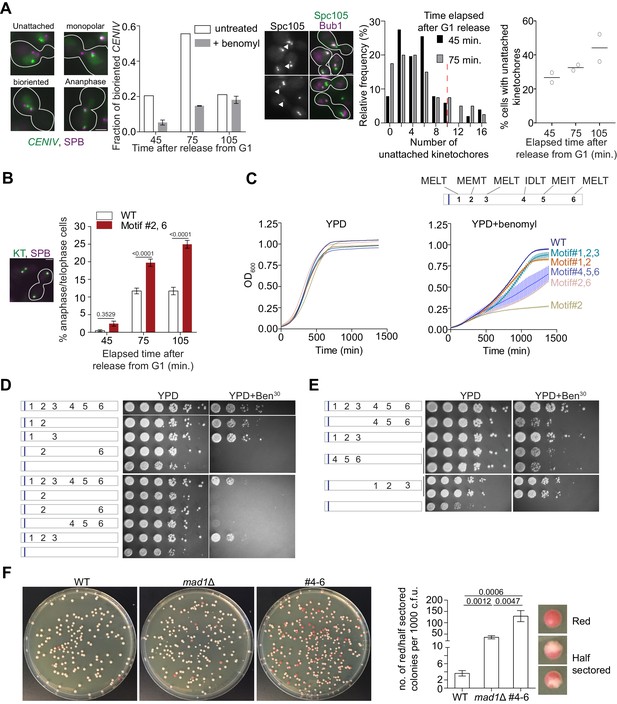
More than one high affinity MELT motifs are necessary to minimize chromosome loss.
(A) Effects of benomyl on chromosome biorientation. Left: Kinetics of chromosome IV biorientation visualized using a centromere-proximal TetO array. Micrographs depict representative images of cells showing fluorescently labeled TetR-GFP bound to the TetO array and Spindle Pole Bodies (SPB). Scale bar ~3.2 µm. Second from the left: Quantification of the fraction of cells with bioriented chromosome IV. Note that the reduced fraction of untreated cells with bioriented chromosomes 105 min after release from G1 is because most cells complete anaphase by this time. (n = 259, 252 and 398 at 45, 75 and 105 min respectively in normal media, one repeat. n = 855, 816 and 626 respectively at 45, 75 and 105 min in benomyl-containing media from three technical repeats). When the two CENIV foci were unresolvable and localized in the vicinity of one of the two SPBs, they were scored as monopolar attachments. When the two CENIV foci were clearly separated from each other and located along the spindle axies, they were considered to be bioriented. Middle: Representative micrographs of cells grown in benomyl media containing unattached kinetochores recruiting Bub1. Arrow heads indicate the unattached kinetochores with Bub1 localizations. Scale bar ~3.2 µm. Right: Quantification of the number and frequency distribution of unattached kinetochores in yeast cells growing in media containing benomyl. The unattached kinetochore number was estimated by comparing the Spc105-GFP fluorescence from the unattached kinetochore cluster (marked by Bub1-mCherry recruitment) with the total fluorescence from the kinetochores along the spindle axis. (n = 686, 501 and 543 at 45, 75 and 105 min respectively, pooled from two technical repeats). Red dashed line in relative frequency plot indicates the average number of unattached kinetochores observed after 90 min nocodazole treatment of wild type yeast cell (Aravamudhan et al., 2016). (B) Left: Micrograph displays representative cells in metaphase and anaphase in benomyl-containing media. Scale bar ~3.2 µm. Right: Quantification of the number of anaphase cells observed after the indicated time following release from a G1 arrest for the indicated strains. (mean+ s.e.m, n = 1029, 889 and 992 at 45, 75 and 105 min respectively for WT cells, pooled from three experimental repeats. n = 890, 864 and 812 at 45, 75 and 105 min time points respectively for #2,6, pooled from three technical replicates). (C) Top: Schematic of the Spc105 phosphodomain with the amino acid sequence indicated at the top. Bottom: Quantification of the evolution of cell density of the indicated strains in rich media (left) and media-containing benomyl (right). (D–E) Assessment of the sensitivity of yeast strains to benomyl using the spotting assay. Schematics on the left show the active motif number and its position. The blue bar represents the ‘basic patch’ in Spc105, which promotes PP1 recruitment. The photographs on the right show the results of spotting a serial dilution of yeast cells on rich media (YPD) and media containing benomyl (20 or 30 μg/ml). (F) Assessment of chromosome loss by colony sectoring assay. Left: Images of WT, mad1Δ and #4–6 colonies grown in YPD plates. Right: Bar graph shows the rate of loss of SUP11 containing chromosome fragment measured as the number of red/half red colonies (example shown on the right) for every 1000 colonies plated. N = 5081, 4406 and 4965 for WT, mad1Δ and #4–6 respectively, pooled from at least six technical repeats.
-
Figure 2—source data 1
Data for graphs depicted in Figure 2A, B, C and F.
Individual pages were created in the excel file for each subfigure.
- https://cdn.elifesciences.org/articles/55096/elife-55096-fig2-data1-v2.xlsx
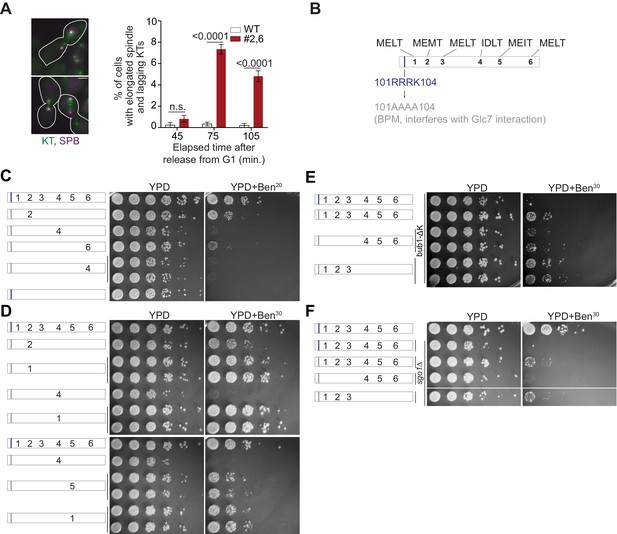
Requirements of the MELT motif number per Spc105 and their affinity for survival on media containing benomyl.
(A) Quantification of cells of indicated strains showing an elongated spindle, which is indicative of anaphase onset. (mean+ s.e.m., n = 1029, 889 and 992 at 45, 75 and 105 min respectively for WT, n = 890, 864 and 812 at 45, 75 and 105 min respectively for #2,6, pooled from three technical repeats). n.s. Not significant. (B) Schematic of the Spc105 phosphodomain with the amino acid sequence indicated at the top. The 101-RRRK-104 span is designated as the anterior basic patch of Spc105. It is implicated in the binding of PP1 to Spc105 (Roy et al., 2019). (C–F) Benomyl-sensitivity of the indicated strains. Schematic on the left displays the number and position of active MELT motifs. The gray bar indicates that the basic patch is replaced by non-polar alanine residues (101-RRRK-104::AAAA). This mutation, either abbreviated as BPM or denoted as Spc105BPM, results in reduced PP1 binding to Spc105 (Roy et al., 2019). It does not increase the strength of SAC signaling from unattached kinetochores, but results in a modest SAC silencing defect. Importantly, reduced PP1 binding to Spc105 significantly improves chromosome biorientation. Consequently, even SAC deficient yeast cells can grow on benomyl (Roy et al., 2019). (C) We previously found that mutants expressing Spc105-5A are inviable in presence of benomyl (Aravamudhan et al., 2016). Mutation of the basic patch (gray bar) suppresses that sensitivity of Spc105-5A variants only if the MELT motif possesses the optimal, consensus sequence. The benomyl sensitivity of the sixth MELT motif in this assay is surprising given that this motif possesses the consensus amino acid sequence. We speculate that the lower activity is due to the absence of the negatively charged residue directly downstream from the MELT motif, which contributes to the interaction of the MELT motif with the Bub3-Bub1 complex (Primorac et al., 2013). (D) The activity of a MELT motif is determined by its sequence, but not position. (E–F), The benomyl sensitivity of strains expressing Spc105BPM with either #1–3 or #4–6 motifs as active MELT motifs along with either bub1Δkinase or sgo1Δ. The basic patch mutation is necessary, because it alleviates partially suppresses the lethality of bub1Δkinase and sgo1Δ in benomyl containing media (Roy et al., 2019).
-
Figure 2—figure supplement 1—source data 1
Data required for the graph shown in Figure 2—figure supplement 1A.
- https://cdn.elifesciences.org/articles/55096/elife-55096-fig2-figsupp1-data1-v2.xlsx
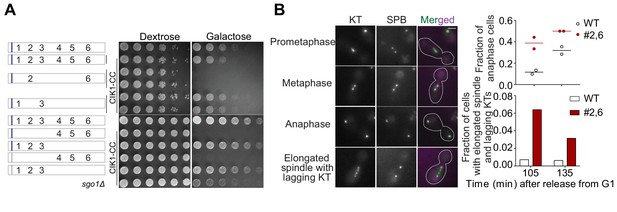
Multiple, strong MELT motifs per Spc105 are essential for cell survival when chromosome biorientation is challenged by conditionally disrupting the mitotic Cik1 functions.
(A) Effect of Cik1-CC overexpression (stimulated by the use of galactose as the carbon source) on the indicated strains. (B) Micrographs on the left show the scoring scheme used to identify cells in anaphase and those with elongated spindles, which are indicative of anaphase onset (but cannot be confirmed as anaphase). Bar ~3.2 µm. Scatter and bar plots on the right show quantification. (n = 694 and 476 at 105 and 135 min respectively for WT and 547 and 317 at 105 and 135 min respectively for #2,6 from two repeats).
-
Figure 2—figure supplement 2—source data 1
Data needed for the plots shown in Figure 2—figure supplement 2B top right and 2B bottom right.
- https://cdn.elifesciences.org/articles/55096/elife-55096-fig2-figsupp2-data1-v2.xlsx
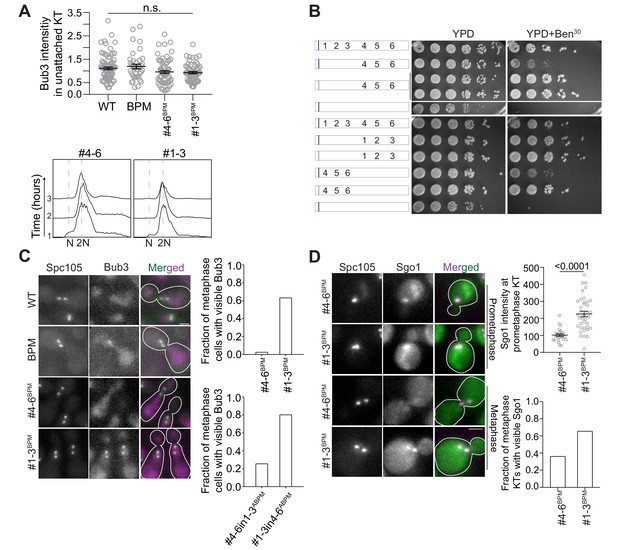
The difference in the SAC signaling activities of the first three and last three MELT motifs in Spc105 is detectable in benomyl-containing media, but not in cells treated with nocodazole.
(A) Top left: Scatter plot revealing the intensity of Bub3-mCherry in unattached kinetochores of indicated strains when the cells were treated with nocodazole for 2 hr. (mean+ s.e.m. n = 82, 39, 67 and 59 for WT, BPM, #4-6BPM and #1-3BPM respectively, accumulated from two repeats). Bottom left: Representative flow cytometry-based quantification of the DNA content of the indicated strains during a prolonged exposure to nocodazole. (B) Benomyl spotting assay of indicated strains. Schematic on the left shows the number and position of active MELT motifs. The basic patch at the N-terminus is indicated by the blue bar; gray bar indicates that the basic residues in this patch are replaced by non-polar alanine residues. Mutation of anterior basic patch alleviates the growth of strains expressing Spc105#4-6, independently of the positions of #4–6 in Spc105 phosphodomain. (C) Representative images show the recruitment of Bub3-mCherry to bioriented kinetochore clusters in the indicated strains. Bar ~3.2 µm. Right: Bar graphs show the fraction of metaphase cells with Bub3 recruited at the kinetochores in CDC20 depleted (top) and cycling (bottom) population of the indicated strains. Top: n = 116 and 106 for #4-6BPM and #1-3BPM respectively, pooled from two biological replicates for each strain. Bottom: n = 90 and 96 for #4-6in1-3 and #1-3in4-6 respectively, accumulated from two biological replicates for each strain. n.s. Not significant. (D) Left: Representative micrographs showing the colocalization of Sgo1-GFP with kinetochore clusters undergoing biorientation and after biorientation (kinetochores visualized by Spc105222:mCherry, scale bar ~3.2 μm). Right: Scatter plot at the top displays the quantification of Sgo1-GFP colocalized with prometaphase kinetochores. n = 26 and 38 for #4-6BPM and #1-3BPM respectively, pooled from two technical repeats, p<0.0001, derived from Mann-Whitney test. Bar plot at the bottom displays the scoring of cells wherein Sgo1-GFP visibly colocalizes with bioriented kinetochore clusters. n = 133 and 84 for #4-6BPM and #1-3BPM respectively, pooled from two experimental repeats, p<0.0001, derived from Fisher’s exact test.
-
Figure 2—figure supplement 3—source data 1
Data required in the graphs mentioned in Figure 2—figure supplement 3A top, C right top, C right bottom, D right top and D right bottom.
Individual pages were created in this file for each subfigure.
- https://cdn.elifesciences.org/articles/55096/elife-55096-fig2-figsupp3-data1-v2.xlsx
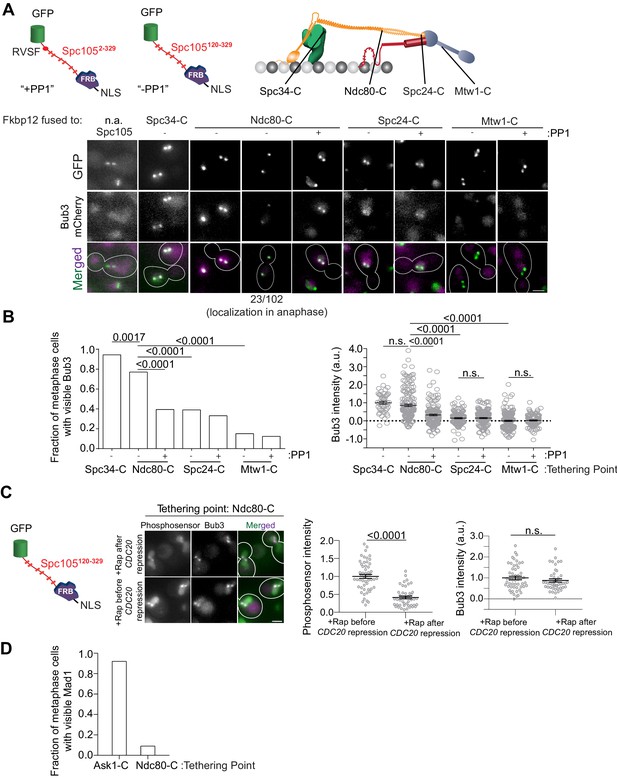
A gradient of Mps1 kinase activity continues to phosphorylate MELT motifs after chromosome biorientation.
(A) Top left: Cartoons show schematics of the two Spc105 phosphodomains tethered to kinetochore subunits using the rapamycin-induced dimerization of Fkbp12 and Frb domains. Top right: The kinetochore subunits (Spc34-C, Ndc80-C, Spc24-C and Mtw1-C) at which, the phosphodomains were tethered by rapamycin induced dimerization. Bottom: Micrographs show representative images of cycling cells wherein the phosphodomain is tethered to the C-terminus of the indicated kinetochore subunit. (n.a. - Not Applicable). (B) Left: The bar plot shows the scoring of cells with bioriented kinetochore clusters based on whether or not they visibly recruit Bub3-mCherry. (n = 71 for Spc34-C, 109 and 94 for Ndc80-C, 95 and 103 for Spc24-C, 106 and 122 for Mtw1-C, accumulated from two experimental and biological repeats (whenever possible). Right: The scatter plot on the right shows the quantification of Bub3-mCherry fluorescence from all cells with bioriented kinetochore clusters. (mean+ s.e.m. n = 47 for Spc34-C, 159 and 153 for Ndc80-C, 114 and 154 for Spc24-C, 173 and 104 for Mtw1-C, accumulated from two experimental and biological repeats wherever possible). n.s. Not significant. (C) Micrographs show the recruitment of Bub3-mCherry by the phosphosensor tethered at the Ndc80 C-terminus in cycling (before CDC20 repression) and metaphase arrested cells (after CDC20 repression). Scatterplots show the relative fluorescence signal from the GFP-tagged phosphosensor and Bub3-mCherry respectively. (mean+ s.e.m. n = 52 and 45 for rapamycin treated samples before and after Cdc20 depletion from two technical replicates). n.s. Not significant. (D) Bar graph shows fraction of cells visibly recruiting Mad1-mCherry as shown in the micrographs (n = 242 and 142 for Ask1-C and Ndc80-C respectively).
-
Figure 3—source data 1
Data that are included in the plots of Figure 3B left and right, 3C left and right and 3D are shown here.
Individual pages were created in this excel file for each subfigure.
- https://cdn.elifesciences.org/articles/55096/elife-55096-fig3-data1-v2.xlsx
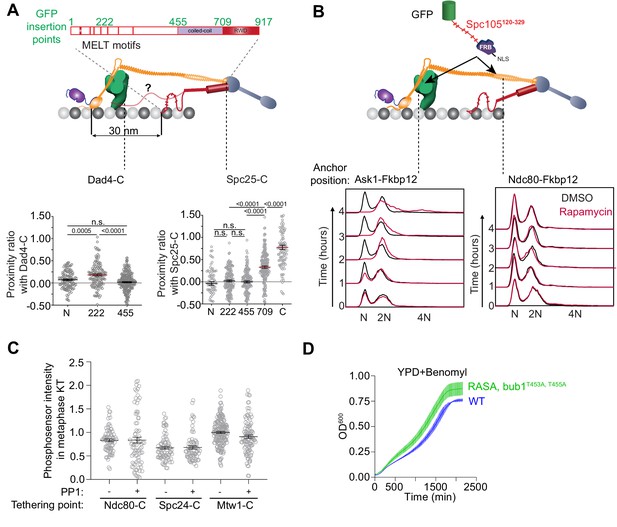
The distribution of the Spc105 phosphodomain in bioriented kinetochores and the position-dependent effects of tethering an additional phosphodomain within the kinetochore.
(A) The assessment of the nanoscale distribution of the unstructured phosphodomain of Spc105 using FRET. Top: Schematic displays the GFP-insertion positions within Spc105. Middle: Positions of the mCherry tagged kinetochore subunits (Dad4-C and Spc25-C) that demarcate the region that was expected to be occupied by the Spc105 phosphodomain. Bottom: Scatter plots show the proximity ratio (which is proportional to FRET) of donor GFP fused at the indicated residue in Spc105 with Spc25 C-terminus (left) and Dad4 C-terminus (right). (mean+ s.e.m. n = 74, 165, 142, 177 and 103 respectively for FRET involving Spc25-mCherry, accumulated from at least three repeats. n = 114, 134 and 224 for N, 222 and 455 respectively from three repeats for FRET involving Dad4-mCherry, accumulated from at least two repeats). n.s. Not significant. (B) Flow cytometry-based quantification of the DNA content in cells treated with rapamycin (prior to sample collection at 0 min) to tether the Spc105 phosphodomain anchored to Ask1-2xFkpb12 (which is proximal to the CH-domains) or Ndc80-2xFkbp12 (which is distal from the CH-domains) (schematic shown at the top). Bottom: Representative histogram from two trials for each strain is shown. (C) Quantification of amount of Spc105 phosphodomain tethered to bioriented kinetochore clusters (mean+ s.e.m. n = 74 and 80 for Ndc80-C, 78 and 76 for Spc24-C, 102 and 173 for Mtw1-C, pooled from at least two experimental replicates). (D) (Supports in Figure 4C) Quantification of population growth for the indicated strains in benomyl-containing media (30 µg/ml) (mean ± s.e.m., n = at least 3).
-
Figure 3—figure supplement 1—source data 1
Data required to prepare the plots included in Figure 3—figure supplement 1A bottom left and right, C and D.
Individual pages were created in this file for each subfigure.
- https://cdn.elifesciences.org/articles/55096/elife-55096-fig3-figsupp1-data1-v2.xlsx
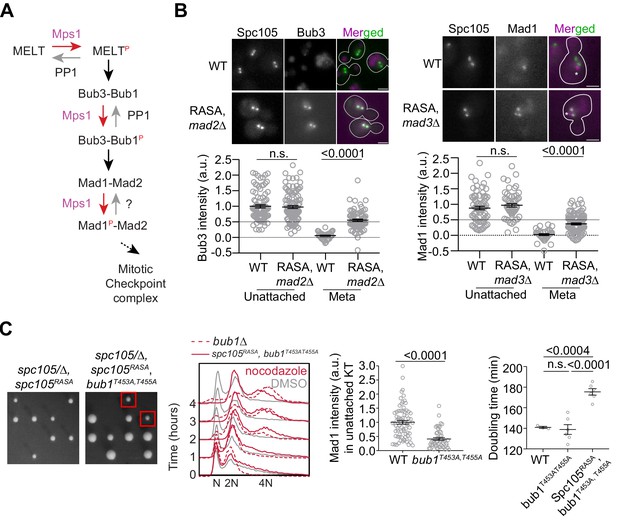
Revealing the determinants of the responsiveness of the SAC to PP1-mediated silencing.
(A) A schematic of the SAC signaling cascade highlighting the potential steps that can be disrupted by Protein Phosphatase I (PP1). (B) Top: Representative micrographs of cells with bioriented kinetochore clusters showing the colocalization of the indicated proteins with fluorescently labeled kinetochores. Note that the Mad1-mCherry puncta marked with an asterisk result from the deletion of the nuclear pore protein Nup60. These puncta are not associated with kinetochores. The ones co-localized with kinetochores are marked with arrowheads. Scale bar ~3.2 µm. Scatter plots at the bottom show the quantification of fluorescence signal of kinetochore colocalized Bub3-mCherry and Mad1-mCherry (mean+ s.e.m., normalized to the respective average signal measured in nocodazole-treated cells). Bub3-mCherry: n = 93 and 37 for WT, 107 and 77 for Spc105RASA, mad2Δ. Mad1-mCherry: n = 74 and 34 for WT, 60 and 111 for Spc105RASA, mad3Δ, pooled from two technical repeats. n.s. Not significant. (C) Left: Tetrad dissection analysis of the indicated strains. Second from the left: Flow cytometry of DNA content of the indicated strains during prolonged exposure to nocodazole. The best of the two technical repeats is shown here. Second from the right: quantitative comparison of Mad1-mCherry recruited by unattached kinetochores in nocodazole-treated cells. (mean+ s.e.m. n = 74 for WT and 49 for bub1453A, 455A, pooled from two experimental repeats). Right: Doubling time of the indicated strains in rich media (horizontal line indicates the mean value, obtained from at least three experimental repeats). n.s. Not significant.
-
Figure 4—source data 1
Data needed to prepare the graphs shown in Figure 4B bottom left and bottom right, 4C 2nd from the right and 4C right.
Individual pages were created in this file for each subfigure.
- https://cdn.elifesciences.org/articles/55096/elife-55096-fig4-data1-v2.xlsx
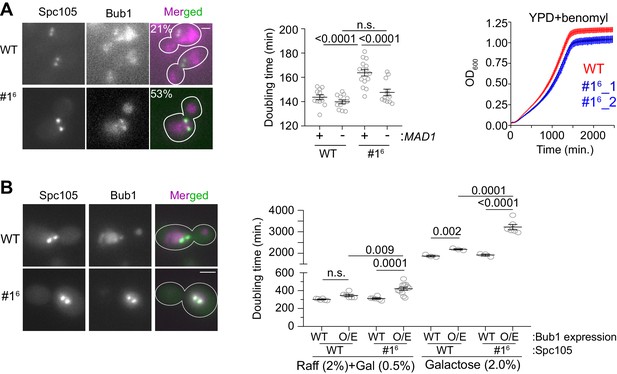
The influence of MELT motif activity and Bub1 expression level on the strength and responsiveness of the SAC.
(A) Left: Representative micrographs show the recruitment of Bub1-mCherry to bioriented kinetochore clusters (%age noted at the top of each micrograph indicates the fraction of cells with Bub1-mCherry visibly recruited to bioriented kinetochore clusters; also see Figure 5—figure supplement 1A) Scale bar ~3.2 µm. Second from the left: Quantification of the growth rate of indicated strains in non-selective media. Second from the right: The scatter plot shows the indicated strains (mean+ s.e.m from at least six repeats). Right: Quantification of the growth rate of indicated strains in benomyl-containing media (20 µg/ml) (mean+/-s.e.m. from n = 3). n.s. Not significant. (B) Left: Representative images show Bub1 recruitment at the bi-oriented kinetochores when Bub1 is overexpressed in indicated strains (n = 2). Scale bar ~3.2 µm. Right: Scatter plot shows the doubling time of the indicated strains, grown in raffinose (2%)+galactose (0.5%) and in galactose (2%). mean+ s.e.m obtained from at least six repeats. Note that the growth rates of all strains are greatly reduced, when raffinose or galactose rather glucose are used as the carbon source. n.s. Not significant.
-
Figure 5—source data 1
The data required for creating the graphs in Figure 5A middle and right and 5B right.
Individual pages were created in this file for each subfigure.
- https://cdn.elifesciences.org/articles/55096/elife-55096-fig5-data1-v2.xlsx
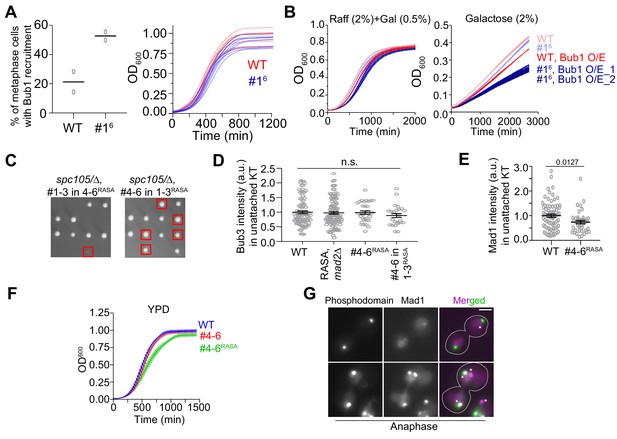
The influence of the affinity of MELT motifs to Bub3-Bub1 expression on the responsiveness of the SAC to silencing mechanisms.
(A) Scatter plot showing the percentage of cells with bioriented kinetochores with Bub1 localization in the indicated strains. (n = 211 and 74 for WT and MELT16 respectively, horizontal line indicates mean value).( B) Growth curves of the indicated strains in media containing raffinose+galactose (left) and just galactose (right) quantified by measuring the absorbance at 600 nm (mean ± s.e.m. shown for each time point from three replicates). (C) Tetrad dissection analysis of the indicated strains. Red squares mark the tetrads of the desirable genotype. The dissections were repeated with two biological replicates. (D) Quantification of Bub3-mCherry intensity values of the indicated strains. (mean+ s.e.m. n = 92, 107, 33 and 26 for WT, spc105RASA, #4-6, mad2Δ, spc105RASA, #4-6 and spc105RASA, #4-6in1-3 respectively, pooled from two repeats of the experiment). n.s. Not significant. (E) Quantitative comparison of the amount of Mad1-mCherry recruited by unattached kinetochores in spc105RASA, #4-6 cells treated with nocodazole. n = 74 and 42 for WT and spc105RASA, #4-6 respectively, pooled from two experimental repeats. D and E support Figure 6B. (F) Left: Growth curve shows quantification of the evolution of cell density of the indicated strains in YPD. (G) Representative micrographs display Mad1-mCherry localization relative to kinetochores of anaphase cells in the indicated strains after treatment with rapamycin. The Mad1-mCherry puncta marked with an asterisk result from the deletion of the nuclear pore protein Nup60. These puncta are not associated with kinetochores. The ones co-localized with kinetochores are marked with arrowheads.
-
Figure 5—figure supplement 1—source data 1
Data needed to prepare the graphs shown in Figure 5—figure supplement 1A left and right, B left and right, D, E and F.
Individual pages were created in this file for each subfigure.
- https://cdn.elifesciences.org/articles/55096/elife-55096-fig5-figsupp1-data1-v2.xlsx
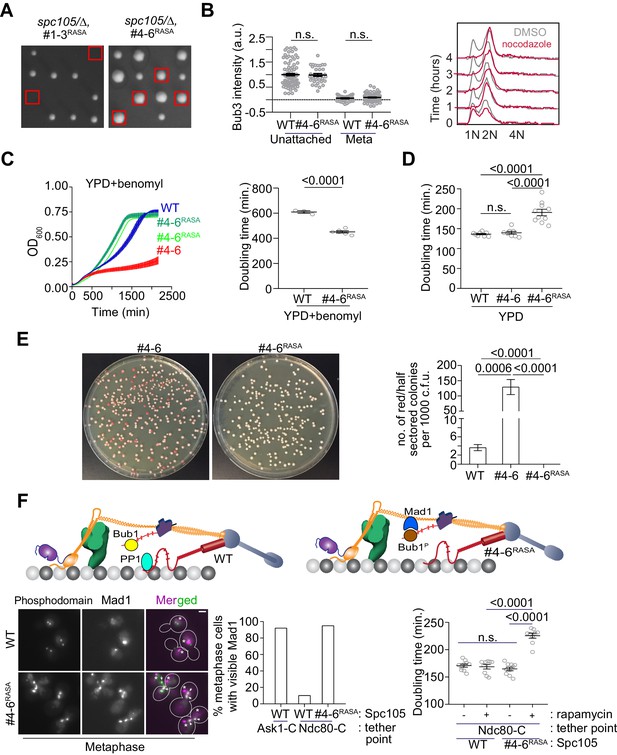
Designing an Spc105 variant optimized for SAC signaling, error correction, and SAC silencing.
(A) Left: Tetrad dissections showing the rescue of Spc105RASA by inactivating the first three MELT repeats. Right: Scatter plot depicting Bub3 intensities in unattached and bioriented kinetochores of indicated strains (mean+ s.e.m. n = 92 and 37 for WT, 33 and 98 for #4-6RASA, pooled from two technical repeats). n.s. Not significant. (B) Representative flow cytometry-based quantification of the DNA content of the indicated strains during a prolonged exposure to nocodazole (n = 2). (C) Quantification of the doubling times of the indicated strain in YPD respectively (horizontal line marks the mean value, minimum number of experimental repeats n = 2).( D) Left: Quantification of population growth for the indicated strains in presence of benomyl (30 µg/ml) (mean ± s.e.m., n = 3). Right: Quantification of the doubling times of the indicated strain in YPD+benomyl (30 µg/ml) respectively (horizontal line marks the mean value, minimum number of experimental repeats n = 2). (E) Estimation of chromosome loss by colony sectoring assay. Left: Plate images of #4–6 and #4-6RASA strains grown in YPD plates. Right: Bar graph displays the number of red or half sectored colonies per 1000 colony forming units. N = 5081, 4965 and 6596 for WT, #4–6 and #4-6RASA respectively, pooled from ≥6 technical repeats. (F) Top: Model explaining why Spc105 phosphodomain tethered in the inner kinetochore does not activate the SAC. Bottom left: Representative micrographs display Mad1-mCherry localization relative to bioriented kinetochores in the indicated strains after treatment with rapamycin. The Mad1-mCherry puncta marked with an asterisk result from the deletion of the nuclear pore protein Nup60. These puncta are not associated with kinetochores. The ones co-localized with kinetochores are marked with arrowheads. Scale bar ~3.2 µM. Bottom middle: Bar graph of the percentage of cells visibly recruiting Mad1-mCherry (n = 242, 142 and 290 for Ask1-C, Ndc80-C and Ndc80-C, #4-6RASA respectively, derived from two technical repeats). Bottom right: Scatter plot presents the doubling time of the indicated strains. mean+ s.e.m from ≥three experimental repeats. n.s. Not significant.
-
Figure 6—source data 1
Data required for graphs shown in Figure 6B and C left and right, 6D, 6E, 6F bottom middle and 6F bottom right.
Individual pages were created in this file for each subfigure.
- https://cdn.elifesciences.org/articles/55096/elife-55096-fig6-data1-v2.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (S. cerevisiae) | AJY# | This paper and previous literatures. | N.A. | Detailed information of strain genotypes is provided inSupplementary file 2. See plasmid and strain construction below for more details. |
| Recombinant DNA reagents | pAJ# | This paper or obtained by requests to other labs | N.A. | Detailed information of the recombinant DNA reagents (plasmids) used in this study is provided in Supplementary file 3. See plasmid and strain construction below for more details. |
| Chemical compound, drug | Benomyl | Millipore Sigma | 45339 | Both of the benomyl powders have been used in spotting and plate reader assays. Please refer to materials and methods for concentration of Benomyl. See benomyl sensitivity assay mentioned below for more details. |
| Chemical compound, drug | Benomyl | Millipore Sigma | 381586 | |
| Chemical compound, drug | Nocodazole | Fisher Scientific | AC358240100 | Used to disrupt spindle and create unattached kinetochores. Please see below for concentration of Nocodazole. |
| Chemical compound, drug | Propidium Iodide (PI) | Millipore Sigma | P4864 | Used in flow cytometry. Please refer to flow cytometry section below for concentration of PI, |
| Chemical compound, drug | Rapamycin | Fisher Scientific | NC9362949 | Used in phosphodomain tethering assays. Please see flow cytometry section for concentration of Rapamycin. |
| Chemical compound, drug | RNase | Fisher Scientific | SIG10109169001 | Used in flow cytometry. Please refer to flow cytometry section for concentration of RNase. |
| Software/algorithm | Graphpad Prism | Graphpad software Inc | Prism Version 8 | Used for making plots and performing statistical analyses |
| Software/algorithm | ImageJ (Fiji) | https://imagej.net/Fiji/Downloads | Used for Spc105, phosphodomain tethering, Bub3, Bub1, Mad1 and Sgo1 intensity measurement. | |
| Software/algorithm | Matlab | Mathworks | For colony counting, we used Colony counter | |
| Software/algorithm | Metamorph | Molecular devices | Used for imaging involving microscopy assays. Please refer to Materials and methods for details of imaging | |
| Software/algorithm | Adobe Illustrator | Adobe creative cloud | Version 2019 | Used to assemble the data and to prepare the figures |
| Other | Gene and protein information resources for S. cerevisiae | Saccharomyces genome database (https://www.yeastgenome.org) | N.A. | Used for recombinant DNA reagents and S. cerevisiae strain construction. |
Additional files
-
Supplementary file 1
Table with detailed dataset obtained from flow cytometry assay depicted in Figure 1D.
- https://cdn.elifesciences.org/articles/55096/elife-55096-supp1-v2.docx
-
Supplementary file 2
Table with details of Saccharomyces Cerevisiae strains used in this study which includes the strain numbers, their background, details of genotype and where they have used in the study.
- https://cdn.elifesciences.org/articles/55096/elife-55096-supp2-v2.docx
-
Supplementary file 3
The details of recombinant DNA reagents or plasmids used in this study are tabulated in this file.
- https://cdn.elifesciences.org/articles/55096/elife-55096-supp3-v2.docx
-
Supplementary file 4
The source code files which were used in MATLAB to count red and white colonies in the chromosome loss assays mentioned in Figures 2F and 6E.
- https://cdn.elifesciences.org/articles/55096/elife-55096-supp4-v2.mlapp
-
Supplementary file 5
The source code files which were used in MATLAB to count red and white colonies in the chromosome loss assays mentioned in Figures 2F and 6E.
- https://cdn.elifesciences.org/articles/55096/elife-55096-supp5-v2.mlapp
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/55096/elife-55096-transrepform-v2.docx





