Cortical anchoring of the microtubule cytoskeleton is essential for neuron polarity
Figures
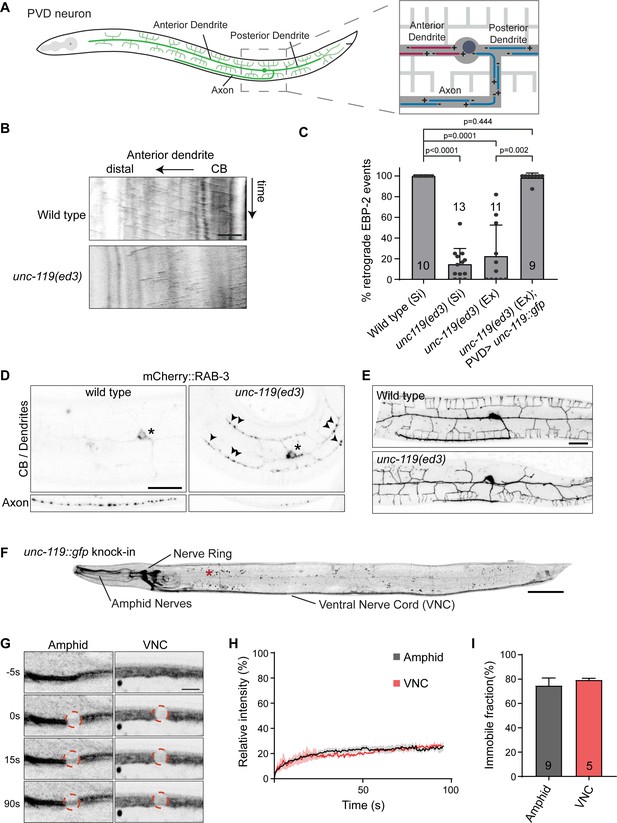
unc-119 is essential for neuronal microtubule organization.
(A) Schematic representation of the PVD neuron and its microtubule organization; it possesses two highly branched sensory dendrites and a single axon which connects to other neurons in the ventral nerve chord. Only the anterior dendrite has minus-end out microtubules (red), whereas the axon and posterior dendrites have plus-end out microtubules (blue) (Harterink et al., 2018). (B–C) Representative kymographs and quantification of the microtubule orientation in the PVD anterior dendrite. EBP-2::GFP was expressed in the PVD to visualize growing microtubules using the Pdes-2 promotor. EBP-2::GFP strains were single copy integration lines (Si) generated using mosSCI or were extrachromosomal lines (Ex). For unc-119 (Si) we used CRISPR to mutate the cb-unc-119 from the mosSCI generated line (wild type (Si)). Number of analyzed animals is indicated. Scale, 5 µm. (D) Representative images of synaptic vesicle localization in the PVD neuron marked by RAB-3::mCherry. Cell body (CB) and dendrites are separately shown from the axon to minimize the autofluorescence. Asterisk, marks the cell body; arrowheads point to RAB-3::mCherry in the dendrites; scale, 20 µm. (E) Representative image of the PVD morphology in wild type and the unc-119 mutant. Scale, 20 µm. (F) unc-119::gfp knock-in animals. Expression is strongest in neurons, but can also be detected at low levels in non-neuronal cells such as the vulva cells and the seam cells. The locomotion of the gfp knock-in animals is not affected (Figure 1—figure supplement 1M), indicating that the fusion protein is functional. The red asterisk marks autofluorescent gut granules (small puncta) seen throughout the animal. Image were straightened using in imageJ. Scale, 50 µm. (G) UNC-119::GFP FRAP in a dendrite bundle (Amphid) and axon bundle (VNC). Scale, 5 µm. (H–I) Average normalized intensity graph of UNC-119::GFP FRAP ± SEM (H) and the percentage immobile fractions after FRAP (I). Number of analyzed animals is indicated. Analyzed animals were from the L4 or young adult stage; error bars represent SD; statistical analysis, Kruskal-Wallis test followed by Dunn’s multiple comparisons test.
-
Figure 1—source data 1
Source data for graphs in Figure 1.
- https://cdn.elifesciences.org/articles/55111/elife-55111-fig1-data1-v1.xlsx
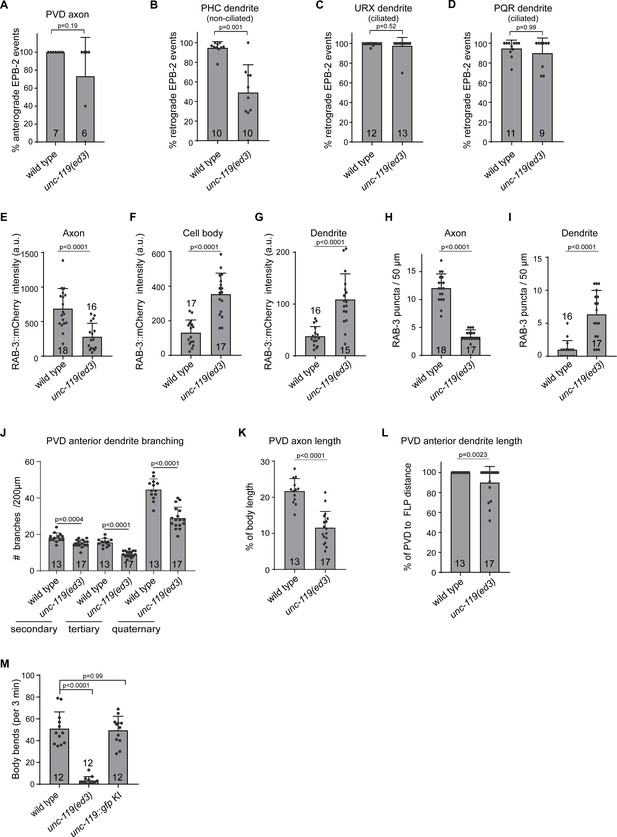
Quantifications of the unc-119 mutant phenotypes.
(A–D) Quantification of microtubule orientation using EBP-2::GFP in the PVD axon (A), the non-ciliated PHC dendrite (B) and the ciliated URX (C) and PQR (D) dendrites. (E–G) Quantification of RAB-3::mCherry fluorescence intensity in the PVD axon (E), the cell body (F) and the proximal anterior dendrite (G). Intensities are arbitrary units (a.u.). (H–I) Quantification of RAB-3::mCherry puncta in the axon (H) and proximal anterior dendrite (I). (J) Quantification of PVD anterior dendrite primary, secondary and quaternary branches, within 200 µm from the cell body. (K) Quantification of the relative axon length in the ventral nerve cord. (L) Quantification of the anterior dendrite outgrowth towards the FLP cell body localized in the head. (M) Quantification of animal locomotion during 3 min of forward motion (KI = knock in). Number of analyzed animals is indicated. Analyzed animals were from the L4 or young adult stage; error bars represent SD; statistical analysis, unpaired student T-test for (A–L) and Kruskal-Wallis test followed by Dunn’s multiple comparisons test for (K).
-
Figure 1—figure supplement 1—source data 1
Source data for graphs in Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/55111/elife-55111-fig1-figsupp1-data1-v1.xlsx
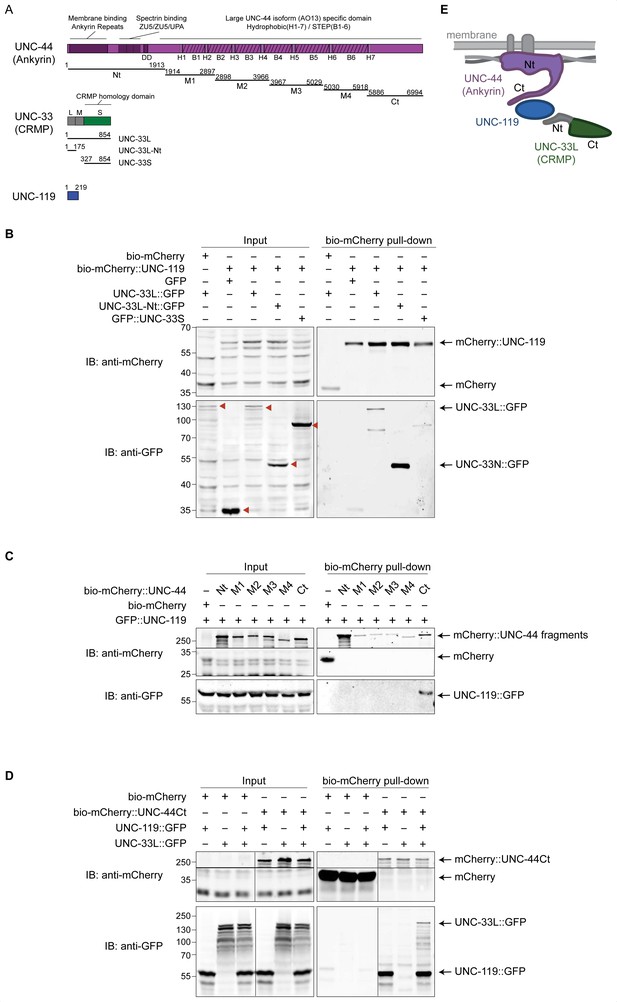
UNC-119 forms a complex with UNC-44 (Ankyrin) and UNC-33L (CRMP).
(A) Schematic representation of the protein and fragments used for pull-down experiments. C. elegans expresses a giant UNC-44 (Ankyrin) isoform (AO13) that is mainly expressed in neurons (Otsuka et al., 2002) and which contains six blocks of serine/threonine/glutamic acid/proline rich (STEP) repeats separated by seven hydrophobic domains (H) in its C-terminus. Three isoforms exist for UNC-33 (CRMP). The N-terminal extensions of UNC-33M and UNC-33L are marked in grey. Nt = N terminus; Ct = C terminus; DD = death domain. (B–D) Streptavidin pull-down assays were performed by incubating streptavidin coated magnetic beads with lysates of HEK293T cells (co)expressing the indicated fusion-constructs for 1 hr at 4°. (B) Pull-down from lysates of cells coexpressing BirA with either bio-mCherry or bio-mCherry-UNC-119 (bait) and the indicated GFP labeled proteins (prey). The GFP labeled proteins are indicated by red arrowheads in the input blot. (C) Pull-down of bio-mCherry labeled UNC-44 fragments and UNC-119::GFP. Proteins were separately expressed in HEK293T cells (UNC-44 fragments required longer incubation time for expression). The lysates were mixed before the pull-down. We noticed that the UNC-44 fragments (M1–M4 and Ct) run at higher Mw than expected, which could be caused by post translational modifications. (D) Pull-down of bio-mCherry or bio-mCherry labeled UNC-44 C-terminus (Ct) with UNC-119::GFP and/or UNC-33L::GFP. mCherry proteins were separately expressed from the GFP proteins. Lysates were mixed before the pull-down. (E) Interaction model based on the pull-down experiments; where UNC-44 (Ankyrin) is drawn associated to membrane proteins (top) and spectrin (side).
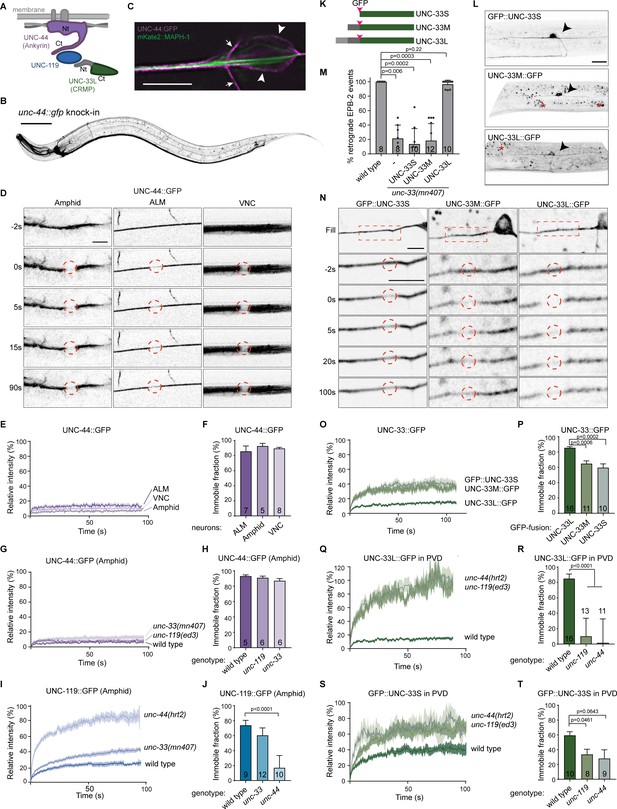
UNC-44 (Ankyrin) anchors UNC-33 (CRMP) via UNC-119.
(A) Schematic representation of the UNC-44/UNC-119/UNC-33 complex (B) unc-44::gfp knock-in animals show neuronal GFP expression. The animals are superficially wild type, but the locomotion somewhat affected (Figure 3—figure supplement 1A and B). Scale, 50 µm. (C) Magnification of the ALM neuron cell body (arrowheads) and axon (extending to the left), expressing the microtubule marker mKate2::MAPH-1 (green) in the unc-44::gfp (magenta) knock-in animal. SRRF was used to improve the resolution (Gustafsson et al., 2016). The small arrows indicate a vertically crossing motor neuron axon. Scale, 5 µm. (D) UNC-44::GFP FRAP in the knock-in animal in amphid nerves (dendrites), the ALM axon or ventral nerve cord (VNC, mainly axons). Scale, 5 µm. (E–J) Average normalized intensity graphs of UNC-44::GFP (D,F) and UNC-119::GFP FRAP (H) ± SEM and the percentage immobile fractions after FRAP (E,G,I) in the indicated tissues and mutants. Number of analyzed animals is indicated. (K) Schematic representation of the three UNC-33 isoforms and GFP insertion. (L) Expression of GFP labeled UNC-33 isoforms in the PVD neuron. Red asterisk marks autofluorescent gut-granules. Scale, 20 µm. (M) Quantification of minus-end out microtubules in the PVD anterior dendrite in wild type or unc-33 mutant with or without PVD specific expression of GFP labeled UNC-33 isoforms. (N) FRAP of GFP labeled UNC-33 isoforms in the PVD anterior dendrite. Scale, 5 µm. (O–T) Average normalized intensity graphs of UNC-33::GFP FRAP ± SEM (N,P,R) and the percentage immobile fractions after FRAP (O,Q,S) in the indicated tissues and mutants. Number of analyzed animals is indicated. All experiments analyzing GFP tagged UNC-44 and UNC-119 proteins were performed in the knock-in animals, whereas all experiments analyzing GFP tagged UNC-33 were done upon mild overexpression in the PVD neuron using single copy integrated strains (MosSCI; Frøkjaer-Jensen et al., 2008), except for the UNC-33M in (M). Analyzed animals were from the L4 or young adult stage; error bars represent SD; statistical analysis, Kruskal-Wallis test followed by Dunn’s multiple comparisons test.
-
Figure 3—source data 1
Source data for graphs in Figure 3.
- https://cdn.elifesciences.org/articles/55111/elife-55111-fig3-data1-v1.xlsx
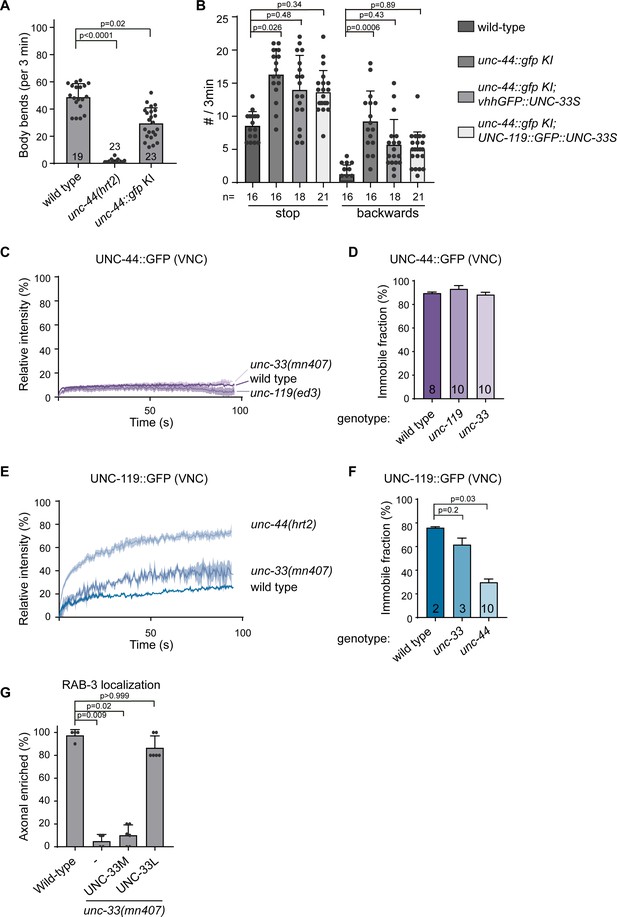
UNC-44 (Ankyrin) anchors UNC-33 (CRMP) via UNC-119 supplement.
(A) Quantification of animal locomotion during 3 min of forward motion (KI = knock in). (B) Quantification of animal stopping or reversing locomotion during 3 min. Animals stopping for longer than 5 s were not included. (C–F) Average normalized intensity graphs of UNC-44::GFP and UNC-119 FRAP in the VNC ± SEM (C,E) and the percentage immobile fractions after FRAP (D,F) in the indicated mutants. (G) Quantification of RAB-3::mCherry localization in the PVD neuron in wild type and unc-33 mutant upon single copy (mosSCI) expression of internally gfp tagged UNC-33M or UNC-33L in the PVD. Number of analyzed animals is indicated. Analyzed animals were from the L4 or young adult stage; error bars represent SD; statistical analysis, Kruskal-Wallis test followed by Dunn’s multiple comparisons test; ns = non significant.
-
Figure 3—figure supplement 1—source data 1
Source data for graphs in Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/55111/elife-55111-fig3-figsupp1-data1-v1.xlsx
FRAP microscopy of UNC-33L::GFP in the PVD neuron in wild type and unc-44 mutant animals.
The FRAP was set at 0 s (regions are indicated); time, min:sec.
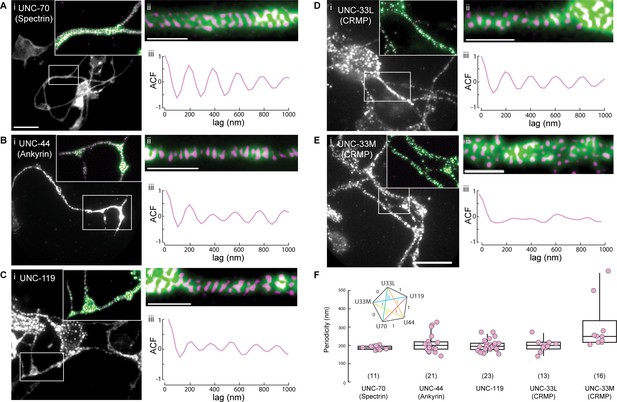
UNC-44 (Ankyrin), UNC-119 and UNC-33L (CRMP) form a periodic pattern along neurites.
(A–E) Superresolution imaging of primary neurons from isolated C elegans cultures. Representative images of cells extracted from animals expressing GFP or GFP derivative tagged (A) UNC-70 (β-spectrin), (B) UNC-44, (C) UNC-119, (D) UNC-33L and (E) UNC-33M with the corresponding (i) widefield, (ii) supersolved images and (iii) autocorrelation function. The inset in (i) shows a magnified region with widefield (green) and superresolved pattern (magenta). Scale bars in (i) = 5 µm; in (ii) = 1 µm. (F) Quantification of the peak frequencies observed after Fourier decomposition of the line profiles, extracted from the superresolved images. ROIs were choses at various locations along the neurites showing periodicity with a minimum length of 2 µm. The inset shows the results from a non-parametric multicomparison test (Wilcoxon with Nemenyi post-hoc) for unequal sample size with H0 that the median is from the same population. If test statistic 0.7 < q < 3.5, H0 = 1, otherwise H0 = 0.
-
Figure 4—source data 1
Source data for graphs in Figure 4.
- https://cdn.elifesciences.org/articles/55111/elife-55111-fig4-data1-v1.xlsx
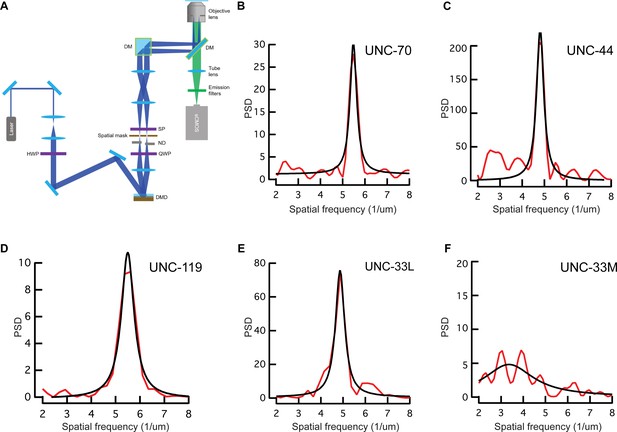
TIRF-SIM imaging setup and power spectral density plots.
(A) TIRF-SIM (structured illumination microscopy) setup used for the imaging of the cultured C. elegans neurons. Details can be found in the Materials and methods. (B–F) Representative power spectral density of the periodicity pattern showing the main frequency of (B) UNC-70, (C) UNC-44, (D) UNC-119, (E) UNC-33L and (F) UNC-33M per µm. Black denotes a fit to a Lorentzian curve.
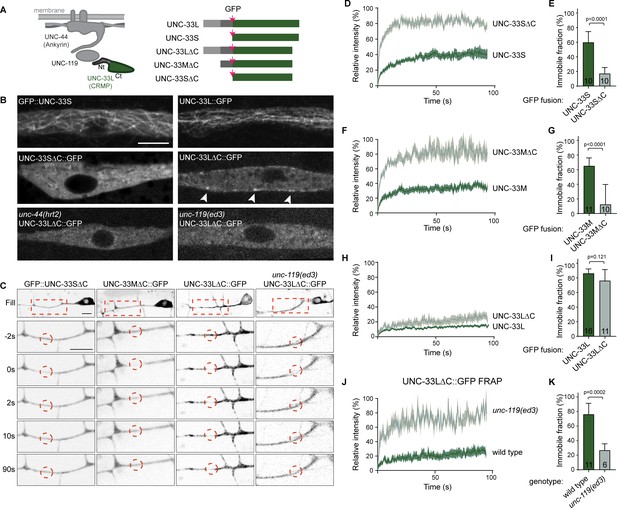
The microtubule binding UNC-33L (CRMP) is anchored to the cortex via its N-terminus.
(A) Schematic representation of the UNC-44/ UNC-119/UNC-33 complex and the different UNC-33 (CMRP) constructs used. (B) Ectopic expression of the indicated GFP tagged UNC-33 constructs in hypodermal seam cells. Arrowheads indicate cortical localized UNC-33LΔC::GFP. Scale, 5 µm. (C) FRAP imaging of the indicated GFP tagged UNC-33 constructs mildly overexpressed in the PVD neuron using single copy integrated strains (MosSCI; Frøkjaer-Jensen et al., 2008). (D–K) Average normalized intensity graphs of FRAP imaging of indicated GFP tagged UNC-33 constructs ± SEM (D,F,H,J) and the percentage immobile fractions after FRAP (E,G,I,K) in the indicated mutants. Number of analyzed animals is indicated. Analyzed animals were from the L4 or young adult stage; error bars represent SD; statistical analysis, unpaired t test.
-
Figure 5—source data 1
Source data for graphs in Figure 5.
- https://cdn.elifesciences.org/articles/55111/elife-55111-fig5-data1-v1.xlsx
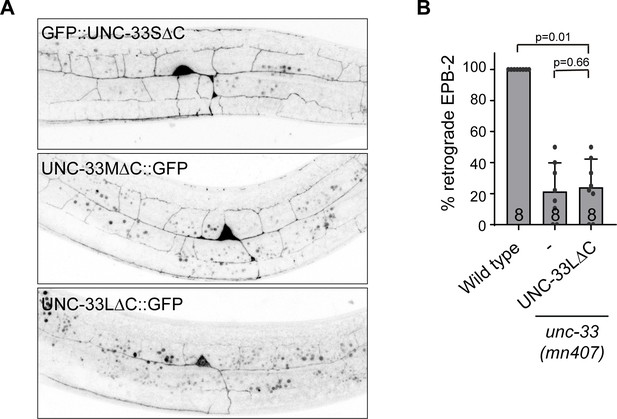
UNC-33L immobilization does not require microtubule binding.
(A) Localization of the three GFP-tagged UNC-33 isoforms lacking the microtubule binding C-terminus expressed in the PVD neuron. (B) Quantification of minus-end out microtubules in the PVD anterior dendrite, in wild type and unc-33 mutant expressing GFP tagged UNC-33L lacking the microtubule binding C-terminus. Number of analyzed animals is indicated. Analyzed animals were from the L4 or young adult stage; error bars represent SD; statistical analysis, Kruskal-Wallis test followed by Dunn’s multiple comparisons test.
-
Figure 5—figure supplement 1—source data 1
Source data for graphs in Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/55111/elife-55111-fig5-figsupp1-data1-v1.xlsx
Expression of various GFP tagged UNC-33 constructs in the seam cells.
Time, min:sec.
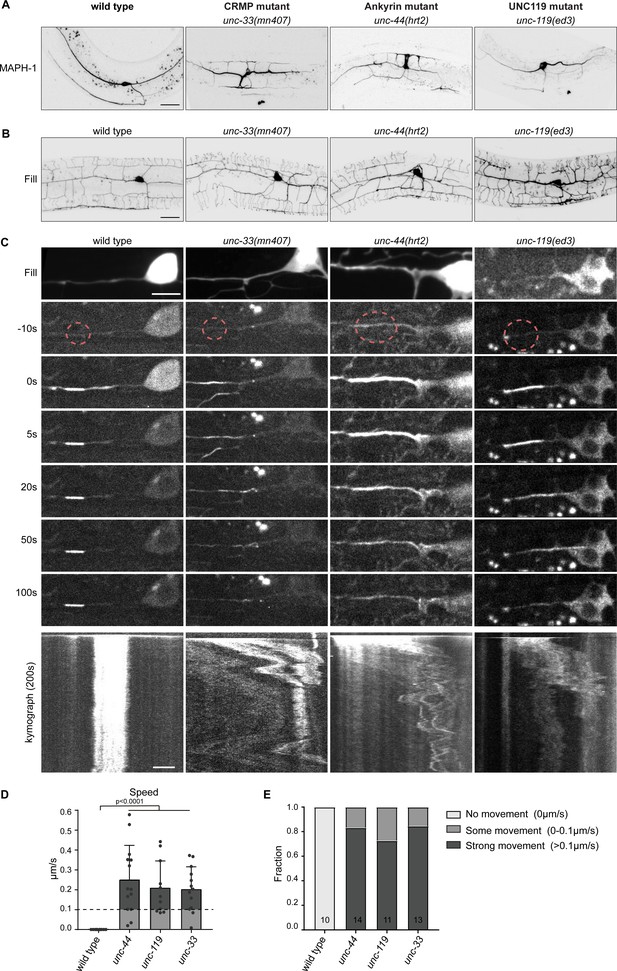
The cortical anchor complex immobilizes the microtubule cytoskeleton.
(A) Representative images of mKate2::MAPH-1 microtubule marker in the PVD in indicated mutants. (see also Figure 6—video 1). Scale, 20 µm. (B) Representative images of PVD neuron morphology visualized with myristoylated GFP of the indicated mutants. Scale, 20 µm. (C) Representative stills and kymograph of microtubule mobility in the indicated mutants, visualized using PA-GFP::TBA-1 (tubulin) expressed in PVD. The photoactivated region is indicated. Scale, 5 µm for the stills and 2 µm for the kymographs. (D–E) Quantification of microtubule sliding. All sliding evens were averaged per animal. We defined an average sliding above 0.1 µm/s as strong sliding. Analyzed animals were from the L4 or young adult stage. Error bars represent SD; statistical analysis, unpaired student T-test.
-
Figure 6—source data 1
Source data for graphs in Figure 6.
- https://cdn.elifesciences.org/articles/55111/elife-55111-fig6-data1-v1.xlsx
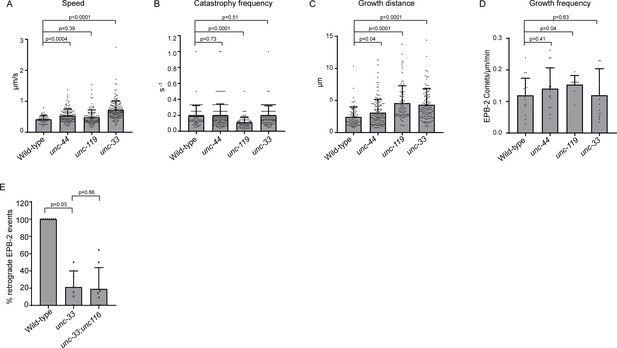
Microtubule plus-end dynamics in mutants for the cortical anchor complex.
(A–C) Quantification of microtubule plus-end dynamics in the anterior PVD dendrite in wildtype and indicated mutants using the plus tip marker EBP-2::GFP. For speed only growth events of >2 µm were considered. (D) EBP-2 growth evens during 2 min acquisitions (E) Quantification of the microtubule orientation in the PVD anterior dendrite. Only acquisitions with ≥5 growth events were considered. For A-D: Animals analyzed: wildtype = 17, unc-44 = 14, unc-119 = 9; unc-33 = 15. For E N = 8 animals. Analyzed animals were from the L4 or young adult stage. Error bars represent SD.
-
Figure 6—figure supplement 1—source data 1
Source data for graphs in Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/55111/elife-55111-fig6-figsupp1-data1-v1.xlsx
Visualization of the microtubule cytoskeleton using the microtubule binding protein MAPH-1, in the indicated genetic backgrounds.
Time, min:sec.
Expression of PA-GFP::TBA-1 (tubulin) expressed in PVD neuron in indicated genetic backgrounds to visualize microtubule dynamics.
Time, min:sec.
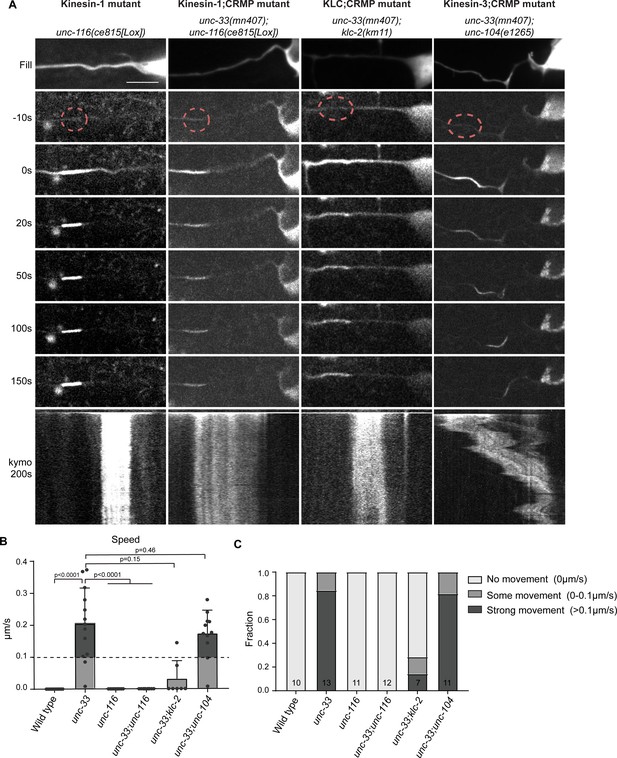
Kinesin-1 is sliding microtubule in the absence of the cortical anchor complex.
(A) Representative stills and kymograph of microtubule mobility in the indicated mutants, visualized using PA-GFP::TBA-1 (tubulin) expressed in PVD. The photoactivated region is indicated. Scale, 5 µm for the stills and 2 µm for the kymographs. (B–C) Quantification of microtubule sliding. All sliding evens were averaged per animal. We defined an average sliding above 0.1 µm/s as strong sliding. Analyzed animals were from the L4 or young adult stage. Error bars represent SD; statistical analysis, unpaired student T-test.
-
Figure 7—source data 1
Source data for graphs in Figure 7.
- https://cdn.elifesciences.org/articles/55111/elife-55111-fig7-data1-v1.xlsx
Expression of PA-GFP::TBA-1 (tubulin) expressed in PVD neuron in indicated genetic backgrounds to visualize microtubule dynamics.
Time, min:sec.
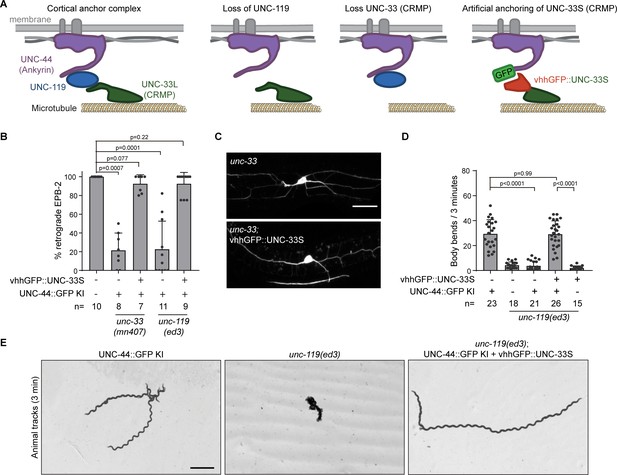
UNC-33 (CRMP) anchoring by UNC-119 is essential for neuron development and animal locomotion.
(A) Model: The microtubule binding UNC-33L (CRMP) is cortically anchored to UNC-44 (Ankyrin) via UNC-119. In the absence of UNC-119 or the long UNC-33 isoform the putative microtubule binding UNC-33 is not anchored to the cortex. By fusing the UNC-33S (CRMP) to the GFP nanobody (vhhGFP) we can artificially anchor UNC-33 to the cortex by expressing the fusion protein in the unc-44::gfp knock-in animal. If the main function of UNC-119 neuron development is to anchor UNC-33L to the cortex, we expect that the artificial attachment of UNC-33 will suppress the unc-119 and unc-33 mutant phenotype. (B–C) Quantification of minus-end out microtubules in the PVD anterior dendrite (B) and examples of PVD neuron morphology (imaged with PVD >EBP-2::mKate2) (C) with or without PVD specific expression of vhhGFP::UNC-33S together with the unc-44::gfp knock-in in indicated genetic backgrounds. Scale, 20 µm. (D) Quantification of the animal locomotion upon expression of pan-neuronal vhhGFP::UNC-33 in the indicated backgrounds. (E) Example tracks of indicated genotype during 3 min (maximum projection). Scale, 1 mm. Analyzed animals were from the L4 or young adult stage; error bars represent SD; statistical analysis, Kruskal-Wallis test followed by Dunn’s multiple comparisons test.
-
Figure 8—source data 1
Source data for graphs in Figure 8.
- https://cdn.elifesciences.org/articles/55111/elife-55111-fig8-data1-v1.xlsx
Direct coupling of UNC-33 to UNC-44 by expressing vhhGFP::UNC-33S using a pan-neuronal Prab-3 promoter in the unc-44::gfp knock-in animals rescues animal locomotion (wild type is the unc-44::gfp knock-in).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Rabbit polyclonal anti-GFP | Abcam | Cat# ab290; RRID:AB_303395 | 1:10000 |
| Antibody | Mouse monoclonal anti-mCherry | Clontech Laboratories | Cat# 632543; RRID:AB_2307319 | 1:10000 |
| Antibody | Goat Anti-Rabbit IgG Antibody, IRDye 680LT Conjugated | LI-COR Biosciences | Cat# 827–11081; RRID:AB_10795015 | 1:3000 |
| Antibody | Goat Anti-Mouse IgG Antibody, IRDye 680LT Conjugated | LI-COR Biosciences | Cat# 827–11080; RRID:AB_10795014 | 1:3000 |
| Antibody | Goat Anti-Rabbit IgG Antibody, IRDye 800CW Conjugated | LI-COR Biosciences | Cat# 827–08365; RRID:AB_10796098 | 1:3000 |
| Antibody | Goat Anti-Mouse IgG Antibody, IRDye 800CW Conjugated | LI-COR Biosciences | Cat# 827–08364; RRID:AB_10793856 | 1:3000 |
| Cell line | Human embryonic kidney 239T (HEK293T) | ATCC | CRL-3216 RRID:CVCL_0063 | |
| Chemicals | PEI | PolySciences | Cat# 24765–2 | |
| Software | ImageJ | NIH | https://imagej.nih.gov/ij/; RRID:SCR_003070 | |
| Software | GraphPad Prism 8 | GraphPad | https://www.graphpad.com/scientific-software/prism/; RRID:SCR_002798 | |
| Software | Kymoreslicewide | Github | https://github.com/ekatrukha/KymoResliceWide | |
| Strain, strain background (C. elegans) | unc-33(mn407); kyIs445[des-2::mCherry::rab-3; des-2::sad-1::gfp; Podr-1::dsRed] | Cross - this paper | STR110 | |
| Strain, strain background (C. elegans) | unc-119(ed3); hrtIs3[Pdes-2::myr::GFP; Punc-122::dsRed] | Cross - this paper | STR174 | |
| Strain, strain background (C. elegans) | unc-44(tm349);hrtIs3[Pdes-2::myristoyl::GFP, Punc-122::DsRed] | Cross - this paper | STR176 | |
| Strain Caenorhabditis elegans | unc-44(hrt2) | Harterink et al., 2018 | STR237 | |
| Strain, strain background (C. elegans) | unc-44(hrt5[GFP-KI]) | Injected - this paper | STR282 | strain was generated using pha-1 co-CRISPR |
| Strain, strain background (C. elegans) | unc-33(mn407); unc-44(hrt5[GFP-KI]) | Cross - this paper | STR292 | |
| Strain, strain background (C. elegans) | unc-119(ed3); hrtSi26[Pdes-2::ebp-2::mKate2 LGII] | Injected - this study | STR316 | |
| Strain, strain background (C. elegans) | unc-119(ed3); hrtSi26[Pdes-2::ebp-2::mKate2 LGII] | Injected - this paper | STR316 | |
| Strain, strain background (C. elegans) | unc-119(ed3); hrtSi28[Pdes-2::mKate2::maph-1.1 LGIV] | Harterink et al., 2018 | STR318 | |
| Strain, strain background (C. elegans) | hrtEx127[Punc-86::ebp-2::gfp; Pmyo-2::tdTomato] | Injected - this paper | STR366 | |
| Strain, strain background (C. elegans) | unc-33(hrt7); unc-119(ed3);hrtSi28[Pdes-2::mKate2::maph-1.1 LGIV] | Injected - this paper | STR367 | hrt7 was generated using CRISPR (11 bps deletion) |
| Strain, strain background (C. elegans) | unc-119(ed3); hrtSi41[Pdes-2::unc-33L::gfp LGI] | Injected - this paper | STR369 | gfp is inserted in unc-33 at the start of the S isoform |
| Strain, strain background (C. elegans) | unc-33(mn407); kyIs445[des-2::mCherry::rab-3; des-2::sad-1::gfp; Podr-1::dsRed]; hrtSi41[Pdes-2::unc-33L::gfp LGI] | Cross - this paper | STR386 | |
| Strain, strain background (C. elegans) | unc-44(hrt2); hrtSi41[Pdes-2::unc-33L::gfp LGI] | Cross - this paper | STR387 | |
| Strain, strain background (C. elegans) | unc-44(hrt8); unc-119(ed3); hrtSi28[Pdes-2::mKate2::maph-1.1 LGIV] | Injected - this study | STR388 | hrt8 was generated using CRISPR (11 bps insertion) |
| Strain, strain background (C. elegans) | unc-33(mn407); hrtSi26[Pdes-2::ebp-2::mKate2 LGII]; hrtSi41[Pdes-2::unc-33L::gfp LGI] | Cross - this paper | STR405 | |
| Strain, strain background (C. elegans) | unc-119(ed3); hrtSi54[Pdes-2::unc-33M::gfp LGI] | Injected - this paper | STR425 | gfp is inserted in unc-33 at the start of the S isoform |
| Strain, strain background (C. elegans) | unc-33(mn407); hrtSi26[Pdes-2::ebp-2::mKate2 LGII] | Cross - this paper | STR431 | |
| Strain, strain background (C. elegans) | unc-33(mn407); hrtSi26[Pdes-2::ebp-2::mKate2 LGII] | Cross - this paper | STR431 | |
| Strain, strain background (C. elegans) | unc-119(ed3); unc-44(hrt5[GFP-KI]) | Cross - this paper | STR434 | |
| Strain, strain background (C. elegans) | unc-44(hrt5[GFP-KI]); hrtSi50[Pmec-4::mKate2::maph-1.1 LGIV] | Injected and Cross - this paper | STR439 | |
| Strain, strain background (C. elegans) | unc-119(ed3); wyEx4828[Pdes-2::ebp-2::gfp; Podr- 1::RFP] | Cross - this paper | STR443 | |
| Strain, strain background (C. elegans) | unc-33(mn407); kyIs445[des-2::mCherry::rab-3; des-2::sad-1::gfp; Podr-1::dsRed]; hrtSi54[Pdes-2::unc-33M::gfp LGI] | Cross - this paper | STR444 | |
| Strain, strain background (C. elegans) | unc-119(ed3); hrtEx142[Pdes-2::unc-119::gfp; Pdes-2::ebp-2::mKate2; Pmyo-2::tdTomato] | Injected - this paper | STR448 | |
| Strain, strain background (C. elegans) | unc-119(ed3); hrtEx143[Pdes-2::ebp-2::mKate2; Pmyo-2::tdTomato (no cb-unc-119)] | Injected - this paper | STR449 | |
| Strain, strain background (C. elegans) | unc-119(hrt13[GFP-KI]) | Injected - this paper | STR485 | |
| Strain, strain background (C. elegans) | unc-33(mn407); unc-119(hrt13[GFP-KI]) | Cross - this paper | STR499 | |
| Strain, strain background (C. elegans) | unc-44(hrt2); unc-119(hrt13[GFP-KI]) | Cross - this paper | STR500 | |
| Strain, strain background (C. elegans) | unc-119(ed3); hrtSi69[Pdes-2::gfp::unc-33S LGI] | Injected - this paper | STR512 | |
| Strain, strain background (C. elegans) | unc-33(mn407); hrtSi26[Pdes-2::ebp-2::mKate2 LGII]; hrtSi69[Pdes-2::gfp::unc-33S LGI] | Cross - this paper | STR532 | |
| Strain, strain background (C. elegans) | unc-44(hrt5[GFP-KI]); hrtSi50[Pmec-4::mKate2::maph-1.1 LGIV] | Injected and Cross - this paper | STR439 | |
| Strain, strain background (C. elegans) | unc-119(ed3); wyEx4828[Pdes-2::ebp-2::gfp; Podr- 1::RFP] | Cross - this paper | STR443 | |
| Strain, strain background (C. elegans) | unc-33(mn407); kyIs445[des-2::mCherry::rab-3; des-2::sad-1::gfp; Podr-1::dsRed]; hrtSi54[Pdes-2::unc-33M::gfp LGI] | Cross - this paper | STR444 | |
| Strain, strain background (C. elegans) | unc-119(ed3); hrtEx142[Pdes-2::unc-119::gfp; Pdes-2::ebp-2::mKate2; Pmyo-2::tdTomato] | Injected - this paper | STR448 | |
| Strain, strain background (C. elegans) | unc-119(ed3); hrtEx143[Pdes-2::ebp-2::mKate2; Pmyo-2::tdTomato (no cb-unc-119)] | Injected - this study | STR449 | |
| Strain, strain background (C. elegans) | unc-119(hrt13[GFP-KI]) | Injected - this paper | STR485 | |
| Strain, strain background (C. elegans) | unc-33(mn407); unc-119(hrt13[GFP-KI]) | Cross - this paper | STR499 | |
| Strain, strain background (C. elegans) | unc-44(hrt2); unc-119(hrt13[GFP-KI]) | Cross - this paper | STR500 | |
| Strain, strain background (C. elegans) | unc-119(ed3); hrtSi69[Pdes-2::gfp::unc-33S LGI] | Injected - this paper | STR512 | |
| Strain, strain background (C. elegans) | unc-33(mn407); hrtSi26[Pdes-2::ebp-2::mKate2 LGII]; hrtSi69[Pdes-2::gfp::unc-33S LGI] | Cross - this paper | STR532 | |
| Strain, strain background (C. elegans) | unc-44(hrt5[GFP-KI]); hrtSi50[Pmec-4::mKate2::maph-1.1 LGIV] | Injected and Cross - this paper | STR439 | |
| Strain, strain background (C. elegans) | unc-119(ed3); wyEx4828[Pdes-2::ebp-2::gfp; Podr- 1::RFP] | Cross - this paper | STR443 | |
| Strain, strain background (C. elegans) | unc-33(mn407); kyIs445[des-2::mCherry::rab-3; des-2::sad-1::gfp; Podr-1::dsRed]; hrtSi54[Pdes-2::unc-33M::gfp LGI] | Cross - this paper | STR444 | |
| Strain, strain background (C. elegans) | unc-119(ed3); hrtEx142[Pdes-2::unc-119::gfp; Pdes-2::ebp-2::mKate2; Pmyo-2::tdTomato] | Injected - this paper | STR448 | |
| Strain, strain background (C. elegans) | unc-119(ed3); hrtEx143[Pdes-2::ebp-2::mKate2; Pmyo-2::tdTomato (no cb-unc-119)] | Injected - this paper | STR449 | |
| Strain, strain background (C. elegans) | unc-119(hrt13[GFP-KI]) | Injected - this paper | STR485 | |
| Strain, strain background (C. elegans) | unc-33(mn407); unc-119(hrt13[GFP-KI]) | Cross - this paper | STR499 | |
| Strain, strain background (C. elegans) | unc-44(hrt2); unc-119(hrt13[GFP-KI]) | Cross - this paper | STR500 | |
| Strain, strain background (C. elegans) | unc-119(ed3); hrtSi69[Pdes-2::gfp::unc-33S LGI] | Injected - this study | STR512 | |
| Strain, strain background (C. elegans) | unc-33(mn407); hrtSi26[Pdes-2::ebp-2::mKate2 LGII]; hrtSi69[Pdes-2::gfp::unc-33S LGI] | Cross - this paper | STR532 | |
| Strain, strain background (C. elegans) | hrtEx161[Pdes-2::PA-GFP::tba-1; Pdes-2::mKate2; Pmyo-2::mCherry] | Injected - this paper | STR536 | |
| Strain, strain background (C. elegans) | unc-119(ed3); hrtEx165[Pwrt-2::unc-33L::gfp] | Injected - this paper | STR544 | gfp is inserted in unc-33 at the start of the S isoform |
| Strain, strain background (C. elegans) | unc-119(ed3); hrtEx166[Pwrt-2::gfp::unc-33S] | Injected - this paper | STR545 | |
| Strain, strain background (C. elegans) | unc-119(ed3); hrtSi70[Pdes-2::unc-33LΔC::gfp LGI] | Injected - this paper | STR546 | gfp is inserted in unc-33 at the start of the S isoform |
| Strain, strain background (C. elegans) | unc-44(hrt2); hrtEx161[Pdes-2::PA-GFP::tba-1; Pdes-2::mKate2; Pmyo-2::mCherry] | Cross - this paper | STR548 | |
| Strain, strain background (C. elegans) | unc-119(ed3); hrtEx167[Pwrt-2::gfp::unc-33SΔC] | Injected - this paper | STR550 | gfp is inserted in unc-33 at the start of the S isoform |
| Strain, strain background (C. elegans) | unc-33(mn407); hrtSi26[Pdes-2::ebp-2::mKate2 LGII]; hrtEx168[Pdes-2::unc-33M::gfp;Pmyo-2::mCherry] | Injected - this paper | STR552 | |
| Strain, strain background (C. elegans) | unc-119(ed3); hrtSi73[Prab-3::unc-33M::gfp] | Injected - this paper | STR553 | gfp is inserted in unc-33 at the start of the S isoform |
| Strain, strain background (C. elegans) | unc-119(ed3); hrtSi74[Prab-3::unc −33L::gfp] | Injected - this paper | STR554 | gfp is inserted in unc-33 at the start of the S isoform |
| Strain, strain background (C. elegans) | unc-33(mn407); hrtSi26[Pdes-2::ebp-2::mKate2 LGII]; hrtSi70[Pdes-2::gfp::unc33LΔC LGI] | Cross - this paper | STR559 | |
| Strain, strain background (C. elegans) | unc-119(ed3);hrtSi75[Pdes-2::unc-33MΔC::gfp LGI] | Injected - this paper | STR561 | gfp is inserted in unc-33 at the start of the S isoform |
| Strain, strain background (C. elegans) | unc-33(mn407); hrtEx161[Pdes-2::PA-GFP::tba-1; Pdes-2::mKate2; Pmyo-2::mCherry] | Cross - this paper | STR563 | |
| Strain, strain background (C. elegans) | unc-116(ce815[LoxP1/unc-116/sup-1/LoxP2]); heSi175[Pscm::CRE]; hrtEx161[Pdes-2::PA-GFP::tba-1; Pdes-2::mKate2; Pmyo-2::mCherry] | Cross - this paper | STR564 | |
| Strain, strain background (C. elegans) | unc-33(mn407); unc-116(ce815[LoxP1/unc-116/sup-1/LoxP2]); heSi175[Pscm::CRE]; hrtEx161[Pdes-2::PA-GFP::tba-1; Pdes-2::mKate2; Pmyo-2::mCherry] | Cross - this paper | STR575 | |
| Strain, strain background (C. elegans) | unc-33(mn407); unc-44(hrt5[GFPKI]); hrtSi26[Pdes-2::ebp-2::mKate2 LGII]; hrtEx173[Pdes-2::BFP-NLS::p2A::vhhGFP::unc-33S::tbb-2UTR; Pmyo-2::mCherry] | Injected - this paper | STR576 | |
| Strain, strain background (C. elegans) | hrtIs3[Pdes-2::myr-GFP; Punc-122::dsRed] | Harterink et al., 2018 | STR58 | |
| Strain, strain background (C. elegans) | unc-44(hrt2); hrtEx175[Pwrt-2::unc33LΔC::gfp; Pmyo-2::mCherry] | Injected - this paper | STR584 | |
| Strain, strain background (C. elegans) | hrtEx175[Pwrt-2::unc33LΔC::gfp; Pmyo-2::mCherry] | Cross - this paper | STR585 | gfp is inserted in unc-33 at the start of the S isoform |
| Strain, strain background (C. elegans) | unc-119(ed3); hrtSi81[Pdes-2::gfp::unc33SΔC LGI] | Injected - this paper | STR588 | |
| Strain, strain background (C. elegans) | unc-33(mn407); unc-44(hrt5[GFPKI]); hrtSi26[Pdes-2::ebp-2::mKate2 LGII] | Cross - this study | STR591 | |
| Strain, strain background (C. elegans) | unc-33(mn407); unc-104(e1265); hrtEx161[Pdes-2::PA-GFP::tba-1;Pdes-2::mKate2;Pmyo-2::mCherry] | Cross - this paper | STR592 | |
| Strain, strain background (C. elegans) | klc-2(km11); unc-33(mn407); hrtEx161[Pdes-2::PA-GFP::tba-1;Pdes-2::mKate2;Pmyo-2::mCherry] | Cross - this paper | STR594 | |
| Strain, strain background (C. elegans) | unc-119(ed3);hrtEx178[Pdes-2::PA-gfp::tba-1::tbb-2UTR; Pdes-2::bfp; Pmyo-2::mCherry (no cb-unc-119)] | Injected - this paper | STR595 | |
| Strain, strain background (C. elegans) | unc-119(ed3); hrtSi87[Pdes-2::mKate2::maph-1.1 (no cb-unc-119) LGIV] | Injected - this paper | STR601 | hrtSi87 was generated using CRISPR to mutate cb-unc-119 in hrtSi28 (17 bps deletion) |
| Strain, strain background (C. elegans) | unc-119(ed3); hrtS86[Pdes-2::unc-33L::gfp (no cb-unc-119) LGI] | Injected - this paper | STR608 | gfp is inserted in unc-33 at the start of the S isoform; hrtSi86 was generated using CRISPR to mutate cb-unc-119 in hrtSi41 (2 bp deletion and 42 bp insertion) |
| Strain, strain background (C. elegans) | unc-119(ed3); unc-44(hrt5[GFPKI]; hrtEx181[Prab-3::BFP-NLS::p2A::vhhGFP::unc-33s;Pmyo-2::mCherry (no cb-unc-119)] | Injected - this paper | STR619 | |
| Strain, strain background (C. elegans) | unc-119(ed3); hrtSi89[Pdes-2::ebp-2::gfp (no cb-unc-119) LGI] | Injected - this paper | STR620 | hrtSi89 was generated using CRISPR to mutate cb-unc-119 in hrtSi5 (10 bps deletion) |
| Strain, strain background (C. elegans) | unc-119; hrtSi90[Pdes-2::unc-33LdeltaC::GFP (no cb-unc-119)] | Injected - this paper | STR621 | gfp is inserted in unc-33 at the start of the S isoform; hrtSi90 was generated using CRISPR to mutate cb-unc-119 in hrtSi70 (seven bps deletion) |
| Strain, strain background (C. elegans) | unc119(ed3); hrtSi91[[Pgcy-36::ebp-2::gfp(no cb-unc-119)] | Injected - this paper | STR622 | hrtSi91 was generated using CRISPR to mutate cb-unc-119 in hrtSi4 (10pbs deletion) |
| Strain, strain background (C. elegans) | unc-119(ed3); hrtSi92[Pdes-2::gfp::unc-33S (no cb-unc-119) LGI] | Injected - this study | STR623 | hrtSi92 was generated using CRISPR to mutate cb-unc-119 in hrtSi69 (10pbs deletion) |
| Strain, strain background (C. elegans) | unc-119(ed3); unc-44(hrt5[GFPKI]); hrtEx182[Pdes-2::BFP-NLS::p2A::vhhGFP::unc-33S;Pdes-2::ebp-2::mKate2;Pmyo-2::mCherry (no cb-unc-119)] | Injected - this paper | STR624 | |
| Strain, strain background (C. elegans) | unc-44(hrt2); hrtSi69[Pdes-2::gfp::unc-33S LGI] | Cross - this paper | STR625 | |
| Strain, strain background (C. elegans) | unc-119(ed3);hrtEx186[Pwrt2::unc-33LΔC::gfp (no cb-unc-119)] | Injected - this paper | STR634 | gfp is inserted in unc-33 at the start of the S isoform |
| Strain, strain background (C. elegans) | unc-119(ed3); hrtEx181[Prab-3::BFP-NLS::p2A::vhhGFP::unc-33s;Pmyo-2::mCherry (no cb-unc-119)] | Cross - this paper | STR635 | |
| Strain, strain background (C. elegans) | hrtSi4[Pgcy-36::ebp-2::gfp LGI] | Harterink et al., 2018 | STR66 | |
| Strain, strain background (C. elegans) | hrtSi5[Pdes-2::ebp-2::gfp LGI] | Harterink et al., 2018 | STR71 | |
| Strain, strain background (C. elegans) | unc-33(mn407); hrtIs3[Pdes-2::myristoyl::GFP, Punc-122::DsRed] | Cross - this paper | STR95 | |
| Strain, strain background (C. elegans) | kyIs445[des-2::mCherry::rab-3; des-2::sad-1::gfp; Podr-1::dsRed] | Maniar et al., 2012 | CX9797 | |
| Strain, strain background (C. elegans) | wyEx4828[Pdes-2::ebp-2::gfp; Podr- 1::RFP] | Yan et al., 2013 | TV11781 | |
| Strain, strain background (C. elegans) | unc-33(mn407); unc-116(ce815[LoxP1/unc-116/sup-1/LoxP2]); heSi175[Pscm::CRE];hrtSi5 | Cross - this paper | STR615 | |
| Strain, strain background (C. elegans) | pgIs22 [unc-70::N-TSmod]. oxIs95 [pdi-2p::unc-70 + myo-2p::GFP] | Krieg et al., 2017 | GN600 | |
| Plasmid | sgRNA targeting sequence to generate the unc-119-GFP knock-in | This paper | pMH645 | GATGCATAATTTCCCGCCGA |
| plasmid | sgRNA targeting sequence to generate the unc-44-GFP knock in hrt8 knock-out | This paper | pMH243 | GACACGTATGAATCCGCCCA |
| plasmid | sgRNA targeting sequence to generate the unc-33(hrt7) mutant | This paper | pMH416 | GATGTCGTCGGCAATGATGG |
| RNA oligos | sgRNA targeting sequence to generate mutate cb-unc-119 in mosSCI lines | IDT | unc-119 CB (RNA guide) | CCTTGTTCGGTGCTTGGTGG |
Additional files
-
Supplementary file 1
Mass spectroscopy identification of co-immunoprecipitated proteins for the unc-119::gfp knock-in immunoprecipitation.
Wildtype (N2) and a strain expressing GFP in neurons (OH441) were used as control strains. Find all identified proteins in separate tabs and the analysis of the data in the combined tab. To analyze Affinity Purification Mass Spectrometry Data, a Fold Change score (FC-Score) is calculated based on computing the ratio of average normalized spectral counts (PSMs) in bait purifications versus negative controls using CRAPome (Mellacheruvu et al., 2013).
- https://cdn.elifesciences.org/articles/55111/elife-55111-supp1-v1.xlsx
-
Supplementary file 2
Overview of all constructs and oligos used in this study.
- https://cdn.elifesciences.org/articles/55111/elife-55111-supp2-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/55111/elife-55111-transrepform-v1.pdf





