Hox-dependent coordination of mouse cardiac progenitor cell patterning and differentiation
Figures
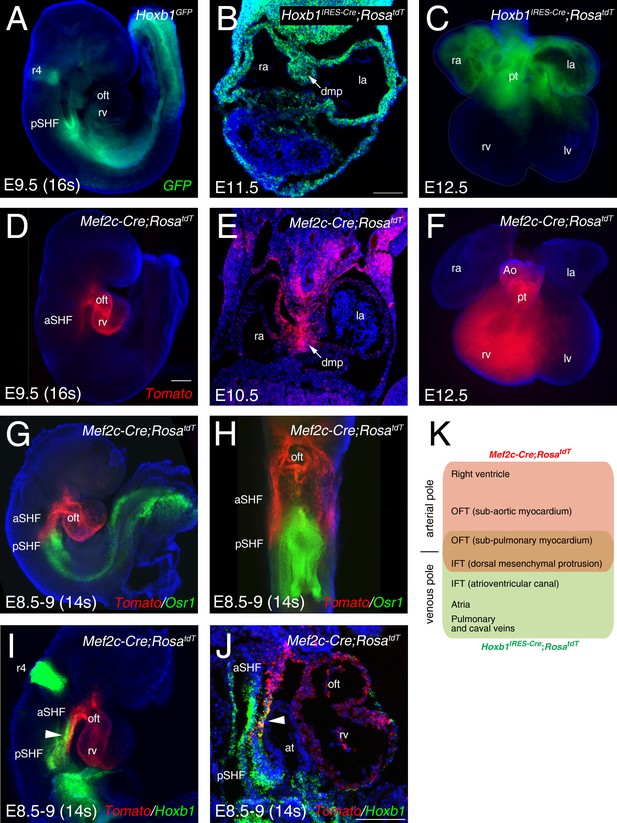
Characterization of two transgenic lines defining complementary domains of the SHF.
(A) Whole-mount fluorescence microscopy of E9.5 (16 somites [s]) Hoxb1GFP embryos. (B) Transverse section at E11.5 heart showing Hoxb1-Cre genetic lineage contribution to atrial myocardium and the dorsal mesenchymal protrusion (DMP). (C) Ventral view of an E12.5 heart showing the Hoxb1-Cre (Hoxb1IRES-Cre;RosatdT - green) genetic lineage contributions to both atria and sub-pulmonary myocardium. (D) E9.5 (16s) Mef2c-Cre;RosatdT embryos showing the contribution of the Mef2c-Cre genetic lineage (Tomato, red) to the outflow tract and the right ventricle. (E) Transverse section at E10.5 showing the Mef2c-Cre genetic lineage contribution to the DMP. (F) Ventral view of an E12.5 heart showing the Mef2c-Cre genetic lineage contribution to the right ventricle and great arteries. (G) RNA-FISH showing the expression of Osr1 (green) and the Mef2c-Cre labeled cells (Tomato; red). (H) Ventral view of the embryo shown in G. Distribution of Osr1 is detected in the posterior domain of Mef2c-Cre. (I,J) RNA-FISH showing a small domain of overlap between Hoxb1 (green) and Mef2c-Cre labeled cells (Tomato; red). (K) Cartoon summarizing the contribution of the Hoxb1-Cre (green) and Mef2c-Cre (red) lineages in the embryo at E9.5. Nuclei are stained with Hoechst. ao, aorta; at, atria; aSHF, anterior second heart field; ift, inflow tract; la, left atria; lv, left ventricle; oft, outflow tract; pt, pulmonary trunk; pSHF, posterior second heart field; r4, rhombomere 4; ra, right atria; rv, right ventricle; Scale bars represent 100 μm (C, J); 200 μm (D).
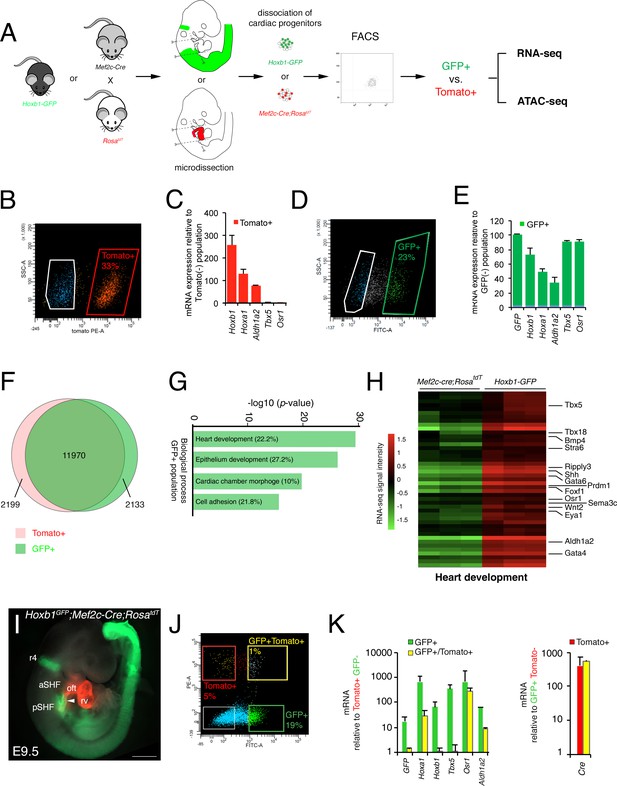
Molecular signature of the posterior SHF.
(A) Scheme of the protocol utilized to characterize the molecular signature of the SHF on isolated Mef2c-Cre;RosatdT (Tomato) and Hoxb1GFP (GFP) positive cells. (B,D) FACS profile of E9.5 cardiac progenitor cells isolated from Mef2c-Cre;RosatdT and Hoxb1GFP embryos. (C,E) Expression of pSHF markers (Hoxa1, Hoxb1, Osr1, Aldh1a2, Tbx5, GFP) was analyzed by real-time qPCR. Data were normalized to HPRT and expressed as folds of increase over negative population. (F) Venn diagram showing transcripts differentially expressed in the GFP+ (green) compared to the Tomato+ (red) populations. (G) Gene ontology (GO) analysis of GFP+ progenitor cells performed with ClusterProfiler system showing enrichment of upregulated genes in the pSHF with ranked by -log10 (p-value). The percentage corresponds to the ‘BG ratio’ obtained in the GO analysis. (H) Example of the heatmap of ‘heart development’ GO term associated genes analyzed by RNA-seq (n = 3 from GFP+ cells, n = 3 from Tomato+ cells). (I) Whole-mount fluorescence microscopy of triple transgenic Hoxb1GFP;Mef2c-Cre;RosatdT embryos at stage E9.5. (J) FACS profile of E9.5 cardiac progenitor cells isolated from Hoxb1GFP;Mef2c-Cre;RosatdT embryos. (K) Expression of pSHF markers (Hoxa1, Hoxb1, Osr1, Aldh1a2, Tbx5), GFP and the Cre recombinase were analyzed with real-time qPCR. Data were normalized to HPRT and expressed as folds of increase over negative population. aSHF, anterior SHF; oft, outflow tract; pSHF, posterior SHF; rv, right ventricle. Scale bars: 500 μm.
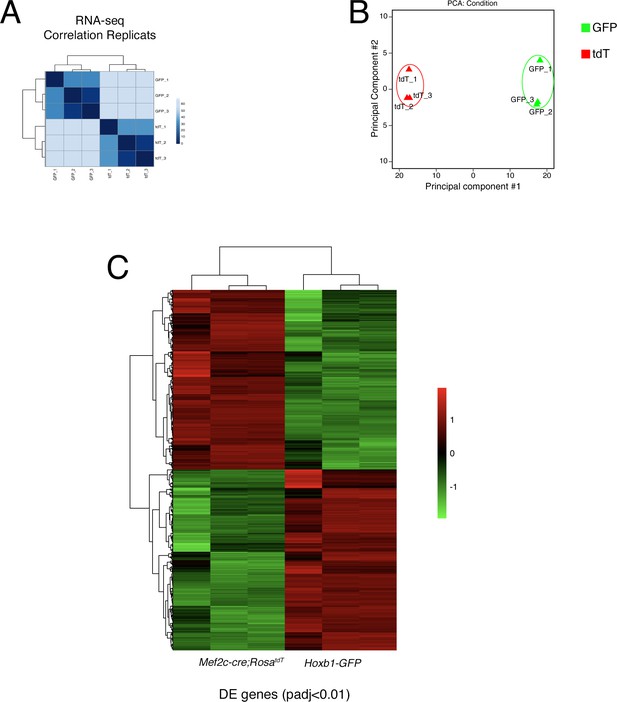
Quality assessment of RNA-seq data performed with purified cardiac progenitor cells.
(A) Replicate correlation for RNA-seq datasets from GFP+ and Tomato+ progenitor cells. (B) Principal component analysis (PCA) of RNA-sequencing datasets from GFP+ and Tomato+ progenitor cells. (C) Unsupervised hierarchical clustering of all differentially expressed genes between GFP+ and Tomato+ progenitor cells.
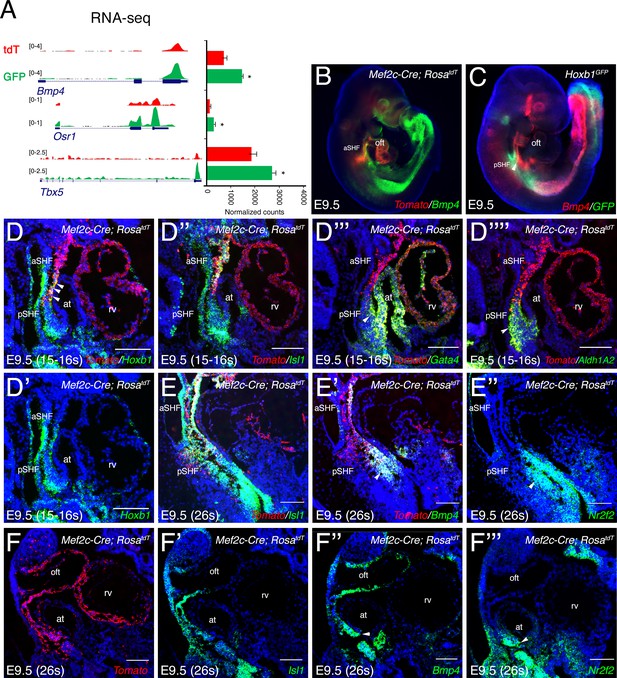
Spatial validation of marker gene expression in cardiac progenitor populations.
(A) RNA-sequencing datasets visualized for the pSHF markers Tbx5, Osr1 and Bmp4. (B) Whole-mount RNA-FISH analysis showing the distribution of Bmp4 transcript (green) in cells overlapping with the posterior border of Mef2c-Cre lineage contribution (red). (C) Bmp4 (red) is expressed in the pSHF overlapping with Hoxb1-GFP (green; arrowhead). (D–F) RNA-FISH analysis on serial sagittal sections of Mef2c-Cre;RosatdT embryos, showing expression of genes enriched in the GFP+ progenitor population (E and F are from the same embryo). Hoxb1, Gata4, Aldh1a2 and Nr2f2 mark the pSHF, whereas Bmp4 expression is enriched in the pSHF compared to the aSHF. Scale bars: 100 μm.
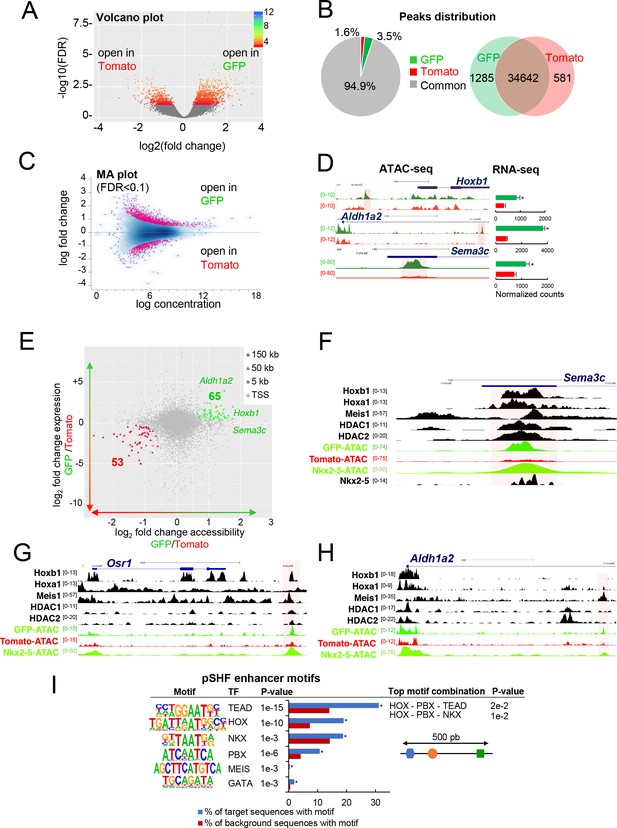
Differential chromatin accessibility in GFP+ and Tomato+ cardiac progenitor cells.
(A) Volcano plot of ATAC-seq performed from GFP+ and Tomato+ cardiac progenitors. (B) Pie chart showing the distribution of the ATAC-seq peaks in the two populations. Venn diagram showing overlap of the ATAC-seq peaks in the two populations. (C) MA plot of ATAC-seq peaks in GFP+ versus Tomato+ cells. (D) Open chromatin profiles correlate with transcriptional expression. Browser views of Hoxb1, Aldh1a2 and Sema3c loci with ATAC-seq profiles of GFP+ pSHF progenitor cells (green) and Tomato+ Mef2c-Cre labeled cells (red). Data represent merged technical and biological replicates for each cell type. The y-axis scales range from 0 to 80 in normalized arbitrary units. The tracks represent ATAC-seq, whereas the bar graphs represent RNA-seq. Boxed regions show cell-type-specific peaks around Aldh1a2, Osr1, and Sema3C gene loci. (E) Change in accessibility versus change in gene expression in GFP+ and Tomato+ cells. For each ATAC-seq peak, the log of the ratio of normalized ATAC-seq reads (GFP/Tomato) is plotted on the x-axis, and the log of the ratio normalized RNA-seq reads corresponding to the nearest gene is plotted in the y-axis. Peaks that are both significantly differentially accessible (FDR < 0.1) and significantly differentially expressed (FDR < 0.1) are colored green (more open in GFP+ cells, higher expression in GFP+ cells; 65 peaks) or red (more open in Tomato population, higher expression in Tomato; 53 peaks). (F–H) Browser views of Sema3c (F), Osr1 (G) and Aldh1a2 (H) gene loci with ATAC-seq profiles of GFP+ pSHF progenitor cells (green) and Tomato+ Mef2c-Cre labeled cells (red). Open chromatin profiles correlate with transcription factor binding at putative enhancers specific for cardiac progenitor cells. (I) pSHF enhancers were enriched in DNA binding motifs for HOX and known cardiac transcription factors such as PBX and MEIS. HOX recognition motifs were strongly enriched in a known motif search in pSHF enhancers. Other enriched matrices match binding sites of known cardiac regulators. HOX binding motifs are highly enriched at genomic regions bound by cardiac transcription factors. p-values were obtained using HOMER (Heinz et al., 2010). Combinations of 3 sequence motifs contained within 500 bp are shown.

Quality assessment of ATAC-seq data performed with purified cardiac progenitor cells.
(A) Replicate correlation for ATAC-seq datasets from GFP+ and Tomato+ progenitor cells. (B) Principal component analysis (PCA) of ATAC-sequencing datasets from GFP+ and Tomato+ progenitor cells. (C) The Gene Ontology (GO) analysis results for the gene loci harboring peaks strictly present in the GFP+ population. (D) Heat maps show the ATAC-seq enrichment (±150 kb region upstream from the annotated TSS). (E) Histograms showing the number of ATAC-seq peaks in the two populations regarding their genomic position with respect to the TSS. (F) ATAC-sequencing on SHF cells identifies previously established SHF REs for Foxf1a (top) and Mef2c (bottom). (G) aSHF enhancers are enriched in DNA binding motifs for RUNX and TEAD transcription factors. p-values were obtained using HOMER (Heinz et al., 2010).
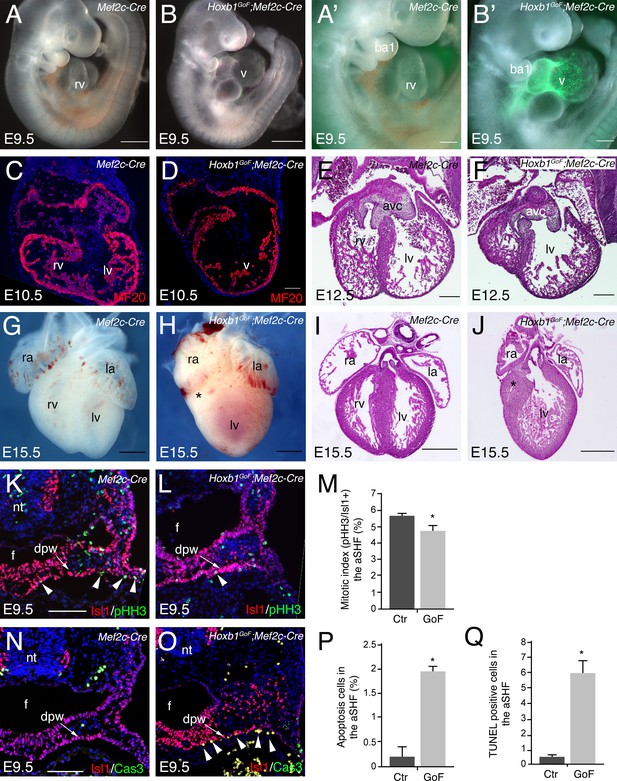
Activation of Hoxb1 expression in aSHF progenitors disrupts the formation of the right ventricle.
(A,B) Macroscopic view of Mef2c-AHF-Cre (Mef2c-Cre, control) and Hoxb1GoF;Mef2c-Cre embryos at E9.5. (A’,B’) High magnification of embryo in A and B showing GFP activity in the Cre-recombinase driven cells. (C,D) Immunofluorescence with MF20 (red) in control (C) and Hoxb1GoF;Mef2c-Cre (D) hearts at E10.5. (E–J) Hematoxylin and Eosin (H and E) staining on transversal section of control (E,I) and Hoxb1GoF;Mef2c-Cre (F,J) hearts at E12.5 and E15.5. The right ventricle (asterisk) in Hoxb1GoF;Mef2c-Cre hearts is hypoplastic compared to control hearts (n = 8). (G,H) Whole-mount views of control (G) and Hoxb1GoF;Mef2c-Cre (H) hearts at E15.5. (K,L) Immunofluorescence with Isl1 (red) and Phospho-histone H3 (pHH3, green) on Mef2c-Cre (K) and Hoxb1GoF;Mef2c-Cre (L) embryos at E9.5. (M) Quantification of pHH3-positive cells in the aSHF Isl1+, showed a reduced of the mitotic index in Hoxb1GoF;Mef2c-Cre (n = 3; GoF) compared to Mef2c-Cre (Ctr) (n = 7) embryos. (N,O) Immunofluorescence with Isl1 (red) and Caspase 3 (Cas3, green) on Mef2c-Cre (N) and Hoxb1GoF;Mef2c-Cre (O) embryos at E9.5. Arrowheads indicate Cas3-positive cells. (P) Quantification of Cas3-positive cells revealed increased cell death in the aSHF of Hoxb1GoF;Mef2c-Cre embryos. (Q) Quantification of TUNEL staining performed on Mef2c-Cre (Ctr) and Hoxb1GoF;Mef2c-Cre embryos (GoF). ao, aorta; avc, atrioventricular canal; ba, branchial arch; dpw, dorsal pericardial wall; f, foregut pocket; la, left atrium; lv, left ventricle; nt, neural tube; oft, outflow tract; PM, pharyngeal mesoderm; pt, pulmonary trunk; ra, right atrium; rv, right ventricle. Scale bars: 100 μm (A’,B’,D,E,F); 200 μm (A’,B’,E,F); 500 μm (A,B,G-J).
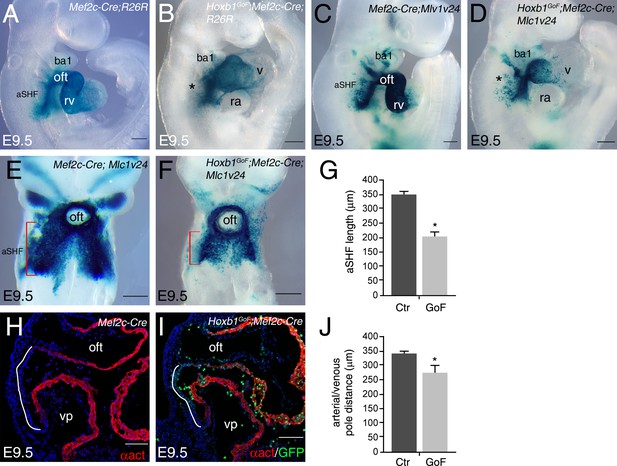
Reduction of SHF length in Hoxb1GoF;Mef2c-Cre embryos.
(A–F) X-gal staining of control and Hoxb1GoF embryos carrying out R26R (A,B) or Mlc1v24 (C–F) transgenes at E9.5. (A,B) Anterior SHF is disrupted in Hoxb1GoF;Mef2c-Cre embryos. (E,F) X-gal staining showing anterior SHF in Mef2c-Cre;Mlc1v24 (control; E) and Hoxb1GoF;Mef2c-Cre;Mlc1v24 (F) embryos at E9.5. (G) Measurement of staining revealed a decrease of anterior SHF length in Hoxb1GoF;Mef2c-Cre;Mlc1v24 compared to control embryos. (H,I) Immunofluorescence with α-actinin (α act, red) and GFP (green) on Mef2c-Cre (control; H) and Hoxb1GoF;Mef2c-Cre (I) embryos at E9.5. (J) Measurement of distance between arterial and venous poles reveals a reduction of dorsal pericardial wall length in Hoxb1GoF;Mef2c-Cre embryos. All measures were calculated from n = 6 embryos for each genotype. Histograms are expressed as mean ± SEM. p-values were determined by Student’s t test. (*p<0.01). Scale bars: 100 μm (H,I) and 200 μm (A–D,E,F).
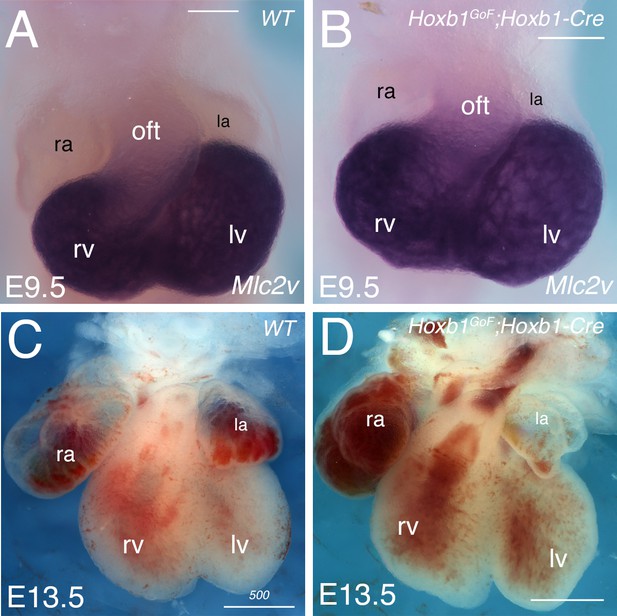
Over-expression of Hoxb1 in the pSHF does not affect the morphology of the embryonic heart.
(A,B) Whole-mount in situ hybridization with Mlc2v riboprobe on E9.5 wild-type (WT; A) and Hoxb1GoF;Hoxb1-Cre (B) embryos. Hearts are similar in WT and Hoxb1GoF;Hoxb1-Cre embryos. (C,D) Frontal views of wild-type (WT; A), and Hoxb1GoF;Hoxb1-Cre (D) hearts at E13.5. No obvious anomalies are detected in Hoxb1GoF;Mef2c-Cre embryos. la, left atria; lv: left ventricle; oft, outflow tract; ra: right atria; rv: right ventricle. Scale bars: 200 μm (A,B) and 500 μm (C,D).
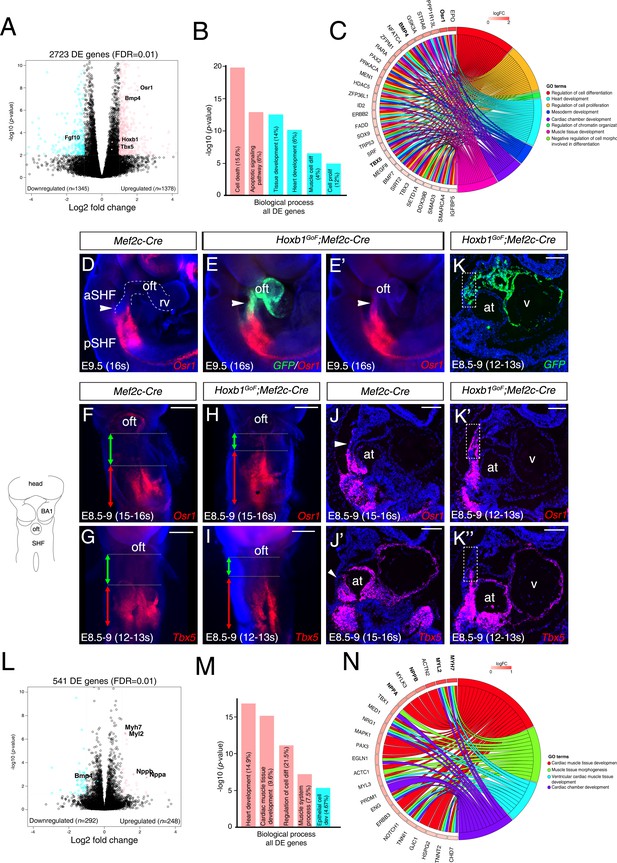
Hoxb1 regulates progenitor identity and differentiation in the pSHF.
(A) Volcano plot of transcriptional profiling results with significantly dysregulated genes between Hoxb1GoF;Mef2c-Cre and control SHF. The y-axis corresponds to the mean expression value of log10 (p-value), and the x-axis displays the log2 fold change value. The colored dots represent the significantly differential expressed transcripts (p<0.05); the black dots represent the transcripts whose expression levels did not reach statistical significance (p>0.05). Differential expression analysis performed using DESeq2 revealed 2,723 genes with Log2-fold changes ≥1 at a False Discovery Rate (FDR) ≤0.01. (B) Gene ontology (GO) analysis of genes deregulated in Hoxb1GoF;Mef2c-Cre embryos performed with ClusterProfiler system. The percentage corresponds to the ‘BG ratio’ obtained in the GO analysis. (C) Chord plot showing a selection of genes upregulated in Hoxb1GoF;Mef2c-Cre embryos present in the represented enriched GO terms. Outer ring shows log2 fold change or GO term grouping (right, key below). Chords connect gene names with GO term groups. (D) Whole-mount RNA-FISH for Osr1 on E9.5 Mef2c-Cre embryos in lateral views. (E, E’) Whole-mount RNA-FISH for GFP (green) and Osr1 (red) on E9.5 Hoxb1GoF;Mef2c-Cre embryos. Whole-mount RNA-FISH for Osr1 in ventral views (F, H) and Tbx5 (G, I) on E8.5-9 Mef2c-Cre and Hoxb1GoF;Mef2c-Cre embryos. An anteriorly shifted expression of Osr1 and Tbx5 is detected in Hoxb1GoF;Mef2c-Cre embryos compared to their control littermates (same somite stage – 15-16s for Osr1 and 12-13s for Tbx5). RNA-FISH against Osr1, Tbx5 and GFP on serial sections in Hoxb1GoF;Mef2c-Cre embryos (K, K’, K’’) compared to control littermates (J, J’; Serial sagittal sections). (L) Volcano plot showing differential expressed genes between Hoxb1-/- and wild-type samples. The y-axis corresponds to the mean expression value of log10 (p-value), and the x-axis displays the log2 fold change value. Colored dots represent the significantly differential expressed transcripts (p<0.05); the black dots represent the transcripts whose expression levels did not reach statistical significance (p>0.05). We identified 249 genes upregulated, and 292 genes downregulated in Hoxb1-/- embryos. (M) GO analysis of genes deregulated in Hoxb1-/- embryos with ranked by -log10 (p-value). (N) Chord plot showing a selection of genes upregulated in dissected pharyngeal mesoderm of Hoxb1-/- embryos present in the represented enriched GO terms. Outer ring shows log2 fold change or GO term grouping (right, key below). Chords connect gene names with GO term groups. Nuclei are stained with Hoechst. at, atria; BA1, branchial arch 1; oft, outflow tract; SHF, second heart field; V, ventricle. Scale bars: 200 μm (F, G, H, I); 100 μm (J, J’, K, K’, K’’).
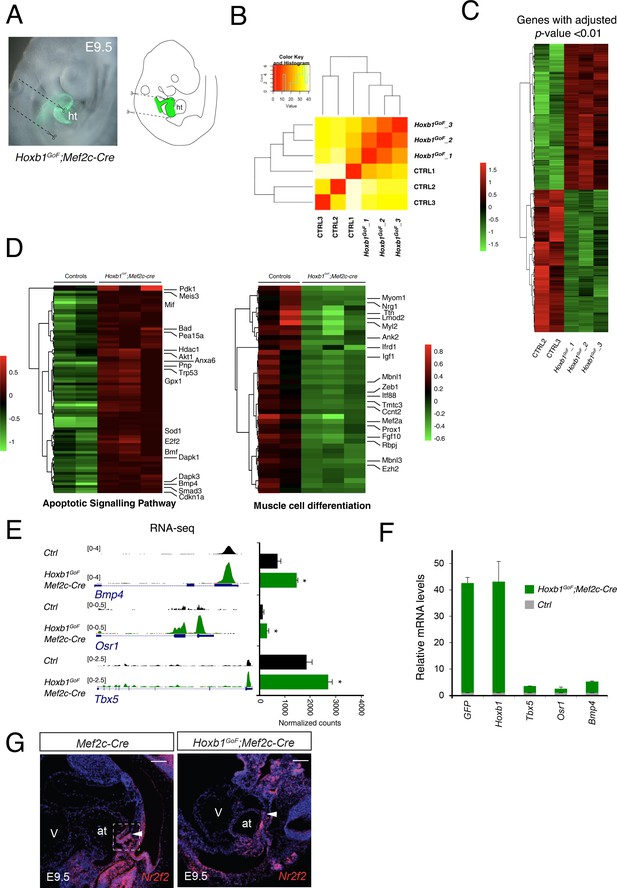
Quality assessment of RNA-seq data performed on the Hoxb1GoF;Mef2c-Cre embryos.
(A) Macroscopic view of Hoxb1GoF;Mef2c-Cre embryos at E9.5 and the micro-dissected region comprising the SHF progenitors and excluding the forming heart. (B) Replicate correlation for RNA-seq datasets from Hoxb1GoF;Mef2c-Cre (GoF) and control (Ctrl) regions. (C) Unsupervised hierarchical clustering of all differentially expressed genes between GoF and control regions. (D) Heatmap of ‘apoptotic signaling pathway’ and ‘muscle cell differentiation’ associated genes analyzed by RNA-seq. (e) RNA-seq datasets visualized for the pSHF markers Tbx5, Osr1 and Bmp4. (F) Real time PCR validation of RNA-seq results. Data were normalized to HPRT and expressed as fold increase over control samples. (G) RNA-FISH against Nr2f2 (red) on section of E9.5 Mef2c-Cre (control) and Hoxb1GoF;Mef2c-Cre embryos. An anterior shift in expression of Nr2f1 is detected in Hoxb1GoF;Mef2c-Cre compared to control embryos. Nuclei are stained with Hoechst. at, atria; V, ventricle. Scale bars: 200 μm.
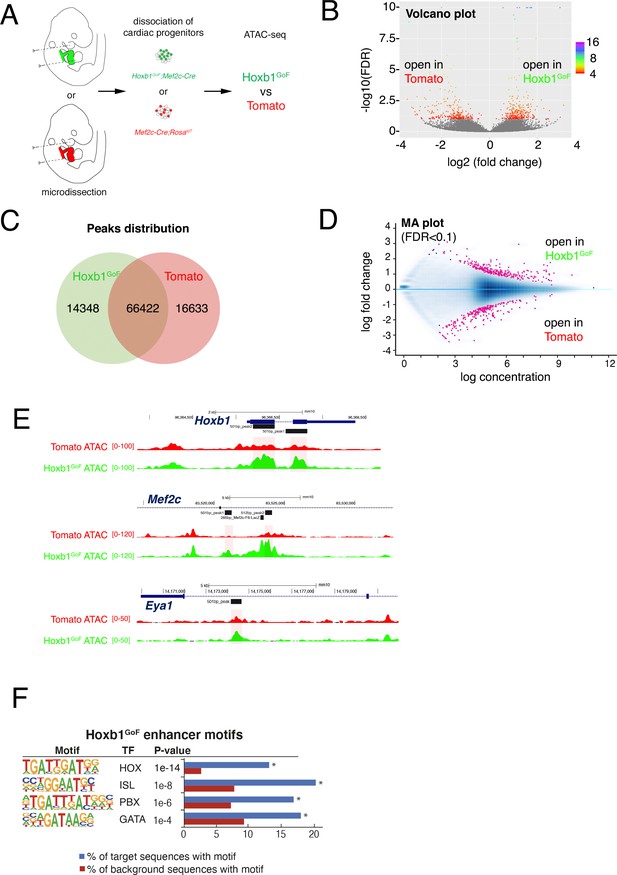
Chromatin accessibility when Hoxb1 is ectopically expressed in the Mef2c-Cre+ cardiac progenitor cells.
(A) Schemes of Hoxb1GoF;Mef2c-Cre (GFP+ named Hoxb1GoF) and Mef2c-Cre;RosatdT (Tomato+) embryos at E9.5 and the micro-dissected regions (GFP+ or Tomato+) comprising aSHF progenitors and the outflow tract of the forming heart. (B) Volcano plot of ATAC-seq performed from Hoxb1GoF and Tomato+ cells. (C) Venn diagram showing overlap of the ATAC-seq peaks in the two populations. (D) MA plot of ATAC-seq peaks in Hoxb1GoF versus Tomato+ cells. (E) Open chromatin profiles correlate with transcriptional expression. Browser views of Hoxb1, Mef2c and Eya1 loci with ATAC-seq profiles of Hoxb1GoF cells (green) and Tomato+ Mef2c-Cre;RosatdT labeled cells (red). Data represent merged technical and biological replicates for each cell type. The y-axis scales range from 0 to 100; 0–120 and 0–50 in normalized arbitrary units. The tracks represent ATAC-seq data. Boxed regions show cell-type-specific peaks around Hoxb1, Mef2c, and Eya1 gene loci. (F) Hoxb1GoF enhancers were enriched in DNA binding motifs for HOX and known cardiac transcription factors including PBX and GATA factors. HOX recognition motifs were strongly enriched in a known motif search of Hoxb1GoF enhancers. p-values were obtained using HOMER.
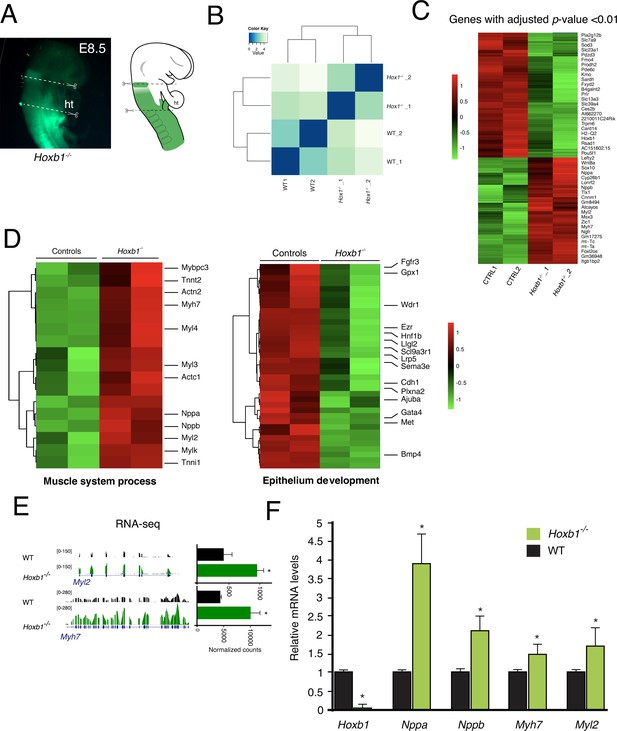
Quality assessment of RNA-seq data performed on the Hoxb1-/- embryos.
(A) Macroscopic view of Hoxb1-/- embryos at E8.5 (6-8ps) and the micro-dissected region comprising the SHF progenitors. (B) Replicate correlation for RNA-seq datasets from Hoxb1-/- and wild-type (WT) regions. (C) Unsupervised hierarchical clustering of all differentially expressed genes between Hoxb1-/- over WT regions. (D) Heatmap of ‘muscle system process’ and ‘epithelium development’ associated genes analyzed by RNA-seq. (E) RNA-sequencing datasets visualized for the cardiac differentiation markers Myl2 and Myh7. (F) Real time PCR validation of RNA-seq results. Data were normalized to HPRT and expressed as fold increase over WT samples.
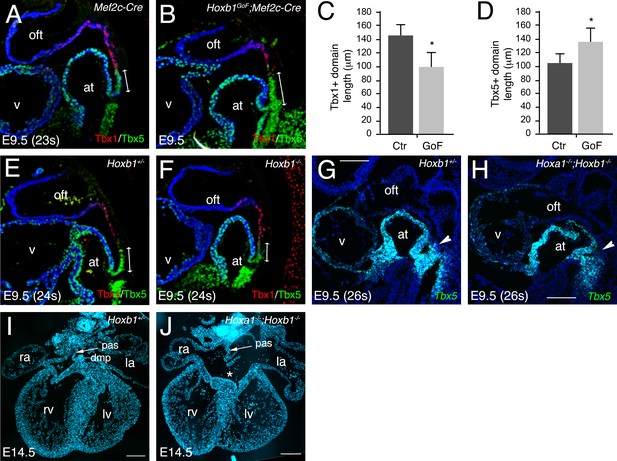
Hoxa1 and Hoxb1 genes are required for atrioventricular septation.
(A–D) Immunofluorescence on medial sagittal sections showing Tbx1 (red) and Tbx5 (green) protein distribution at E9.5 (23–24 s). (A,B) At E9.5, a boundary is observed in Mef2c-Cre embryos between Tbx1+ cells close to the arterial pole of the heart and Tbx5+ cells in the pSHF. In Hoxb1GoF;Mef2c-Cre embryos the Tbx1+ domain appears reduced although the boundary is maintained. (C) Measurement of the Tbx1+ domain confirmed a decrease of aSHF length in Hoxb1GoF;Mef2c-Cre (GoF) compared to control (Ctr) embryos. (D) Measurement of the Tbx5+ domain revealed a shift of the boundary since the length of the pSHF was increased in Hoxb1GoF;Mef2c-Cre (GoF) compared to control (Ctr) embryos. (E,F) The Tbx5+ domain appears reduced in Hoxb1-/- embryos compared to Hoxb1+/- littermates. (G,H) RNA-FISH on sagittal sections showing the reduction of Tbx5+ domain in the pSHF of Hoxa1-/-;Hoxb1-/- embryos compared to Hoxb1+/- littermates. (I,J) DAPI stained sections of a Hoxb1+/- (I) and a Hoxa1-/-;Hoxb1-/- (J) heart at E14.5 showing the primary atrial septum (pas, arrow). Note the AVSD and absence of the DMP in J, n = 3. at, atria; la, left atrium; lv, left ventricle; ra, right atrium; rv, right ventricle; SHF, second heart field; v, ventricle; la, left atrium; lv, left ventricle; ra, right atrium; rv, right ventricle. Scale bars: 100 μm (E,F); 200 μm (G,H).
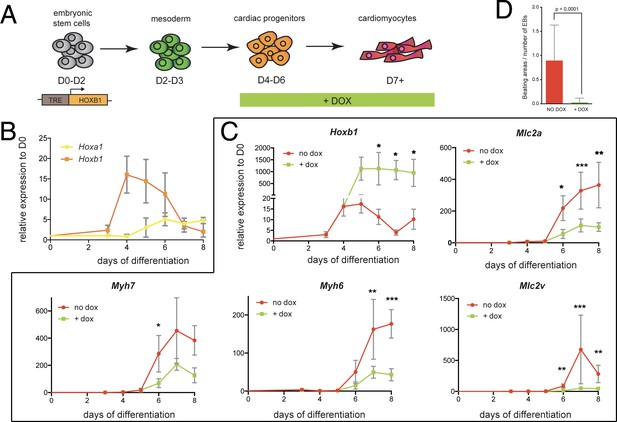
Hoxb1 overexpression in mES leads to arrested cardiac differentiation.
(A) Scheme of the experiment. Using the tet-ON/Hoxb1 mouse embryonic stem cell line (Gouti and Gavalas, 2008), Hoxb1 expression was induced by addition of 1 μg/ml of doxycycline (DOX) at the cardiac progenitor stage from day 4 (D4) during mES cell differentiation into cardiac cells. (B) Kinetics of Hoxa1 and Hoxb1 during mES cell differentiation as measured by real time qPCR. Results are normalized for gene expression in undifferentiated mES cells (D0). (C) Kinetics of expression for Hoxb1 and Myh6, Myh7, Mlc2v (or Myl2), Mlc2a during mES cell differentiation after induction of Hoxb1 expression (+ dox) or in control condition (no dox). Results are normalized for gene expression in undifferentiated mES cells (D0). Paired, one-sided t-test was performed based on relative transcript expression between control (no dox) and doxycycline treatment (+ dox). * indicates a significance level of p<0.05, ** indicates p<0.005, *** indicates p<0.0005. (D) Quantification of beating areas relative to the number of embryonic bodies (EBs) at D8 of mES cell differentiation with or without doxycycline addition. Error bars indicate mean + / - SEM; n = 4 experiments.
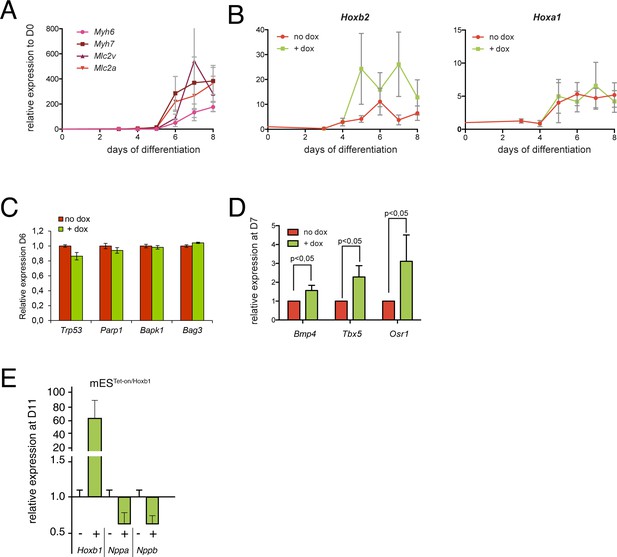
Expression analysis using the mES cells overexpressing model.
(A) Kinetics of myocardial genes (Myh6, Myh7, Myl2 (Mlc2v), Mlc2a) during mES cell differentiation as measured by real time qPCR. (B) Kinetics of Hoxa1 and Hoxb2 during mES cell differentiation after induction of Hoxb1 expression (+ dox) or in control condition (no dox). Results are normalized for gene expression in undifferentiated mES cells (D0). (C) Relative expression of apoptosis genes (Trp53, Parp1, Bapk1, Bag3) at D6 of mES cell differentiation as measured by real time RT-PCR in mES cells induced or not for Hoxb1 expression (+ dox vs no dox). (D) Relative expression of posterior markers of the SHF (Bmp4, Tbx5, Osr1) at D7 of mES cell differentiation as measured by real time PCR in mES cells induced or not for Hoxb1 expression (+ dox vs no dox). (E) Relative expression of Hoxb1, Nppa and Nppb at D11 of mES cell differentiation as measured by real time RT-PCR in mES cells induced or not for Hoxb1 expression (+ dox vs no dox). Results are normalized for mRNA expression in untreated cells (no dox). Error bars indicate mean + / - SEM; n = 4 experiments.
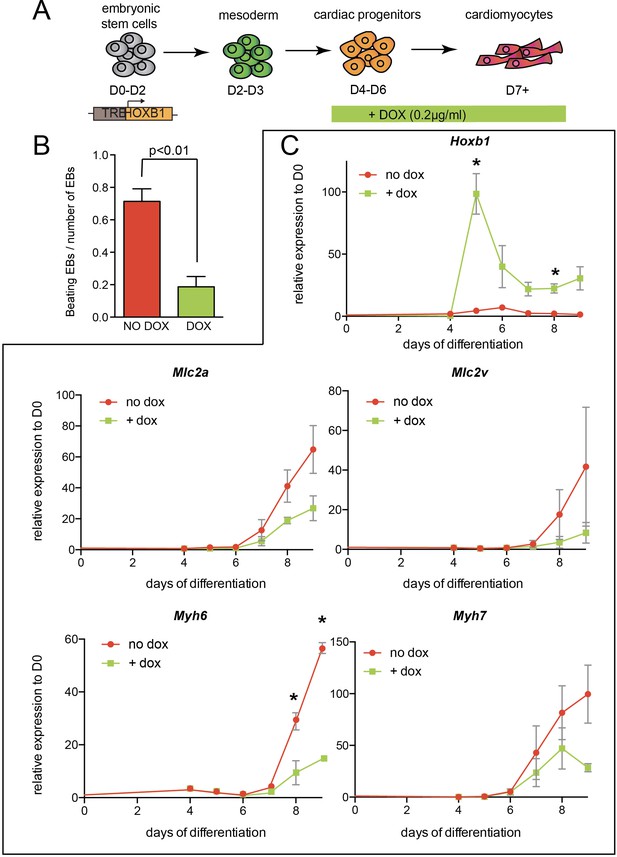
Arrested cardiac differentiation in mES cells using a lower induction of Hoxb1.
(A) Scheme of the experiment. Using the tet-ON/Hoxb1 mouse embryonic stem cell line (Gouti and Gavalas, 2008), Hoxb1 expression was induced by addition of 0.2 µg/ml of doxycycline (DOX) from day 4 (D4) during mES cell differentiation into cardiac cells. (B) Quantification of beating embryoid bodies (EBs) relative to the total number of embryoid bodies. (C) Kinetics of expression for Hoxb1 and Myh6, Myh7, Mlc2v (or Myl2), Mlc2a during mES cell differentiation after induction of Hoxb1 expression (+ dox) or in control condition (no dox). Results are normalized for gene expression in undifferentiated mES cells (D0). Error bars indicate mean + / - SEM; n = 2 experiments. Paired, one-sided t-test was performed based on relative transcript expression between control (no dox) and doxycycline treatment (+ dox). * indicates a significance level of p<0.05.
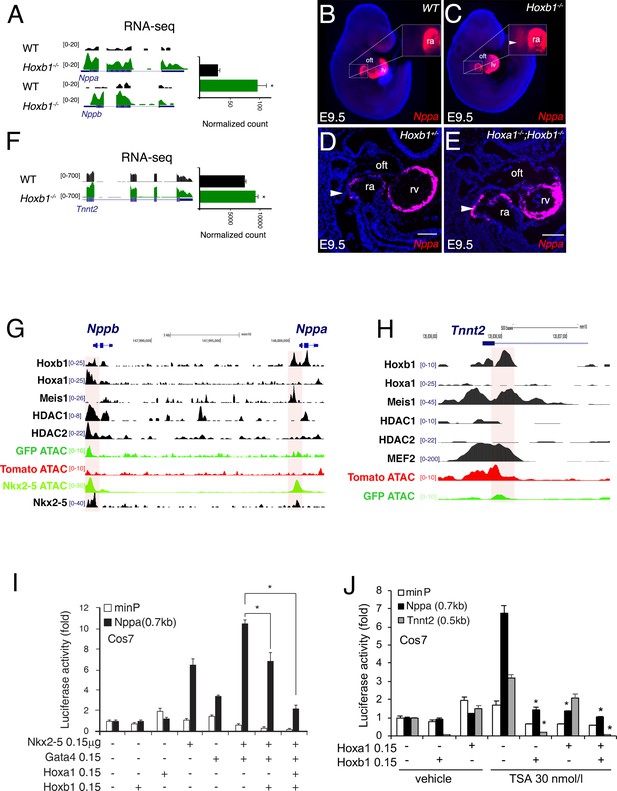
Hoxb1 regulates cardiac differentiation through transcriptional repression of myocardial genes.
(A) Browser views of Nppa and Nppb gene loci with RNA-seq profiles of Hoxb1-/- (green) and wild-type (WT) population (black). Data represent the merge of technical and biological replicates for each cell type. The y-axis scales range from 0 to 20 in normalized arbitrary units. (B,C) Whole mount RNA-FISH for Nppa in WT (B) and Hoxb1-/- (C) E9.5 embryos. Inset displays higher magnification of the posterior heart region. Nuclei are stained with Dapi. RNA-FISH on sagittal sections for Nppa in Hoxb1+/- (D) and Hoxa1-/-;Hoxb1-/- (E) at stage E9.5. (F) Browser view of Tnnt2 locus with RNA-seq profiles of Hoxb1-/- (green) and wild-type (WT) population (black). Data represent the merge of technical and biological replicates for each cell type. The y-axis scales range from 0 to 700 in normalized arbitrary units. (G,H) Browser view of Nppa, Nppb (G) and Tnnt2 (H) loci with ATAC-seq on purified cardiac cells and ChIP-seq profiles of Hoxb1, Hoxa1 (De Kumar et al., 2017), Meis1 (Losa et al., 2017), Nkx2-5 (van den Boogaard et al., 2012), Mef2 (Akerberg et al., 2019), HDAC1 and HDAC2 (Whyte et al., 2012). (I) Constructs were co-transfected with Nkx2-5, Gata4, Hoxa1 and Hoxb1 expression vectors into Cos-7 cells. Luciferase activity was determined and normalized as fold over the reporter alone (mean ± SEM, n = 3, *p<0.05 for Nkx2-5 and Hoxb1 versus Nkx2-5, using ANOVA). (J) Luciferase reporter assays on the −633/+87 bp region of the Nppa promoter and the −497/+1 bp region of the Tnnt2 promoter. Cos-7 cells co-transfected with Hoxa1 and Hoxb1 expressing vector or not were treated in the absence or presence of 30 nmol/l TSA. Bars represent mean ± SEM (n = 3). Statistical test was conducted using ANOVA (*p<0.05 for Hoxa1, Hoxb1 and TSA treatment versus Hoxb1 or TSA treatment). oft, outflow tract; lv, left ventricle; ra, right atrium.
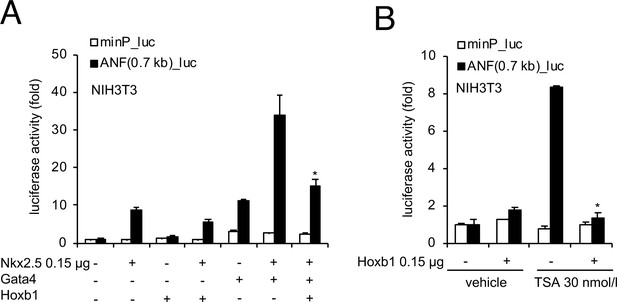
The 0.7 kb Nppa promoter is a functional target for Hoxb1.
(A) Transient transfections were carried out with the 0.7 kb Nppa (ANF) promoter in NIH3T3 cells. Constructs were co-transfected with Nkx2-5, Gata4 and Hoxb1 expression vectors. Luciferase activity was determined and normalized as fold over the reporter alone (mean ± SEM, n = 3, *p<0.05 for Nkx2-5 and Hoxb1 versus Nkx2-5, using ANOVA). (B) Luciferase reporter assays on the 0.7 kb ANF promoter in presence of TSA. NIH3T3 cells co-transfected with and without a Hoxb1 expression vector were treated in the absence or presence of 30 nmol/l TSA. Bars represent mean ± SEM (n = 3). Statistical test was conducted using ANOVA (*p<0.05 for Hoxb1 and TSA treatment versus Hoxb1 or TSA treatment).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background Mus musculus | C57Bl/6J | Charles River | ||
| Genetic reagent Mus musculus | Hoxb1GFP | PMID:11076756 | RRID:MGI:1928090 | Mario Capecchi (HHMI) |
| Genetic reagent Mus musculus | Hoxb1IRES-Cre | PMID:12815623 | RRID:MGI:2668513 | Mario Capecchi (HHMI) |
| Genetic reagent Mus musculus | Tg(CAG-Hoxb1-EGFP)1Sza | PMID:30134070 | ||
| Genetic reagent Mus musculus | Mef2c-AHF-Cre | PMID:15253934 | RRID:MGI:4940068 | Brian Black (UCSF) |
| Genetic reagent Mus musculus | Gt(ROSA)26Sortm1Sor(R26R) | PMID:9916792 | RRID:MGI:1861932 | |
| Genetic reagent Mus musculus | Gt(ROSA)26Sortm9(CAG-tdTomato)Hze | PMID:20023653 | RRID:MGI:3809523 | |
| Genetic reagent Mus musculus | Mlc1v-nlacZ-24 or Fgf10Tg(Myl3-lacZ)24Buck | PMID:15217909 | RRID:MGI:3629660 | |
| Genetic reagent Mus musculus | Mlc3f-nlacZ-2E or Tg(Myl1-lacZ)1Ibdml | PMID:15217909 | RRID:MGI:5449224 | |
| Gene (Mus musculus) | Hoxb1 | Mus musculus genome resource | RRID:MGI:96182 | |
| Antibody | Rabbit anti-GFP Polyclonal Antibody | Thermo Fisher Scientific | Cat# A-11122, RRID:AB_221569 | IF (1:500) |
| Antibody | Rabbit anti-Hoxb1 Polyclonal Antibody | Covance | Cat# PRB-231P-100, RRID:AB_291592 | IF (1:200) |
| Antibody | Mouse anti-αactinin Monoclonal Antibody | Sigma | A7732, RRID:AB_2221571 | IF (1:500) |
| Antibody | Mouse anti-myosin Monoclonal Antibody | DHSB | Cat# MF 20, RRID:AB_2147781 | IF (1:100) |
| Antibody | Rabbit anti- cleaved Caspase3 Polyclonal Antibdoy | Cell signaling Technology | Cat# 9661, RRID:AB_2341188 | IF (1:300) |
| Antibody | Rabbit anti-phospho-Histone H3 Polyclonal Antibody | Millipore | Cat# 06–570, RRID:AB_310177 | IF (1:400) |
| Antibody | Mouse anti-Islet1 Monoclonal Antibody | DSHB | Cat# 39.4D5, RRID:AB_2314683 | IF (1:100) |
| Antibody | Rabbit anti-Tbx1 Polyclonal Antibody | LifeSpan | Cat# LS-C31179-100, RRID:AB_911118 | IF (1:100) |
| Antibody | Goat anti-Tbx5 Polyclonal Antibody | Santa Cruz | Cat# sc-17866, RRID:AB_2200827 | IF (1:250) |
| Antibody | Donkey anti-rat IgG (H+L) Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-21208, RRID:AB_141709 | IF (1:500) |
| Antibody | Donkey anti-Mouse IgG (H+L) Alexa Fluor 555 | Thermo Fisher Scientific | Cat# A-31570, RRID:AB_2536180 | IF (1:500) |
| Antibody | Goat anti-rabbit IgG (H+L) Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-21244, RRID:AB_2535812 | IF (1:500) |
| Sequence-based reagent | mm-Hoxb1-C1 Mus musculus | Acdbio | 541861 | RNA-FISH probe |
| Sequence-based reagent | mm-Nppa Mus musculus | Acdbio | 418691 | RNA-FISH probe |
| Sequence-based reagent | mm-Bmp4-C1 Mus musculus | Acdbio | 401301 | RNA-FISH probe |
| Sequence-based reagent | mm-Gata4 Mus musculus | Acdbio | 417881 | RNA-FISH probe |
| Sequence-based reagent | mm-Aldh1a2 Mus musculus | Acdbio | 447391 | RNA-FISH probe |
| Sequence-based reagent | mm-eGFP-C3 | Acdbio | 400281 | RNA-FISH probe |
| Sequence-based reagent | mm-Isl1 Mus musculus | Acdbio | 451931 | RNA-FISH probe |
| Sequence-based reagent | mm-Nr2f2-C3 Mus musculus | Acdbio | 480301 | RNA-FISH probe |
| Sequence-based reagent | mm-tdTomato-C3 | Acdbio | 317041 | RNA-FISH probe |
| Sequence-based reagent | mm-Tbx1-C1 Mus musculus | Acdbio | 481911 | RNA-FISH probe |
| Sequence-based reagent | mm-Tbx5-C2 Mus musculus | Acdbio | 519581 | RNA-FISH probe |
| Sequence-based reagent | mm-Osr1-C2 Mus musculus | Acdbio | 496281 | RNA-FISH probe |
| Commercial assay or kit | RNAscope Multiplex Fluorescent v2 Assay | Acdbio | 323110 | |
| Commercial assay or kit | Ovation RNAseq v2 kit | NuGEN | 7102–08 | RNA-seq |
| Commercial assay or kit | AffinityScript Multiple Temperature cDNA synthesis kit | Agilent | 600107 | cDNA synthesis kit |
| Commercial assay or kit | Master Mix PCR Power SYBR Green | ThermoFisher | 4334973 | qPCR |
| Commercial assay or kit | NucleoSpin RNA XS | Machery-Nagel | 740902.50 | RNA extraction |
| Recombinant DNA reagent | pNppa(−633/+87)-Luc (plasmid) | PMID:12023302 | Vincent Christoffels (Amsterdam University Medical Center) | |
| Recombinant DNA reagent | pTnnT2(−497/+1)-Luc (plasmid) | PMID:12023302 | Vincent Christoffels (Amsterdam University Medical Center) | |
| Cell line (Cercopithecus aethiops) | Cos-7 | ATCC | RRID:CVCL_0224 CRL-1651 | |
| Cell line Mus musculus | NIH3T3 | ATCC | RRID:CVCL_0594 CRL-1658 | |
| Cell line Mus musculus | mESTet-On/Hoxb1 | PMID:18499896 | Anthony Gavalas (Technische Universitat Dresden) | |
| Sequence-based reagent | Bmp4-FW | This paper | qPCR primers | TTCCTGGTAACCGAATGCTGA |
| Sequence-based reagent | Bmp4-Rev | This paper | qPCR primers | CCTGAATCTCGGCGACTTTTT |
| Sequence-based reagent | Bag3-Fw | This paper | qPCR primers | GCCCTAAGGACACTGCATCTT |
| Sequence-based reagent | Bag3-Rev | This paper | qPCR primers | GCTGGGAGTAGGCATGGAAA |
| Sequence-based reagent | Bak1-Fw | This paper | qPCR primers | CCTTCTGAACAGCAGGTTGC |
| Sequence-based reagent | Bak1-Rev | This paper | qPCR primers | GACCCACCTGACCCAAGATG |
| Sequence-based reagent | Dapk1-Fw | This paper | qPCR primers | GAGGTGGTGGCTGCGTC |
| Sequence-based reagent | Dapk1-Rev | This paper | qPCR primers | CGCAGACCTCCGGTCC |
| Sequence-based reagent | GFP-Fw | This paper | qPCR primers | CGACGTAAACGGCCACAAGTT |
| Sequence-based reagent | GFP-Rev | This paper | qPCR primers | TTGATGCCGTTCTTCTGCTTGT |
| Sequence-based reagent | Hoxa1-Fw | This paper | qPCR primers | AGAAACCCTCCCAAAACAGG |
| Sequence-based reagent | Hoxa1-Rev | This paper | qPCR primers | TTGTTGAAGTGGAACTCCTTCTC |
| Sequence-based reagent | Hoxb1-Fw | This paper | qPCR primers | AAGAGAAACCCACCTAAGACAGC |
| Sequence-based reagent | Hoxb1-Rev | This paper | qPCR primers | TGAAGTTTGTGCGGAGACC |
| Sequence-based reagent | Hprt-Fw | This paper | qPCR primers | TGTTGGATATGCCCTTGACT |
| Sequence-based reagent | Hprt-Rev | This paper | qPCR primers | GATTCAACTTGCGCTCATCT |
| Sequence-based reagent | Isl1-Fw | This paper | qPCR primers | GCAACCCAACGACAAAACTAA |
| Sequence-based reagent | Isl1-Rev | This paper | qPCR primers | CCATCATGTCTCTCCGGACT |
| Sequence-based reagent | Nppa-Fw | This paper | qPCR primers | CACAGATCTGATGGATTTCAAGA |
| Sequence-based reagent | Nppa-Rev | This paper | qPCR primers | CCTCATCTTCTACCGGCATC |
| Sequence-based reagent | Nppb-Fw | This paper | qPCR primers | GTCCAGCAGAGACCTCAAAA |
| Sequence-based reagent | Nppb-Rev | This paper | qPCR primers | AGGCAGAGTCAGAAACTGGA |
| Sequence-based reagent | Nr2f2-Fw | This paper | qPCR primers | CCTCAAAGTGGGCATGAGAC |
| Sequence-based reagent | Nr2f2-Rev | This paper | qPCR primers | TGGGTAGGCTGGGTAGGAG |
| Sequence-based reagent | Osr1F-Fw | This paper | qPCR primers | AGAAGCGTCAGAAGTCTAGTTCG |
| Sequence-based reagent | Osr1R-Rev | This paper | qPCR primers | GGAACCGCAATGATTTCAA |
| Sequence-based reagent | Parp1-Fw | This paper | qPCR primers | AGGGCTGCCTGGAGAAGATA |
| Sequence-based reagent | Parp1-Rev | This paper | qPCR primers | TCGTCCCGCTTCTTGACAAA |
| Sequence-based reagent | Raldh2-Fw | This paper | qPCR primers | CATGGTATCCTCCGCAATG |
| Sequence-based reagent | Raldh2-Rev | This paper | qPCR primers | GCGCATTTAAGGCATTGTAAC |
| Sequence-based reagent | Tbx5-Fw | This paper | qPCR primers | CCCGGAGACAGCTTTTATCG |
| Sequence-based reagent | Tbx5-Rev | This paper | qPCR primers | TGGTTGGAGGTGACTTTGTG |
| Sequence-based reagent | Trp53-Fw | This paper | qPCR primers | AGTATTTCACCCTCAAGATCCGC |
| Sequence-based reagent | Trp53-Rev | This paper | qPCR primers | GGAGCTAGCAGTTTGGGCTT |
Additional files
-
Supplementary file 1
Excel file containing the list of deregulated genes identified by RNA-seq analysis (sheet one, upregulated genes; sheet two, downregulated genes).
- https://cdn.elifesciences.org/articles/55124/elife-55124-supp1-v2.xlsx
-
Supplementary file 2
Excel file containing the list of regions of open chromatin identified by ATAC-seq analysis (sheet one, upregulated genes; sheet two, downregulated genes).
- https://cdn.elifesciences.org/articles/55124/elife-55124-supp2-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/55124/elife-55124-transrepform-v2.docx





