Mechanisms of nucleotide selection by telomerase
Figures
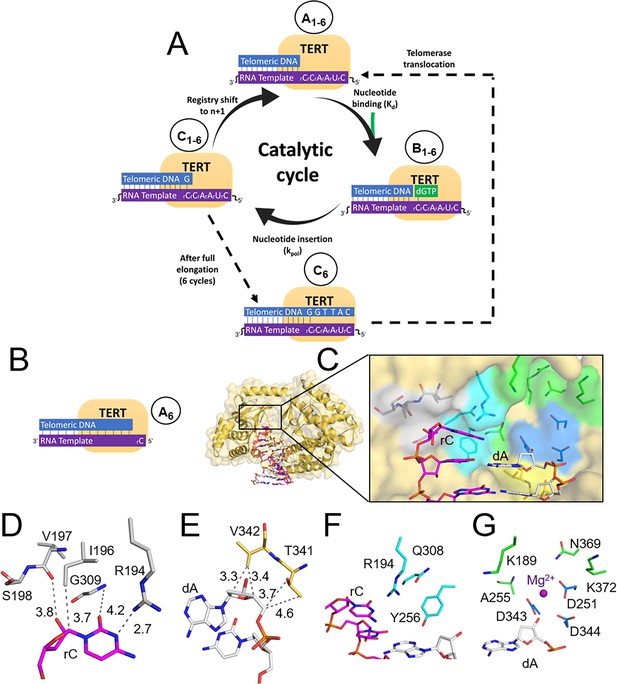
The telomerase catalytic cycle, and its first structural state.
(A) Overview of the telomerase catalytic cycle. Telomerase forms a prenucleotide bound binary complex (State A1). Then, it binds the incoming nucleotide triphosphate to form a ternary complex (State B1), chemically links it to the telomere terminus (State C1), and then shifting registry to bind the next incoming nucleotide (State A2). After this cycle completes six times (State C6), telomerase will either disassociate or undergo translocation (dotted line), which places it back into State A1. (B) The tcTERT prenucleotide binary complex. tcTERT (pale orange cartoon and surface) encircles the DNA (white) and RNA (purple) substrate. (C) Active site pocket of the prenucleotide binary complex. rC binding (gray), dG binding residues (yellow), nucleoside residues (cyan), catalytic residues (blue), and triphosphate binding (green) are shown as sticks. (D) Closeup views of the rC binding residues, (E) terminal dG binding residues, (F) nucleoside binding residues, and (G) the tcTERT catalytic residues and triphosphate coordinating residues. A Mg2+ ion is shown as a purple sphere and DNA is presented as white sticks.
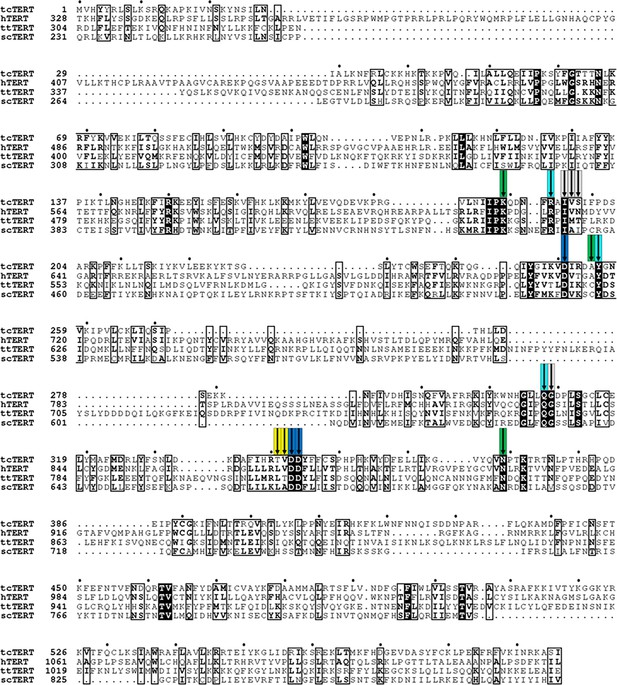
Alignments of several telomerase reverse transcriptase homologs.
A multiple sequence alignment is shown from several established TERT models, including Tribolium castaneum (tc), Homo sapiens (h), Tetrahymena thermophila (tt), andSaccharomyces cerevisiae(sc). The active site residues discussed are highly conserved throughout all four species. Colored arrows mark the positions of each group as in Figure 1, with residues coordinating the RNA terminus shown in gray, the primer terminus in yellow, the catalytic residues in blue, the nucleoside coordinating residues in cyan, and the triphosphate coordinating residues in green.
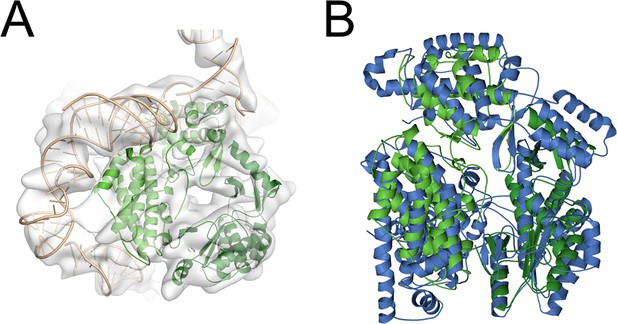
Overlay of TERT from Tribolium castaneum with other TERT structures.
(A) An overlay of tcTERT with the density from a cryo-EM structure of human telomerase. tcTERT is shown as a green cartoon, density is a gray surface, and hTR is shown as a beige cartoon (taken from Nguyen et al., 2018). (B) An overlay of tcTERT with a cryo-EM structure of TERT from Tetrahymena thermophila. tcTERT is shown as a green cartoon, and ttTERT is shown as a blue cartoon. ttTERT structure takin from Jiang et al., 2018; PDB code 6D6V.
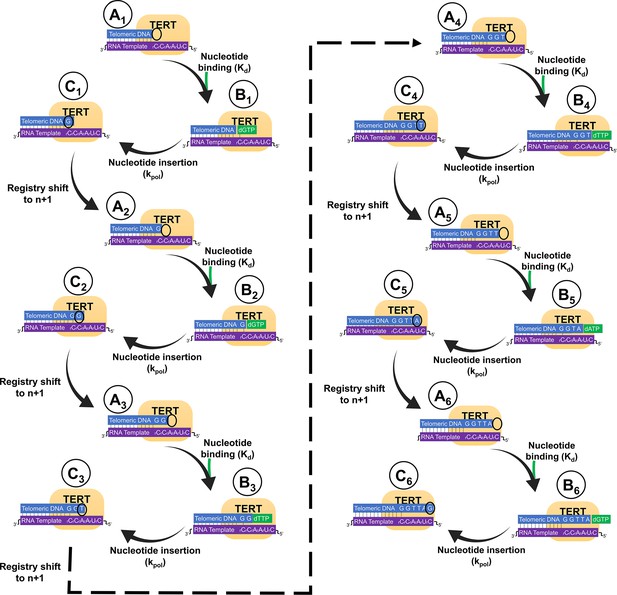
All 18 structural states involved in the extension of a telomeric repeat.
To extend a full telomeric repeat of 6 nucleotides, 18 structural states are sampled, from A1 to C6. Telomeric DNA is shown in blue, telomerase RNA in purple, the TERT protein in yellow, and nucleotide triphosphates in green. For states without a nucleotide triphosphate bound, the position of the active site is shown with a black circle. The A category represents prenucleotide states, the B category represents ternary states, and the C category represents product states. The structures presented in this manuscript are A6, B6, and C6.
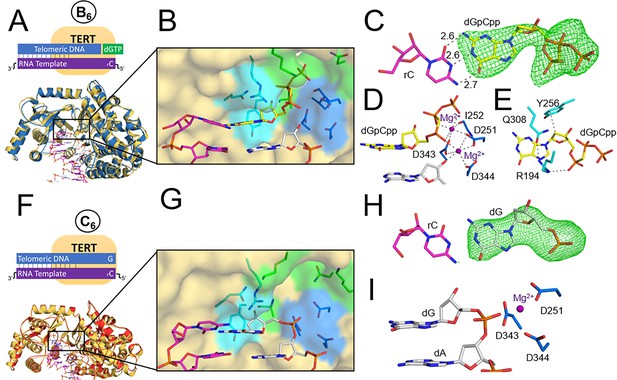
TERT ternary and product structures.
(A) The tcTERT ternary structure, overlayed with the prenucleotide binary complex. DNA (white), RNA (purple), binary tcTERT (yellow cartoon), and ternary tcTERT (blue cartoon) are shown. (B) The tcTERT active site. Nucleoside residues (cyan), triphosphate residues (green), and catalytic residues (blue) are shown. The dGpCpp (yellow), DNA, and RNA are represented as sticks. (C) Closeup view of dGpCpp with a polder OMIT map contoured at σ = 3.0 (green mesh). (D, E) dGpCpp contacts are shown with Mg2+ (purple), catalytic residues (marine) and nucleoside residues (cyan) indicated. (F) The tcTERT product structure (red cartoon), overlaid with the prenucleotide binary complex (yellow cartoon). (G) An active site view of the tcTERT product structure. (H) A display of a polder OMIT map contoured at σ = 3.3 around the incoming dGpCpp (green mesh). (I) Catalytic residues (marine) coordinate the inserted dG (white).
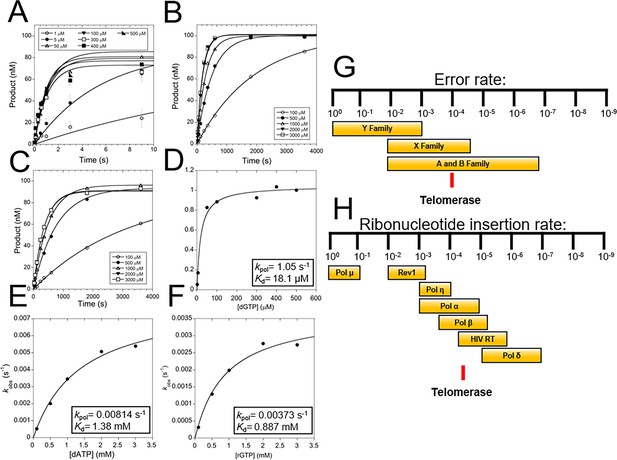
TERT fidelity and sugar discrimination.
(A) Pre-steady-state kinetics of WT tcTERT inserting dGTP opposite rC. Data was fit to Equation 1 (Supplementary file 3, Table 3a and b). Error bars represent the standard deviation of the mean. These experiments were also performed with WT tcTERT inserting dATP across from rC (B) and rGTP across from rC (C). Replots of the data and fits to Equation 2 were performed for (D) dGTP across from rC, (E) dATP across from rC, and (F) rGTP across from rC. (G) A comparison of TERT nucleobase fidelity (red line) compared to other DNA polymerase families. (H) TERT’s rNTP discrimination rates (red line) compared to select DNA polymerases is shown (Brown and Suo, 2011; McCulloch and Kunkel, 2008).
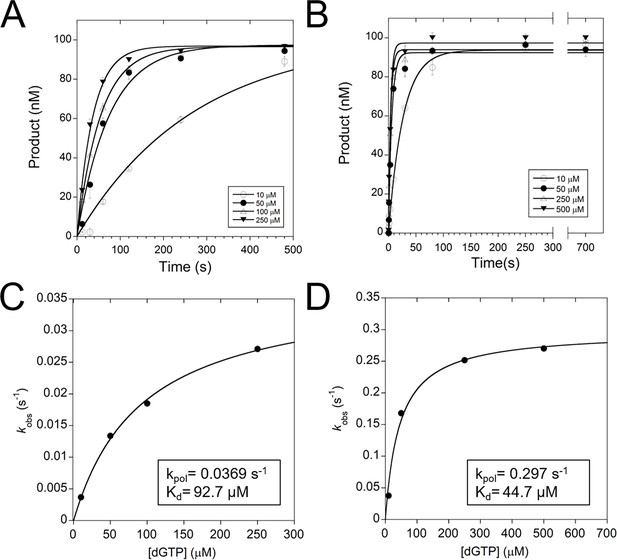
Pre-steady-state kinetics of additional TERT active site mutants.
(A) Pre-steady-state kinetics of tcTERT with an R194A substitution. Error bars represent the standard deviation of the mean. Data was fit to the exponential equation 1 to determine the kobs (rate) and target engagement. Pre-steady-state parameters were also calculated for (B) tcTERT with a Q308A substitution. (C) A replot of the fits from panel A, showing the kobs values obtained from each concentration of dGTP. Data was fit to equation 2. (D) A replot of tcTERT Q308A.
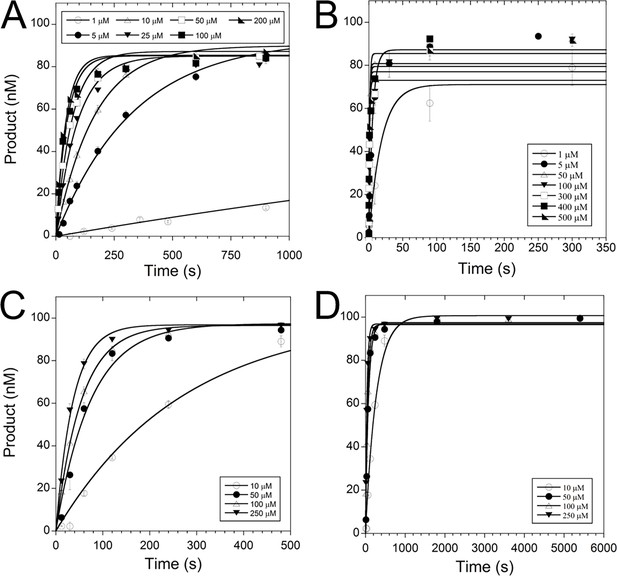
Complete graphs of selected TERT pre-steady-state kinetics.
Due to space considerations, only a zoomed in portion of pre-steady-state kinetic data was shown in certain portions of the text. Both the zoomed and complete curves were shown for (A, B) WT TERT inserting dGTP across from dC, (C, D) TERT R194A inserting dGTP across from dC.
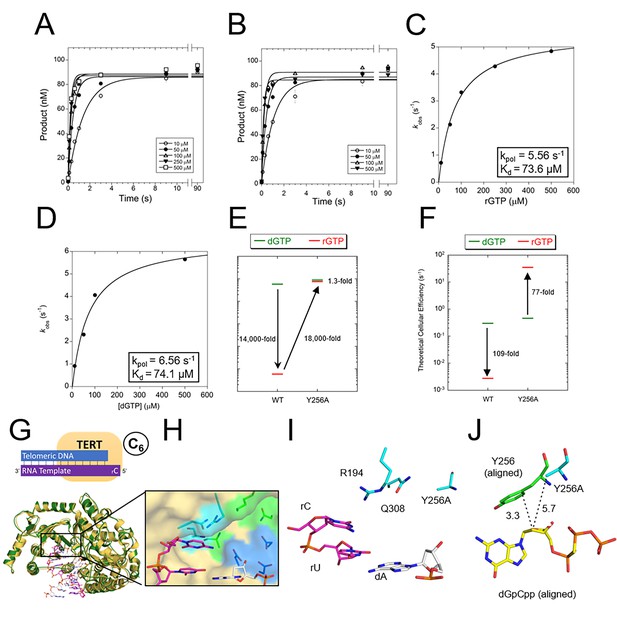
The steric gate of TERT prevents insertion of rNTPs.
Pre-steady-state kinetics of tcTERT with its steric gate removed (i.e. Y256A) for both (A) rGTP insertion and (B) dGTP insertion (Supplementary file 3, Table 3a). Error bars represent standard deviation of the mean. These graphs were replotted and fit to Equation 2 for (C) tcTERT Y256A inserting rGTP and (D) tcTERT Y256A inserting dGTP (E) A comparison of catalytic efficiencies (kpol/Kd) of both WT TERT and TERT Y256A for the insertion of dGTP (green) versus rGTP (red). (F) A comparison of TERT efficiencies adjusted for cellular nucleotide concentrations. (G) The prenucleotide binary complex of TERT Y256A (green) overlaid with the WT TERT prenucleotide binary complex structure (yellow). (H, I) The active site pocket of TERT Y256A, with DNA (white), RNA (purple), catalytic residues (blue), and nucleoside coordinating residues (cyan) shown. (J) The closest contacts to the C2 position of dGpCpp from the ternary structure (aligned and shown with yellow and green sticks) compared to Y256A tcTERT (cyan).
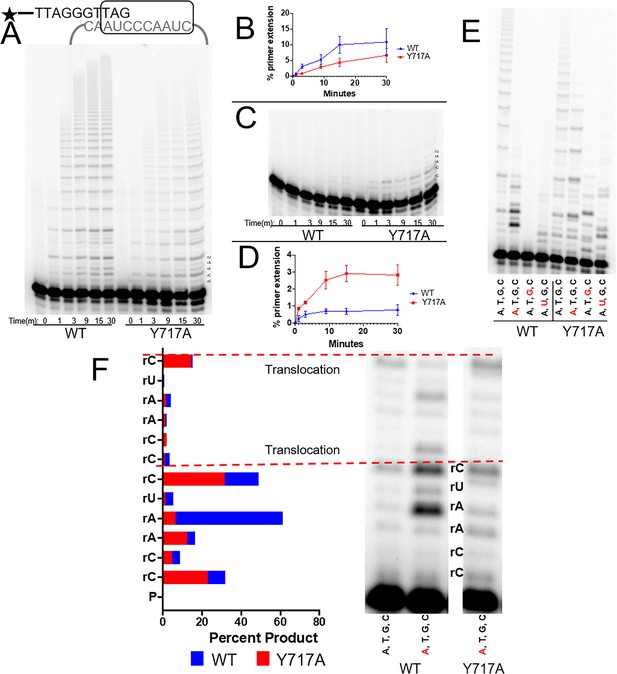
The steric gate of human telomerase.
(A) Timecourse of primer extension with all four dNTPs by WT (left) and Y717A (right) human telomerase. (B) Quantification of percent primer extension, with WT shown as a blue line and the Y717A mutant shown in red. (C) Timecourse of primer extension with all four rNTPs for WT telomerase (left) and Y717A telomerase (right). (D) Results from the gel in panel C, quantified with the mutant telomerase shown in red and WT telomerase shown in blue. (E) Primer extension from both WT (left) and Y717A telomerase. Each lane contains either all four dNTPs, or three dNTP and one dNTP marked in red. (F) A close-up view of the first two telomeric repeats from selected lanes in panel E. rNTPs present in the mix are shown in red. Quantifications of each band in terms of percent product are also shown and labeled by the position of the telomerase templating base. The base marked by an asterisk is complementary to the rNTP in the solution. Error bars represent standard deviations of four biological replicates.
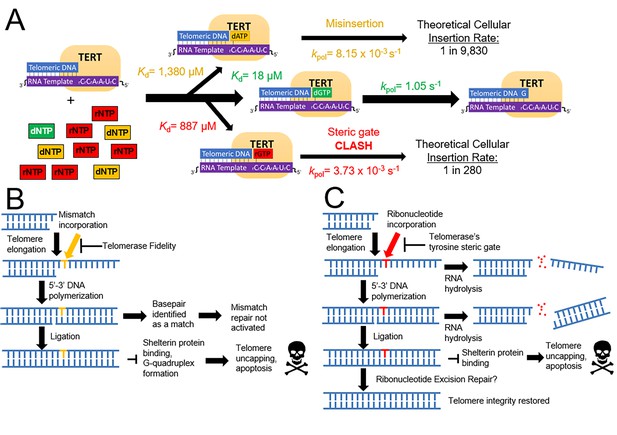
A model for how telomerase preserves telomeric integrity and biological implications.
(A) The telomerase catalytic cycle, with matched dNTP (green), mismatched dNTPs (orange), and matched rNTPs (red). For each nucleotide path, kinetic parameters are labeled for each step and the insertion probability. (B) Downstream consequences of mismatch insertion by telomerase may include evasion of mismatch repair, reduced binding affinity for shelterin proteins, and altered stability of telomeric G quadruplexes. (C) Eventual consequences of rNTP insertion by telomerase. After insertion into telomeres, ribonucleotides could cause harsh consequences via hydrolysis, or disrupting telomere capping and stability. Ribonucleotides may also be removed by ribonucleotide excision repair.
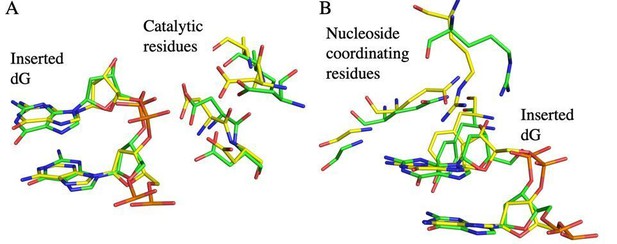
A comparison of ttTERT active site residues with tcTERT.
A superposition of the tcTERT product structure active site (state C6, yellow) with the active site from the product structure from ttTERT (State C3, green, from Jiang et al., 2018, PDB code: 6D6V). Key active site amino acid residues including catalytic residues (in A), and nucleoside coordinating residues (B) align closely, exhibiting a Root Mean Squared Deviation (RMSD) of 0.83.
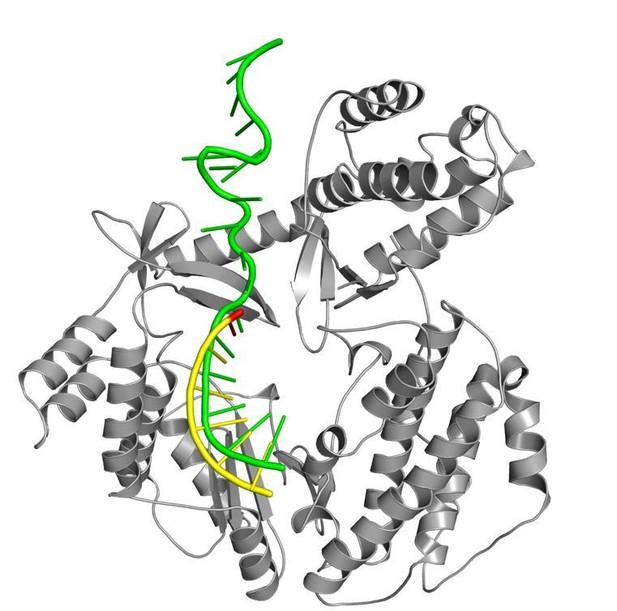
RNA used with tcTERT structures vs TR from Tetrahymena thermophila.
A superposition of the RNA component from our product structure (yellow, state C6) and the recent cryo-EM structure of T. thermophila (green, state C3, Jiang et al., 2018, PDB code 6d6v). tcTERT protein is shown as gray ribbons for reference.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Tribolium castaneum) | tcTERT | GenScript | ||
| Gene(Homo sapiens) | hTR | Gift from Dr. Tom Cech | ||
| Gene (Homo sapiens) | hTERT | Gift from Dr. Tom Cech | ||
| Strain, strain background (Escherichia coli) | One Shot BL21(DE3)pLysS Chemically Competent E. coli | Invitrogen | Cat# C606010 | |
| Strain, strain background (Escherichia coli) | One Shot TOP10 Chemically Competent E. coli | Invitrogen | Cat# C606010 | |
| Cell line (Homo sapiens) | HEK 293 T Cells, female | ATCC | RRID:CVCL_0063 | Cells were acquired from ATCC, and have not since been tested for mycoplasma, as they were used for protein generation not biological assays |
| Transfected construct (Homo sapiens) | pSUPER-hTR | Gift from Dr. Tom Cech | N/A | |
| Transfected construct (Homo sapiens) | pVan107 3X FLAG hTERT | Gift from Dr. Tom Cech | N/A | |
| Recombinant DNA reagent | pET-28a(+) with Tribolium castaneum TERT | Genscript | N/A | |
| Sequence-based reagent | Primer for telomerase activity assays | IDT | N/A | 5’-GGTCAGGTCAGGTCA-3’ |
| Sequence-based reagent | RNA template for tcTERT kinetics | IDT | N/A | 5’-rCrUrGrArCrCrUrGACCUGACC-3’ |
| Sequence-based reagent | DNA primer for tcTERT kinetics | IDT | N/A | 5’-/6-FAM/CCAGCCAGGTCAG-3’ |
| Sequence-based reagent | RNA template for tcTERT crystallogarphy | IDT | N/A | 5’- rUrGrArCrCrUrGrArCrCrUrGrG rCrUrGrG-3’ |
| Sequence-based reagent | DNA primer for tcTERT crystallogarphy | IDT | N/A | 5'-GGTTAGGGTTAGGGTTAG-3' |
| Peptide, recombinant protein | T4 polynucleotide kinase | NEB | Cat# M0201S | |
| Peptide, recombinant protein | 3X FLAG Peptide | Sigma Aldrich | Cat# F4799-4MG | |
| Chemical compound, drug | 2'-deoxyguanosine-5'-[(α,β)-methyleno]triphosphate (dGpCpp) | Jena Biosciences | Cat# NU-431S | |
| Chemical compound, drug | γ−32P ATP | Perkin-Elmer | Cat# BLU002Z250UC | |
| Chemical compound, drug | 2-methyl-2,4-pentanediol | Hampton Research | Cat# HR2-627 | |
| Software, algorithm | COOT | Emsley and Cowtan (2004) | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ | RRID:SCR_014222 |
| Software, algorithm | Phenix | Adams et al., 2010 | https://www.phenix-online.org/ | RRID:SCR_014224 |
| Software, algorithm | XDS | Kabsch (2010) | http://xds.mpimf-heidelberg.mpg.de/ | RRID:SCR_015652 |
| Software, algorithm | MolProbity | Chen et al. (2010) | http://molprobity.biochem.duke.edu/ | RRID:SCR_014226 |
| Software, algorithm | ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ | RRID:SCR_003070 |
| Software, algorithm | Kaleidagraph | Synergy Software | http://www.synergy.com/wordpress_650164087/kaleidagraph/ | RRID:SCR_014980 |
| Software, algorithm | ImageQuant TL v8.1 | GE Healthcare Life Sciences | http://www.gelifesciences. com/en/us | RRID:SCR_014246 |
| Software, algorithm | PyMol | Schrödinger LLC | https://pymol. org/2/ | RRID:SCR_000305 |
| Other | Large scale expresion (LEX-48) bioreactor | Epiphyte | https://www.epiphyte3.com/LEX | |
| Other | HisTrap HP 5 mL column | GE healthcare Life Sciences | Cat# 17524801 | |
| Other | POROS HS strong cation ion exchange resin | Thermo scientific | Cat# 1335906 | |
| Other | G-25 spin columns | GE Healthcare Life Sciences | Cat #27532501 | |
| Other | Sephacryl 16/60 S-200 HR Size Exclusion Chromatography column | GE Healthcare Life Sciences | Cat # 17116601 | |
| Other | ANTI-FLAG M2 affinity gel agarose beads | Sigma Aldrich | Cat #A2220 |
Additional files
-
Supplementary file 1
Table 1a.
The conservation and function of TERT active site residues. Table 1b. Telomerase architecture of five different TERTs, including Homo sapiens, Tetrahymena thermophila,Saccharomyces cerevisiae and Tribolium castaneum. Data from this table used from: (Garforth et al., 2006; Gillis et al., 2008; Jiang et al., 2018; Nguyen et al., 2018; Niederer and Zappulla, 2015).
1TR = Telomerase RNA component
*TEN = N terminal domain
*TRBD = Telomerase RNA binding domain
*RT = Reverse transcriptase domain
*CTE = C terminal extension
*IFD = Insertion in fingers domain
2 This IFD is truncated compared to other TERTs.
- https://cdn.elifesciences.org/articles/55438/elife-55438-supp1-v1.xlsx
-
Supplementary file 2
Table 2a.
Data collection and refinement statistics for tcTERT structures.
- https://cdn.elifesciences.org/articles/55438/elife-55438-supp2-v1.xlsx
-
Supplementary file 3
Table 3a.
Kinetic parameters and errors for tcTERT pre-steady-state kinetics of single nucleotide incorporation. Table 3b. Full kinetic parameters of each curve used to generate Kobs and target engagement values in tcTERT pre-steady-state kinetics
- https://cdn.elifesciences.org/articles/55438/elife-55438-supp3-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/55438/elife-55438-transrepform-v1.docx





