An atlas of cell types in the mouse epididymis and vas deferens
Figures
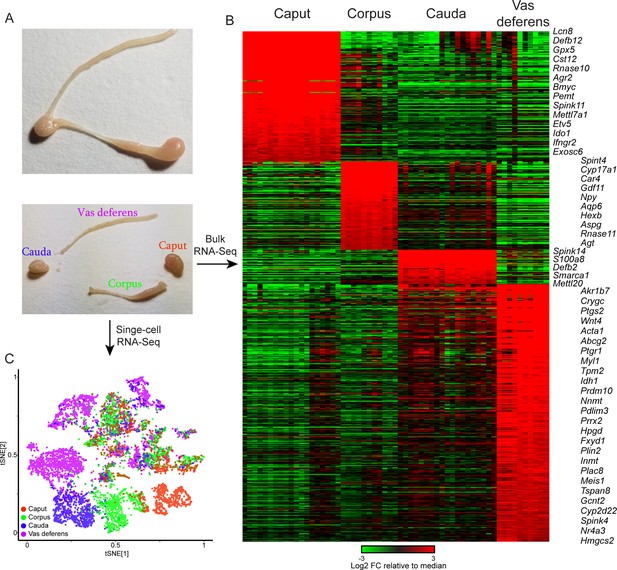
Overview of dataset.
(A) Tissue samples surveyed in this study. Top image shows a single mouse epididymis prior to dissection, while bottom shows a typical dissection into the four anatomical regions – the caput, corpus, and cauda epididymis, as well as the vas deferens –characterized by bulk RNA-Seq and single-cell RNA-Seq. See also Figure 1—figure supplement 1. (B) Bulk RNA-Seq dataset, showing region-enriched genes (log2 fold change relative to dataset median of at least 2, with a maximum mRNA abundance in one of the four dissections of at least 20 ppm). See also Figure 1—figure supplement 2. (C) Single-cell RNA-Seq dataset, clustered by t-SNE and annotated according to the four anatomical regions in (A).
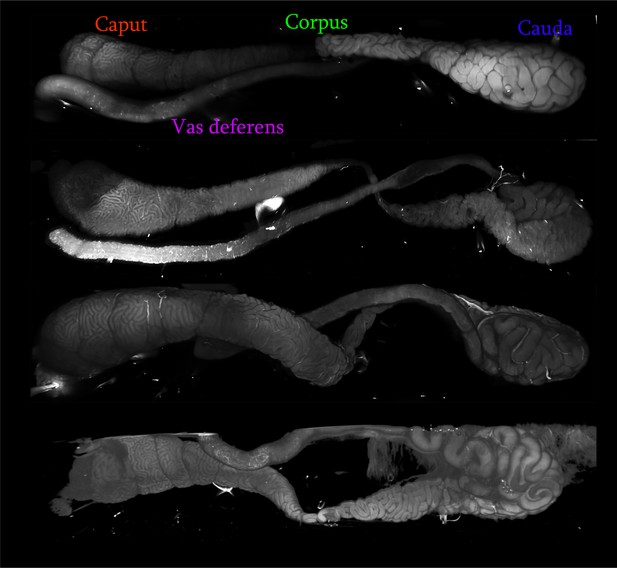
Light sheet imaging of mouse epididymis.
Four typical examples of the mouse epididymis and vas deferens imaged by light sheet microscopy, revealing the extensively coiled tube confined by a connective tissue sheath. Epididymal regions are indicated for the top sample, and all four samples are oriented analogously (caput to the left, cauda to the right, with the vas deferens then proceeding leftwards from the cauda).
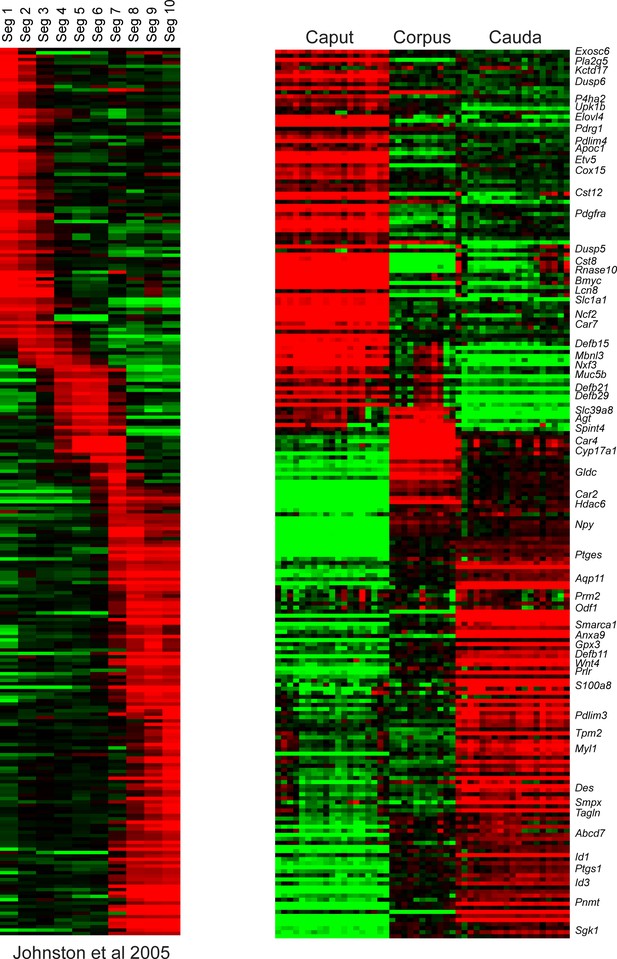
Comparison of RNA-Seq to prior microarray analysis of segmental gene expression.
Left panel shows microarray data from Johnston et al., 2005, sorted according to the epididymal segment exhibiting maximal expression. Right panel shows data for the same genes in the bulk RNA-Seq data from this study, for caput, corpus, and cauda dissections.
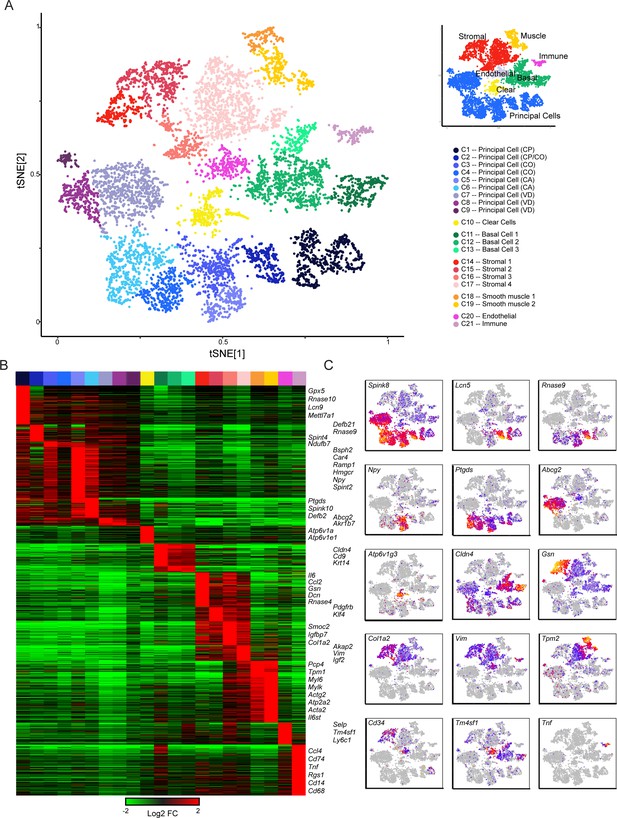
Single-cell decomposition of the epididymis.
(A) Single-cell cluster (expanded from Figure 1C) with 21 clusters annotated according to predicted cell types. Inset shows coarser cell type groupings used for downstream subclustering. (B) Heatmap of genes enriched in each cluster. Among the genes exhibiting significant variation across the 21 clusters, 1241 genes enriched at least fourfold in at least one cluster were selected for visualization. Heatmap shows these genes sorted according to the cluster where they exhibit maximal expression. (C) Expression of key marker genes across the entire dataset. See also Figure 2—figure supplement 1. For this and other single-cell images, color scale runs from gray (no expression) through blue, then red and finally orange (highly expressed), with heatmap for each panel scaled according to expression level of the gene in question.

Additional images of gene expression levels across the entire single-cell dataset.
(A) Gpx genes, including classic markers of caput (Gpx5) and cauda (Gpx3) principal cells, shown as in Figure 2C. (B) Several well-known sperm mRNAs, shown as in (A). We find no evidence for any sperm being captured, consistent both with our efforts to remove as many sperm as possible, and with the expectation that the unusually elongated dimensions of sperm would be incompatible with the droplet sizes used in the 10X Chromium platform.
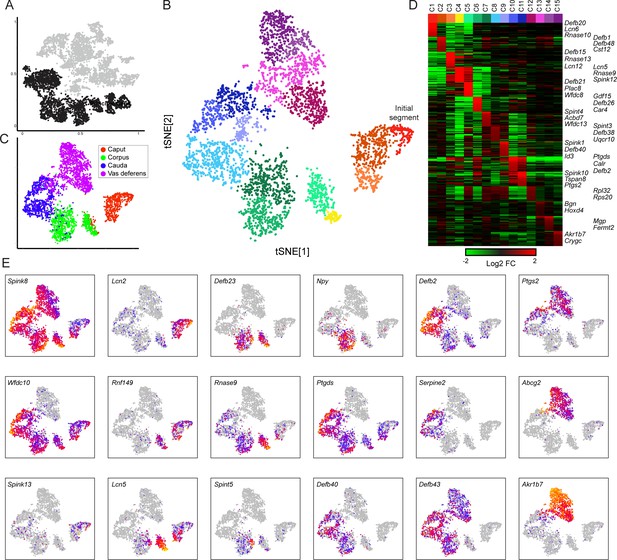
Modulation of the principal cell transcriptome across the male reproductive tract.
(A) Clusters from the overall dataset (Figure 2A), with the principal cell clusters extracted for reclustering highlighted in black. (B) Reclustering of extracted principal cells, visualized by t-SNE. Distinct colors highlight the 15 resulting principal cell clusters. See also Figure 3—figure supplements 1–4. (C) Reclustered principal cells, as in panel (B), with cells colored according to anatomical origin. (D) Heatmap showing the top 20 genes enriched in each of the principal cell clusters. Based on highly enriched marker genes, our epididymis clusters from 1 to 11 (ignoring the four vas deferens clusters) correspond to the following segments from Johnston et al., 2005: segment 1 (initial segment), segments 1–2, segments 3–4, segment 5 (early), late segment 5/segment 6, segment 7, late segment 7, segments 8–9, segment 9, late segment 9 (some segment 10 markers), segment 10. (E) Expression of key marker genes across all principal cells, visualized as in Figure 2C.
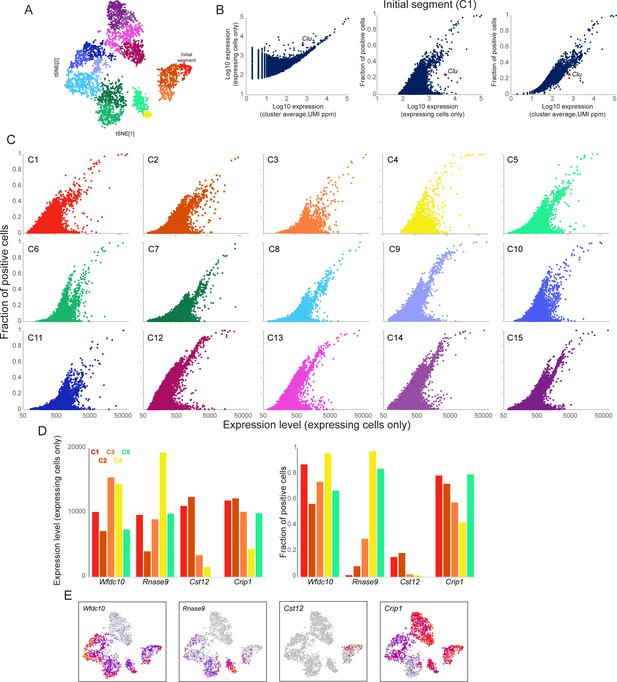
Gene expression heterogeneity within principal cell subclusters.
In order to explore cell-to-cell variability in mRNA abundance between individual cells comprising any given principal cell subcluster, we set out to identify the fraction of cells in each subcluster expressing any given gene. However, the low efficiency of mRNA capture in scRNA-Seq makes it difficult to distinguish ‘true zeroes’ – cases where a given cell does not express a gene – from technical dropouts resulting from failure to capture any molecules of the gene in question. To address this, we used DESingle (Miao et al., 2018) to estimate the true fraction of expressing cells for any given gene by inferring statistical likelihood of technical dropouts. Full dataset for this analysis is included as Supplementary file 3. (A) Principal cell clusters 1-15, reproduced from Figure 3B. (B) Using the output of DESingle, three plots show the relationship between gene expression level and fraction of expressing cells, using the principal cell subcluster corresponding to the initial segment (C1) for illustration. Left panel shows average expression level (across all cells in the cluster) on the x axis, expressed as UMIs normalized to parts per million, compared to expression level only in the subset of cells that detectably express the gene. At high expression levels, these values converge, indicating that extremely highly expressed genes are generally consistently detected throughout the cell population, whereas at lower average expression many genes fall above the diagonal, highlighting genes expressed in only a subset of cells. Middle and right panels compare a gene’s expression level (either confined only to expressing cells, or across all cells in the cluster, as indicated) to the fraction of expressing cells. Clu is shown in all three panels to highlight an example of high cell-to-cell variation for a gene known to exhibit patchy expression in the caput epididymis (Hermo et al., 1991). (C) Scatterplots for all 15 principal cell subclusters, comparing gene expression level only in expressing cells (x axis) to fraction of expressing cells (y axis). Substantial cell-to-cell heterogeneity can be observed across all subclusters – in general, genes located in the lower right area of these graphs represent highly expressed genes with unusually high cell-to-cell variability. See also Supplementary file 3. (D-E) Illustrative examples of genes exhibiting similar abundance in expressing cells (plotted in left panel of (D) for the first five principal cell clusters), but with different behaviors across cells (right panel, (D)). Expression across all principal cell subclusters is illustrated in (E) for all four genes. Wfdc10 and Crisp1 are both consistently expressed in the majority of cells in all five subclusters, and are typical examples of highly expressed genes. Rnase9 is a marker of clusters 4–5 and is penetrantly-expressed in these clusters. Importantly, a small number of Rnase9-positive cells can be detected in the more proximal clusters 1–3, and, interestingly, these rare cells express Rnase9 at similar levels to cells in clusters 4–5. Finally, Cst12 is one of the markers of cluster 2, yet even in this cluster only a small subpopulation of cells express this gene.
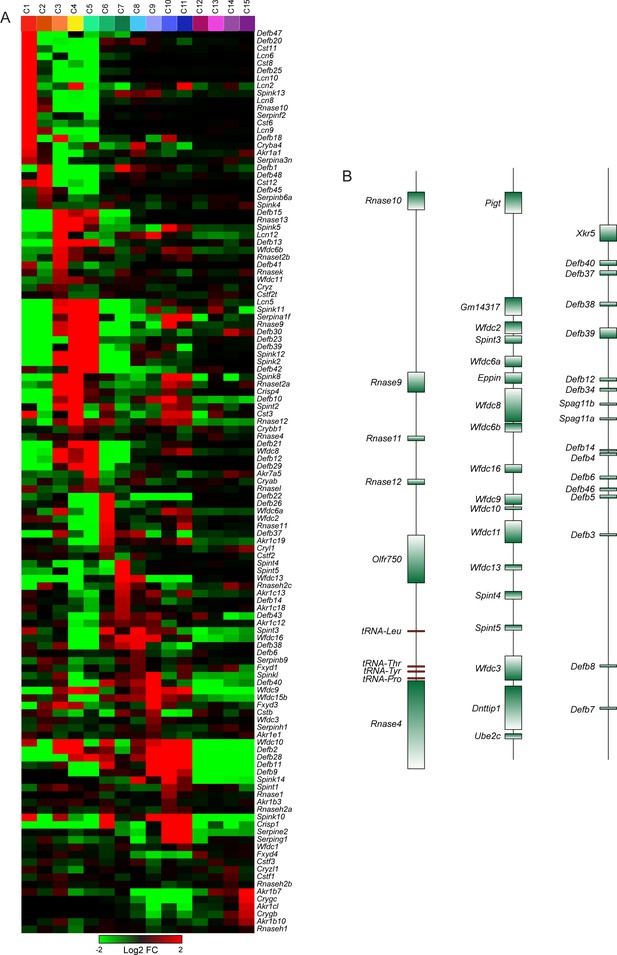
Region-specific expression of multi-gene family members.
(A) Heatmap showing expression of individual members of several multi-gene families (Rnase, Lcn, Cst, Defb, Wfdc, Spint, Spink, Serpin) across the 15 principal cell subclusters from Figure 3. (B) Three examples of relevant multigene clusters exhibiting region-specific expression of individual cluster members.
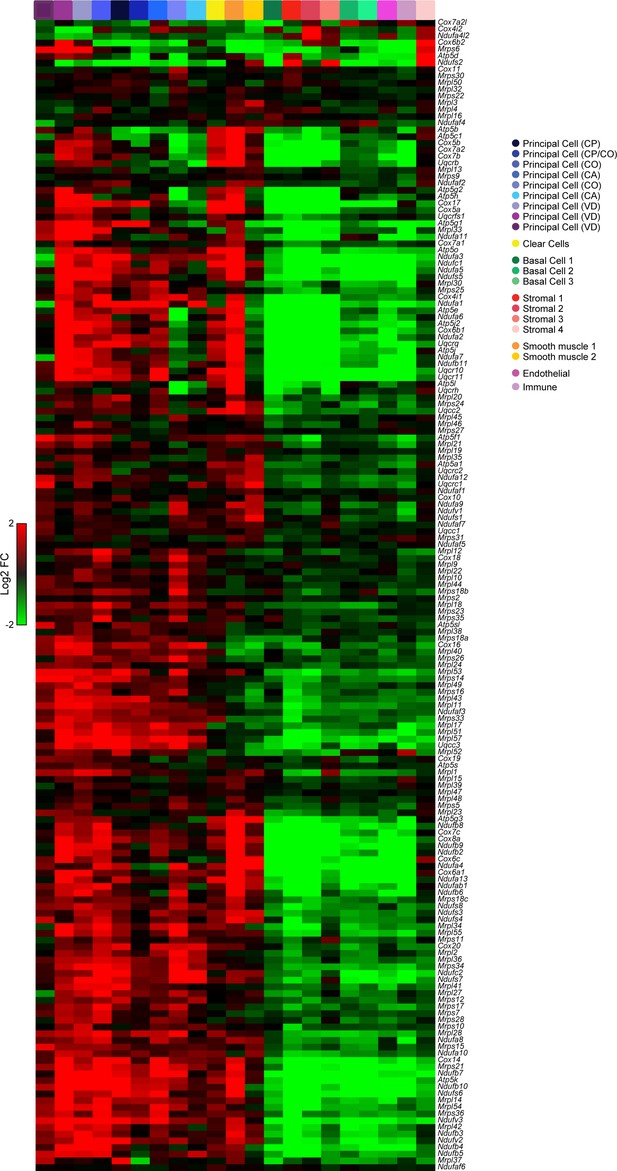
High levels of oxidative energy production in principal, clear, and muscle cells.
Heatmap shows expression of various nuclear-encoded mitochondrial genes (Mrpl, Mrps, Ndufa, Cox, Uqcr gene families) across the entire 21 cluster dataset from Figure 2A. Clusters expressing high levels of nuclear-encoded mitochondrial genes included all principal cell clusters, both muscle clusters, and the clear cells (leftmost 12 columns), while low expression of these genes was observed in basal, immune, and stromal cell types (rightmost nine columns).
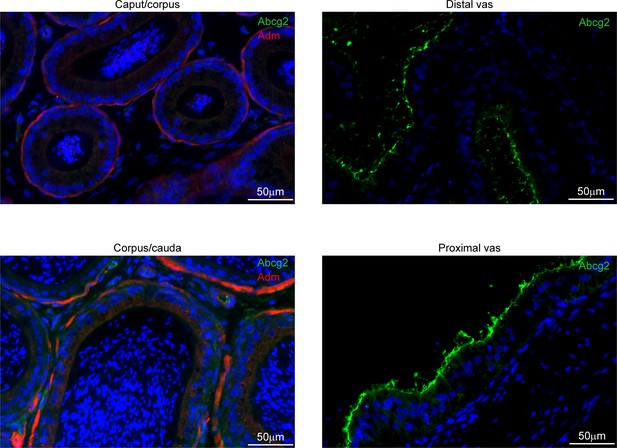
Apical localization of Abcg2 in the vas deferens.
Left panels: immunostaining for Adm (a basal cell marker, in red) and Abcg2 (specific to vas deferens principal cells, in green), and counterstained with DAPI, in the epididymis. Images from the epididymis show Adm-positive basal cells, with no Abcg2-positive cells, as expected. Along with secondary only controls (not shown), the absence of Abcg2 staining in the epididymis provides additional negative controls for the other immunostaining studies in this manuscript. Right panels: vas deferens images show Abcg2 and DAPI staining, confirming widespread principal cell expression and predicted apical staining of Abcg2.
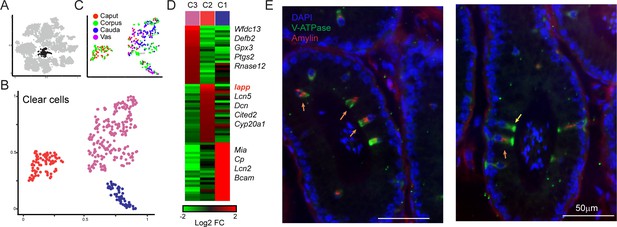
Amylin is a marker for a distinct subset of clear cells.
(A-C) Reclustering of clear cells. Panels are analogous to Figure 3A–C. (D) Heatmap showing marker genes for the three clear cell clusters. (E)Amylin marks a subset of epididymal clear cells. Immunofluorescence image of a tubule of the caput epididymis, stained for DAPI, V-ATPase-G3 (marker for clear cells), and Amylin, as indicated. Yellow arrow shows an Amylin-negative clear cell, while orange arrows show Amylin-positive clear cells. See also Figure 4—figure supplement 1.
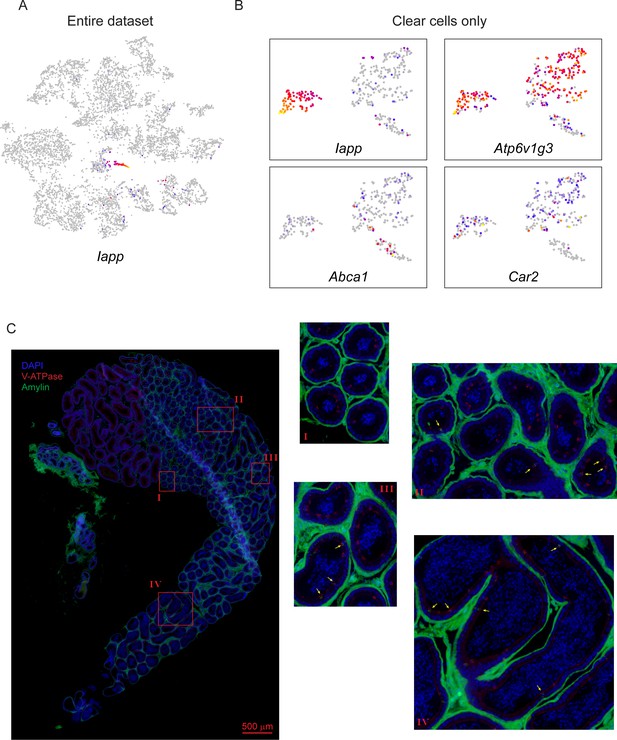
Clear cell subspecialization.
(A) Expression of Iapp across all cells in our dataset, highlighting its expression confined to the clear cell cluster in the center. (B) Gene expression across the reclustered clear cells (Figure 4B), with expression of the indicated marker genes illustrated as in Figures 2C and 3E. Marker genes here include: Iapp, which highlights a well-demarcated cluster of caput and corpus-enriched clear cells; Atp6v1g3, a pan-clear cell marker (Breton et al., 1996; and Car2 and Abca1, previously described as markers of apical and narrow cells of the initial segment (Adamali and Hermo, 1996). (C) Localization and heterogeneity of Amylin-positive clear cells. Immunofluorescence images for the indicated regions of the caput and corpus epididymis, stained with V-ATPase (red) and Amylin (green), and counterstained with DAPI, as in Figure 2E. Left panel shows broad view of first 5–6 segments of the epididymis, with four zoomed in images indicated. Not that this staining experiment was carried out with less stringent washing conditions than for the images shown in Figure 4E, and was contaminated by nonspecific interstitial staining. However, in addition to this background staining in the interstitium, we observed specific Amylin signal in a small subset of clear cells, with a handful of examples indicated by yellow arrows. Importantly, Amylin-positive cells were observed in both caput and corpus epididymis but not in the cauda. In addition, even in the caput and corpus we noted the presence of both Amylin-positive and Amylin-negative clear cells. These two observations argue against Amylin being a marker for narrow or apical cells.
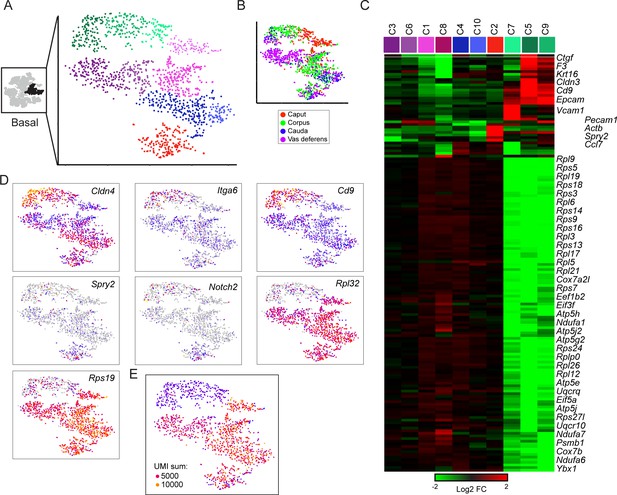
Subclustering of basal cells.
(A-B) Subclustering of basal cells; (A) shows 10 clusters annotated by color, while (B) shows basal cells colored by anatomic origin. (C) Expression of marker genes (minimum fold-enrichment > 4-fold in at least one cluster) across the 10 basal cell subclusters, grouped by hierarchical clustering. Notable here are two major populations, distinguished by expression of ribosomal protein genes. A third minor cluster (C2) was marked by elevated expression of several genes including Notch2 and Spry2. (D) Expression of individual genes across the basal cell subclusters, as in Figure 2C. (E) UMI counts for the basal cell subclustering, revealing a strong correspondence between the RPG high/low divisions and cells with high/low UMI counts.
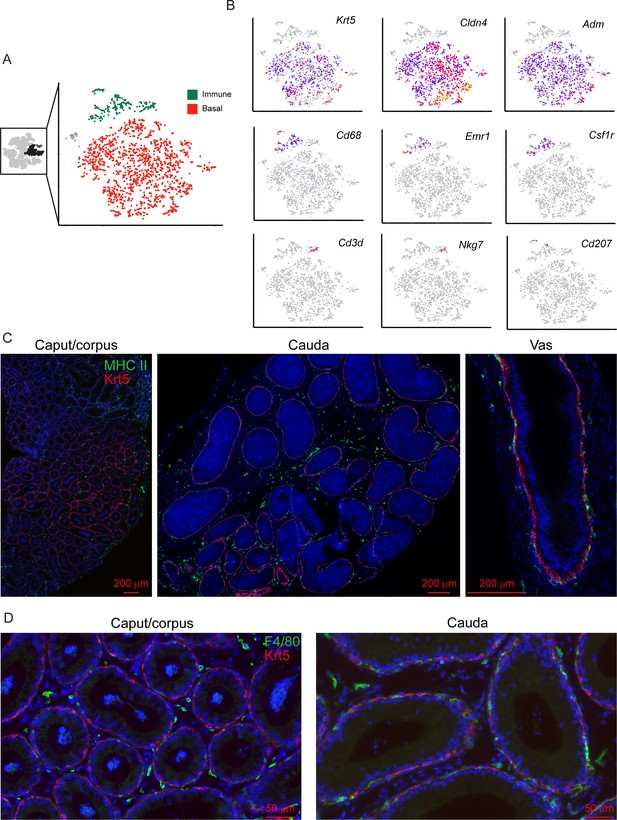
Clear distinctions between basal cells and various immune cell populations.
(A) Subclustering of basal cells and immune cells. Left panel highlights the three clusters of basal cells, and the immune cell cluster, that were extracted from the full dataset for reclustering. Right panel shows subclustering result, with basal and immune cells highlighted and exhibiting clear separation. (B) Marker genes for basal cells (Krt5, Cldn4, Adm) and various immune subpopulations, indicated as in Figure 2C. (C-D) Immunofluorescence confirms distinctions between mature basal cells (using Krt5 as a marker) and antigen presenting cells (MHC Class II, panel (C)) or macrophages (F4/80, panel (D)).
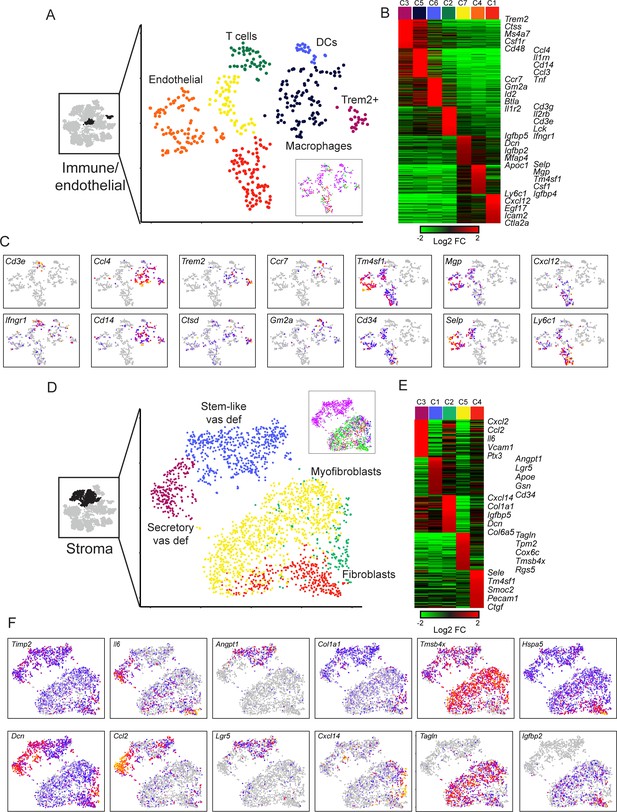
Diverse stromal cell populations in the epididymis.
(A) Reclustering of immune and endothelial cells. Left panel highlights the clusters from full dataset used for reclustering, while right panels show reclustered data. Inset shows cells colored according to the anatomical region where they were found. (B) Heatmap of the top 100 markers for each of the immune and endothelial subclusters in (A). (C) Expression of individual marker genes across immune and endothelial cell subpopulations, as in Figure 2C. (D-F) Reclustering of stromal cells, arranged as in panels (A-C). Heatmap (E) shows the top 50 genes for each subcluster. See also Figure 6—figure supplement 2.
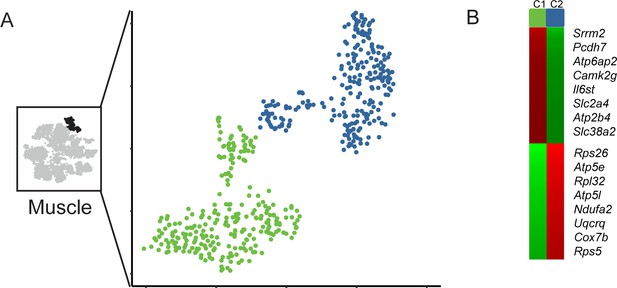
Subclustering of muscle cells.
(A) Subclustering of muscle cells. (B) Markers distinguishing the two muscle cell subtypes.
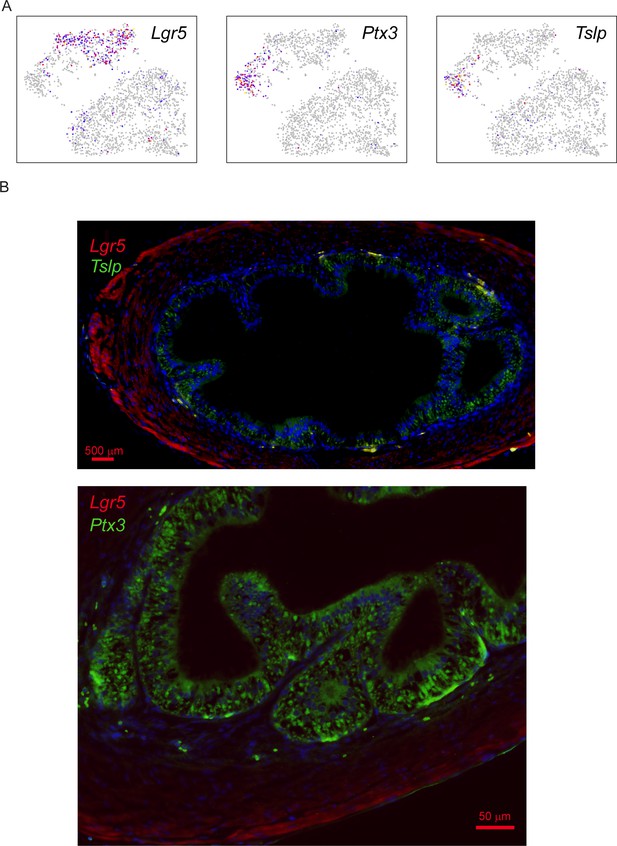
Validation of secretory and stem-like stromal cells in the vas deferens.
(A) Expression of Lgr5, Ptx3, and Tslp in the stromal cell subclustering from Figure 6D, highlighting the distinction between 'stem-like' (Lgr5-positive) and 'secretory' (Ptx3/Tslp-positive). (B) Images show vas deferens sections stained with Lgr5 (red) and either Ptx3 or Tslp (green), as indicated, and counterstained with DAPI. Lgr5 stains a layer of stromal cells at the periphery of the vas deferens, with strongest staining just inside the capsule. In contrast, Ptx3 and Tslp are brightest in the cells immediately outside the epithelium, with abundant staining also visible throughout the epithelium. Together with our RNA-Seq data showing expression of these genes in collagen-enriched stromal cells, our data suggest that the stromal cells underlying the epithelium express and secrete Ptx3 and Tslp, with secreted proteins also being visible in the epithelial layer itself. Future localization studies focused on RNA rather than protein will be needed to more precisely define the Ptx3/Tslp-positive “secretory” stroma; regardless, it is clear in these images that Lgr5 and Ptx3/Tslp do not colocalize.
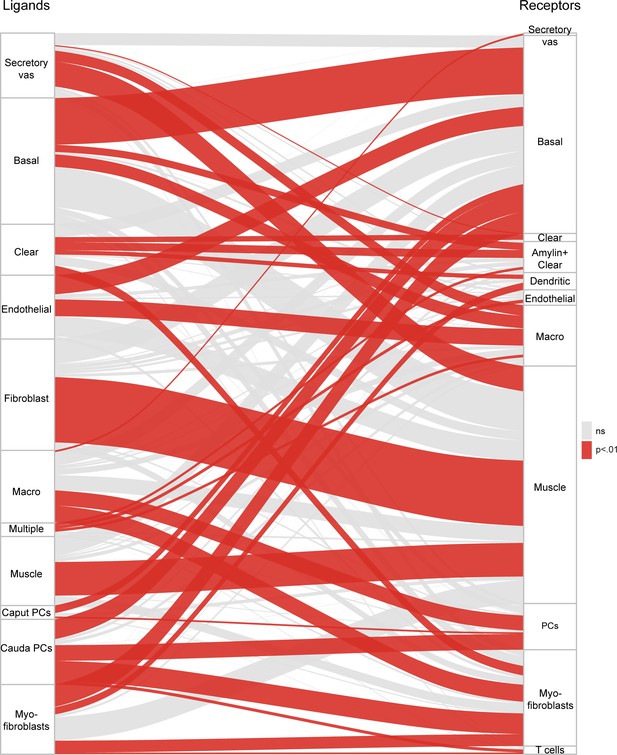
Potential signaling interactions between epididymal cell types.
Bars on left and right show clusters of ligands and receptors (see Supplementary file 4) named according to the cell population with highest expression levels. For brevity, principal cells were abbreviated as ‘PC’. Rectangle lengths represent the number of putative connections between ligands (left panels) and receptors (right panels) expressed in different cell types. Lines connecting rectangles represent connections between matching ligand-receptor pairs expressed. If the number of interactions between a cell type is equal or less than the number expected by chance (based on the number of expressed ligands and receptors in the pair of cell types, hypergeometric test), lines are grey, while lines are colored red for cell type pairs with a significant enrichment of matching ligand-receptor pairs. See Supplementary file 4 for ligand and receptor expression in the 34 cell populations.
Additional files
-
Supplementary file 1
Bulk RNA-Seq dataset.
RNA-Seq for caput (CP), corpus (CO), and cauda (CA) epididymis, and for vas deferens (VD). All data are normalized to parts per million.
- https://cdn.elifesciences.org/articles/55474/elife-55474-supp1-v3.xlsx
-
Supplementary file 2
Marker gene expression across 21 cell clusters.
Log2 fold enrichment for 11089 genes across the 21 cell clusters (Figure 2A) from the full dataset.
- https://cdn.elifesciences.org/articles/55474/elife-55474-supp2-v3.xlsx
-
Supplementary file 3
Principal cell subclustering and cell to cell heterogeneity.
Output of DESingle for the 15 principal cell subclusters (Figure 3B). For each subcluster, table includes fraction of cells not expressing the gene in question (‘theta_2’, or estimated true zeroes), average expression across all cells (‘cluster_UPM’, expressed as UMIs per million), and average expression level only in positive cells (‘expressing_cell_meanUPM’).
- https://cdn.elifesciences.org/articles/55474/elife-55474-supp3-v3.xlsx
-
Supplementary file 4
Ligand and receptor expression across epididymal cell types.
Relative expression levels for all ligands or receptors expressed (>=1 UMI) in at least 20% of cells from one or more of the 34 different cell types listed.
- https://cdn.elifesciences.org/articles/55474/elife-55474-supp4-v3.xlsx
-
Supplementary file 5
Antibodies used for immunofluorescence studies.
List of antibodies used in IF studies throughout the manuscript.
- https://cdn.elifesciences.org/articles/55474/elife-55474-supp5-v3.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/55474/elife-55474-transrepform-v3.docx





